

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476b.10
(527 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
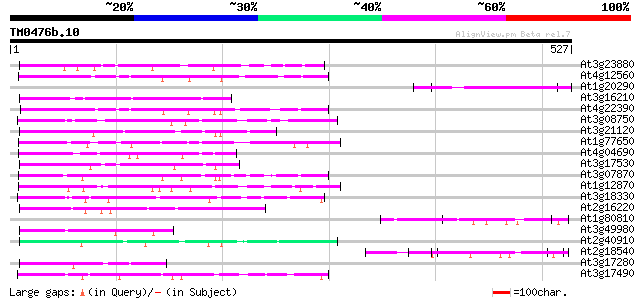
Score E
Sequences producing significant alignments: (bits) Value
At3g23880 unknown protein 114 2e-25
At4g12560 putative protein 95 8e-20
At1g20290 hypothetical protein 94 1e-19
At3g16210 hypothetical protein 89 5e-18
At4g22390 unknown protein 80 3e-15
At3g08750 hypothetical protein 79 8e-15
At3g21120 hypothetical protein 76 4e-14
At1g77650 hypothetical protein 71 2e-12
At4g04690 69 5e-12
At3g17530 hypothetical protein 68 1e-11
At3g07870 unknown protein 67 2e-11
At1g12870 hypothetical protein 67 3e-11
At3g18330 hypothetical protein 65 9e-11
At2g16220 hypothetical protein 65 1e-10
At1g80810 unknown protein 64 2e-10
At3g49980 putative protein 63 4e-10
At2g40910 unknown protein 63 5e-10
At2g18540 putative vicilin storage protein (globulin-like) 62 8e-10
At3g17280 hypothetical protein 62 1e-09
At3g17490 hypothetical protein 61 1e-09
>At3g23880 unknown protein
Length = 364
Score = 114 bits (284), Expect = 2e-25
Identities = 94/310 (30%), Positives = 150/310 (48%), Gaps = 50/310 (16%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESH---FDLNAAPTHRLL---L 63
LP E++ EILLRLPV+SL RFKCVC+SW LIS+ FA H + + A T +
Sbjct: 14 LPLEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYGV 73
Query: 64 PCLDKNKLESLDLES----SSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQV 119
+ L+S + S S+++V+ + RD+ + +G+C G + DY++ +
Sbjct: 74 ITTSRYHLKSCCIHSLYNASTVYVSEH-DGELLGRDYYQV--VGTCHGLVCFHVDYDKSL 130
Query: 120 IVWNPSTGFYKQI----LSFSDFMLDSLYGFGYDNSTDDYFLVLIGLIWVKAIIQAFSVK 175
+WNP+ +++ L SD YGFGYD S DDY +V + + Q VK
Sbjct: 131 YLWNPTIKLQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVAL-------LQQRHQVK 183
Query: 176 TNSCDFKYVNAQYRD---------LGYHYRHGVFLNNSLHWLVTTTDTSFSNDTSFLVVI 226
+ + +R + R G+++N +L+W T++ +S++ +I
Sbjct: 184 IETKIYSTRQKLWRSNTSFPSGVVVADKSRSGIYINGTLNWAATSSSSSWT-------II 236
Query: 227 AYDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSL-CYKGGRRDRAEIWVMK 285
+YD+ E+P PV G + L GCLS+ CY G A++WVMK
Sbjct: 237 SYDMSRDEFKELP------GPVCCGRGCFTM-TLGDLRGCLSMVCYCKGA--NADVWVMK 287
Query: 286 EYKVQSSWTK 295
E+ SW+K
Sbjct: 288 EFGEVYSWSK 297
>At4g12560 putative protein
Length = 408
Score = 95.1 bits (235), Expect = 8e-20
Identities = 85/316 (26%), Positives = 144/316 (44%), Gaps = 40/316 (12%)
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDK 68
T+P +++ +I LRLP ++L+R + + + LI+DP F ESH L++
Sbjct: 3 TIPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRGA 62
Query: 69 NKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNPSTGF 128
+L S+DL+ SL ++ P K ++ GS G + L+ + + V+NPST
Sbjct: 63 LRLYSVDLD--SLDSVSDVEHPMKRGGPTEVF--GSSNGLIGLS-NSPTDLAVFNPSTRQ 117
Query: 129 YKQILSFSDFMLDS-------LYGFGYDNSTDDYFLVLIGLIWVKA----------IIQA 171
++ S + D YG GYD+ +DDY +V + + + ++
Sbjct: 118 IHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKIDSEDELGCSFPYEVKV 177
Query: 172 FSVKTNSCD-FKYVNAQYRDLGYHYRH-------GVFLNNSLHWLVTTTDTSFSNDTSFL 223
FS+K NS + V + + L Y Y H GV NSLHW++ +F
Sbjct: 178 FSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSLHWVLPRRPGLI----AFN 233
Query: 224 VVIAYDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKGGRRDRAEIWV 283
+++ +DL + + +A + + + VL GCL L + ++W+
Sbjct: 234 LIVRFDLALEEFEIVRFPEAVANGNVDIQ-----MDIGVLDGCLCLMCNYD-QSYVDVWM 287
Query: 284 MKEYKVQSSWTKAFVV 299
MKEY V+ SWTK F V
Sbjct: 288 MKEYNVRDSWTKVFTV 303
>At1g20290 hypothetical protein
Length = 497
Score = 94.4 bits (233), Expect = 1e-19
Identities = 54/148 (36%), Positives = 79/148 (52%), Gaps = 10/148 (6%)
Query: 380 FPSEQLHLRVTSLPHGFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAE 439
F +H+ ++ +E E EE ++EEEE E +E EE E E
Sbjct: 348 FTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEE----------EEEEEEEEEEEEE 397
Query: 440 DEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHE 499
+EEEE E+E+ +++E+E EE E+ E E+EE E+ E +EEEE E EE + E
Sbjct: 398 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 457
Query: 500 EVESEKESNKDEEATEDEEEESSEDDYI 527
E E E+E ++EE E+EEEE E Y+
Sbjct: 458 EEEEEEEEEEEEEEEEEEEEEEEEKKYM 485
Score = 71.6 bits (174), Expect = 1e-12
Identities = 41/118 (34%), Positives = 62/118 (51%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E E EE ++EEEE + E +E EE E E+EEEE E+E+ +++E
Sbjct: 380 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 439
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEAT 514
+E EE E+ E E+EE E+ E +EEEE E EE + ++ K+ K +T
Sbjct: 440 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKKRRST 497
>At3g16210 hypothetical protein
Length = 360
Score = 89.4 bits (220), Expect = 5e-18
Identities = 62/199 (31%), Positives = 96/199 (48%), Gaps = 7/199 (3%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP EL EIL+RL ++ L RF+CVC +W LI+DP F E++ D++ A + DKN
Sbjct: 5 LPEELAIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAK----FVSFYDKN 60
Query: 70 KLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNPSTGFY 129
LD+E +T L P + + C G + + N ++VWNP + +
Sbjct: 61 -FYMLDVEGKHPVITNKLDFPLDQSMIDESTCVLHCDGTLCVTLK-NHTLMVWNPFSKQF 118
Query: 130 KQILSFSDFMLDSLYGFGYDNSTDDYFLVLIGLIWVKAIIQAFSVKTNSCDFKYVNAQYR 189
K + + + ++ GFGYD DDY +V + F +T S + + Y
Sbjct: 119 KIVPNPGIYQDSNILGFGYDPVHDDYKVVTFIDRLDVSTAHVFEFRTGSWG-ESLRISYP 177
Query: 190 DLGYHYRHGVFLNNSLHWL 208
D Y R G FL+ L+W+
Sbjct: 178 DWHYRDRRGTFLDQYLYWI 196
>At4g22390 unknown protein
Length = 401
Score = 80.1 bits (196), Expect = 3e-15
Identities = 80/314 (25%), Positives = 136/314 (42%), Gaps = 44/314 (14%)
Query: 11 PHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKNK 70
P +LI E+ LRL +L++ + + F LI P+F SH L++
Sbjct: 5 PTDLINEMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMILLRGPRL 64
Query: 71 LESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNPST-GFY 129
L +++L+S ++P P ++ ++ GS G + L + + ++NPST +
Sbjct: 65 LRTVELDSPE--NVSDIPHPLQAGGFTEVF--GSFNGVIGLC-NSPVDLAIFNPSTRKIH 119
Query: 130 KQILSFSDFMLDSL------YGFGYDNSTDDYFLVLIGLIWVK---------AIIQAFSV 174
+ + DF + YG GYD+ DD+ +V I +K ++ FS+
Sbjct: 120 RLPIEPIDFPERDITREYVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFPCPVEVKVFSL 179
Query: 175 KTNSCDFKYVNAQYRDL--GYHYR------HGVFLNNSLHWLVTTTDTSFSNDTSFLVVI 226
K NS + +++ L Y+Y +GV +NN LHW++ +F +I
Sbjct: 180 KKNSWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNHLHWILPRR----QGVIAFNAII 235
Query: 227 AYDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSL-CYKGGRRDRAEIWVMK 285
YDL + + EL + VL GC+ L CY ++WV+K
Sbjct: 236 KYDLASDDIGVLSFPQELY--------IEDNMDIGVLDGCVCLMCY--DEYSHVDVWVLK 285
Query: 286 EYKVQSSWTKAFVV 299
EY+ SWTK + V
Sbjct: 286 EYEDYKSWTKLYRV 299
>At3g08750 hypothetical protein
Length = 369
Score = 78.6 bits (192), Expect = 8e-15
Identities = 79/312 (25%), Positives = 142/312 (45%), Gaps = 41/312 (13%)
Query: 8 PTLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLD 67
P+LP ELI EIL ++P SL+RFK C W+ LI++ +F +H D + +P + D
Sbjct: 10 PSLPFELIEEILYKIPAESLIRFKSTCKKWYNLITEKRFMYNHLD-HYSPER--FIRTYD 66
Query: 68 KNKLESL-DLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFML-LAYDYNRQVIVWNPS 125
+ ++ + ++ S +L P + RD +Y + C G ML ++ + VWNP
Sbjct: 67 QQIIDPVTEILSDALI-------PDEFRDLYPIYSMVHCDGLMLCTCRKWDNSLAVWNPV 119
Query: 126 TGFYKQILSFSDFMLDSLYGFGYDN--STDDY-FLVLIGLI----WVKAIIQAFSVKTNS 178
K I ++ G GYD+ S D+Y L L+G + + + K++S
Sbjct: 120 LREIKWIKPSVCYLHTDYVGIGYDDNVSRDNYKILKLLGRLPKDDDSDPNCEIYEFKSDS 179
Query: 179 CDFKYVNAQYR-DLGYHYRHGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLLEKSLSE 237
+K + A++ D+ +GV + ++W+ + +I +D ++ E
Sbjct: 180 --WKTLVAKFDWDIDIRCNNGVSVKGKMYWIAKKKED--------FTIIRFDFSTETFKE 229
Query: 238 IPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKGGRRDRAEIWVMKEYKVQ-SSWTKA 296
I + P ++T G G LSL +G E+W+ + + S+++
Sbjct: 230 ICVCP---YTLVTRLGC-------FDGDRLSLLLQGEESQGIEVWMTNKLSDKVVSFSQY 279
Query: 297 FVVTDCDIPCIH 308
F VT D+P +H
Sbjct: 280 FNVTTPDLPALH 291
>At3g21120 hypothetical protein
Length = 367
Score = 76.3 bits (186), Expect = 4e-14
Identities = 64/259 (24%), Positives = 116/259 (44%), Gaps = 28/259 (10%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP +L+ EIL ++P SL RF+ C W L+ D FA+ H+ +++ +
Sbjct: 3 LPEDLVLEILSKVPAVSLARFRSTCRRWNALVVDGSFAKKHYAYGPRQYPIVIMLIEFRV 62
Query: 70 KLESLDLE---------SSSLFVTLNLPPP-CKSRDHNSLYFLGSCRGFMLLAYDYNRQV 119
L S+DL S+ L +L P S + + C G +LL +R++
Sbjct: 63 YLVSIDLHGINNNNGAPSAKLTGQFSLKDPLSNSSEEVDIRNAFHCDG-LLLCCTKDRRL 121
Query: 120 IVWNPSTGFYKQILSFSDFMLDSLYGFGYDNSTDDYFLVLIGLIWVKAIIQAFSVKTNSC 179
+VWNP +G K I + + LY GYDN + Y ++ + + F +++
Sbjct: 122 VVWNPCSGETKWIQPRNSYKESDLYALGYDNRSSSY-----KILRMHPVGNPFHIESEVY 176
Query: 180 DFKYVNAQYRDLG----YHYR----HGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLL 231
DF + +R +G +H + +G+ + + +W + D S+D FL +++D
Sbjct: 177 DF--ASHSWRSVGVTTDFHIQTNESYGMNVKGTTYWFALSKDWWSSDDRRFL--LSFDFS 232
Query: 232 EKSLSEIPLSPELAKPVLT 250
+ +PL ++ LT
Sbjct: 233 RERFQCLPLPADVKNLHLT 251
>At1g77650 hypothetical protein
Length = 383
Score = 70.9 bits (172), Expect = 2e-12
Identities = 82/332 (24%), Positives = 142/332 (42%), Gaps = 61/332 (18%)
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDK 68
+LP +++ E L + P+ SL+ K C + L +D +F +H DL+ RL+ LDK
Sbjct: 5 SLPSDVVEEFLFKTPIESLVLCKPTCKQLYALCNDKRFIYNHLDLS---EERLMRIYLDK 61
Query: 69 NKL---ESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAY-----DYNRQVI 120
K+ +LD+ + L +P S N ++ C G +L + D ++
Sbjct: 62 IKIINPVTLDI------LCLPVPAEFDSVTFNVIH----CDGLLLCRWTTRGLDRYNKLA 111
Query: 121 VWNPSTGFYKQILSFSDFMLDSLYGFGY-DNSTDDYFLVLIGLIWVKAIIQAFSVKTNSC 179
VWNP +G K + SF + D LYGFGY +N D + +L W K + + +K+
Sbjct: 112 VWNPISGQLKFVESFFHGVTD-LYGFGYANNGPRDSYKILRVSYWRKE-CEIYDLKSKL- 168
Query: 180 DFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLLEKSLSEIP 239
++ +A + + VF+N +++W+ T + ++ E +
Sbjct: 169 -WRAFSATLDWVVNTPQQNVFMNGNMYWIADT--------------LGDNIRETFIQSFD 213
Query: 240 LSPELAKPVLTAEGAPKFYHVRVLGGC----------------LSLCYKGGRRDR---AE 280
S E KP+ + K ++ G LSL + D+ E
Sbjct: 214 FSKETFKPICSFPYENKIIDTLIMNGIHRIADSVILSSFRVDRLSLFRQQRLEDKKYEME 273
Query: 281 IWVMKEYKVQ-SSWTKAFVVT-DCDIPCIHFY 310
+WV + + SWTK F VT D+P ++ Y
Sbjct: 274 VWVTNKVTDEVVSWTKYFNVTHHPDLPILNPY 305
>At4g04690
Length = 378
Score = 69.3 bits (168), Expect = 5e-12
Identities = 56/215 (26%), Positives = 101/215 (46%), Gaps = 20/215 (9%)
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDK 68
+LP EL+ EIL + P SL RFK C W+ +I+ +F +H D + R +D
Sbjct: 10 SLPFELVEEILKKTPAESLNRFKSTCKQWYGIITSKRFMYNHLDHSPERFIR-----IDD 64
Query: 69 NKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLL-----AYDYNRQ--VIV 121
+K + + +F + P P R +S + C G ML +Y+ R+ + V
Sbjct: 65 HKTVQIMDPMTGIF--SDSPVPDVFRSPHSFASMVHCDGLMLCICSDSSYERTREANLAV 122
Query: 122 WNPSTGFYKQILSFSDFMLDSLYGFGYDNS-TDDYFLVLIG--LIWVKAIIQAFSVKTNS 178
WNP T K I + +G GYDN+ ++Y +V + + + + K++S
Sbjct: 123 WNPVTKKIKWIEPLDSYYETDYFGIGYDNTCRENYKIVRFSGPMSFDDTECEIYEFKSDS 182
Query: 179 CDFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTD 213
++ ++ +Y D+ Y GV + +++W+ T +
Sbjct: 183 --WRTLDTKYWDV-YTQCRGVSVKGNMYWIADTKE 214
>At3g17530 hypothetical protein
Length = 388
Score = 68.2 bits (165), Expect = 1e-11
Identities = 56/225 (24%), Positives = 95/225 (41%), Gaps = 26/225 (11%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LPH+L +EIL R+P +SL ++K C W+ L DP F + +FD +L+ +
Sbjct: 6 LPHDLESEILSRVPAKSLAKWKTTCKRWYALFRDPSFVKKNFDKAGGREMIVLMNSRVYS 65
Query: 70 KLESL---------DLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVI 120
+L +E + + LN K D ++++ C G +L + ++
Sbjct: 66 NSVNLQGINNRFDPSMEVTGKLIKLN---DSKGVDISAIF---HCDGLILCTTTESTGLV 119
Query: 121 VWNPSTGFYKQILSFSDFMLDSLYGFGYDNSTD---DYFLVLIGLIWVKAIIQAFSVKTN 177
VWNP TG + I + + Y GY NS Y ++ +V + + +
Sbjct: 120 VWNPCTGEIRCIKPRIFYRCNDRYALGYGNSKSSCHSYKILRSCCYYVDQNLSLMAAEFE 179
Query: 178 SCDFKYVNAQYRDLG------YHYRHGVFLNNSLHWLVTTTDTSF 216
DF +RDLG Y GV L + +W+ + + F
Sbjct: 180 IYDFS--TDSWRDLGDITRDMIVYSSGVSLKGNTYWVSGSKEKGF 222
>At3g07870 unknown protein
Length = 417
Score = 67.0 bits (162), Expect = 2e-11
Identities = 84/323 (26%), Positives = 139/323 (43%), Gaps = 53/323 (16%)
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLD- 67
+LP ++IA+I RLP+ S+ R VC SW +++ S +++PT LL D
Sbjct: 27 SLPEDIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSSS---SSSPTKPCLLLHCDS 83
Query: 68 --KNKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYD-YNRQVIVWNP 124
+N L LDL + + +GSC G + L+ YN + ++NP
Sbjct: 84 PIRNGLHFLDLSEEEKRIKTKKFTLRFASSMPEFDVVGSCNGLLCLSDSLYNDSLYLYNP 143
Query: 125 STGFYKQILSFSDFMLDS--LYGFGYDNSTDDYFLVLI-----------------GLIWV 165
T ++ S+ D ++GFG+ T +Y ++ I G I
Sbjct: 144 FTTNSLELPECSNKYHDQELVFGFGFHEMTKEYKVLKIVYFRGSSSNNNGIYRGRGRIQY 203
Query: 166 K-AIIQAFSVKTNSCDFKYVNAQYRDLG---YHY---RHGVFLNNSLHWLVTTTDTSFSN 218
K + +Q ++ + + D + +R LG Y + +N LH++ T
Sbjct: 204 KQSEVQILTLSSKTTD---QSLSWRSLGKAPYKFVKRSSEALVNGRLHFV--TRPRRHVP 258
Query: 219 DTSFLVVIAYDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRV-LGGCL-SLCYKGGRR 276
D F +++DL ++ EIP KP G + H V L GCL ++ Y G
Sbjct: 259 DRKF---VSFDLEDEEFKEIP------KP--DCGGLNRTNHRLVNLKGCLCAVVY--GNY 305
Query: 277 DRAEIWVMKEYKVQSSWTKAFVV 299
+ +IWVMK Y V+ SW K + +
Sbjct: 306 GKLDIWVMKTYGVKESWGKEYSI 328
>At1g12870 hypothetical protein
Length = 416
Score = 66.6 bits (161), Expect = 3e-11
Identities = 80/333 (24%), Positives = 135/333 (40%), Gaps = 57/333 (17%)
Query: 9 TLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDL--NAAPTHRLLLPCL 66
+LP +++ EI L+LPV++L+RFK + W + F++ H + + H ++
Sbjct: 32 SLPDDVVEEIFLKLPVKALMRFKSLSKQWRSTLESCYFSQRHLKIAERSHVDHPKVMIIT 91
Query: 67 DK-----------NKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDY 115
+K LES+ SS+LF N P H+ +Y SC G +
Sbjct: 92 EKWNPDIEISFRTISLESVSFLSSALF---NFP----RGFHHPIYASESCDGIFCIHSPK 144
Query: 116 NRQVIVWNPSTGFYKQI--LSFSDFM------LDSLYGFGYDN-----STDDYFLVLIGL 162
+ + V NP+T +++Q+ F FM LD+L N DY LV +
Sbjct: 145 TQDIYVVNPATRWFRQLPPARFQIFMHKLNPTLDTLRDMIPVNHLAFVKATDYKLVWLYN 204
Query: 163 IWVKAI--IQAFSVKTNSCDFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTDTSFSNDT 220
+ + F K N+ + YR YH + N +L+W T +
Sbjct: 205 SDASRVTKCEVFDFKANAWRYLTCIPSYRI--YHDQKPASANGTLYWFTETYNAE----- 257
Query: 221 SFLVVIAYDLLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCY---KGGRRD 277
+ VIA D+ + +P KP L A P + ++ SLC +G ++
Sbjct: 258 --IKVIALDIHTEIFRLLP------KPSLIASSEPSHIDMCIIDN--SLCMYETEGDKKI 307
Query: 278 RAEIWVMKEYKVQSSWTKAFVVTDCDIPCIHFY 310
EIW +K + +W K + + HF+
Sbjct: 308 IQEIWRLK--SSEDAWEKIYTINLLSSSYCHFH 338
>At3g18330 hypothetical protein
Length = 376
Score = 65.1 bits (157), Expect = 9e-11
Identities = 84/309 (27%), Positives = 131/309 (42%), Gaps = 39/309 (12%)
Query: 8 PTLPHELIAEILLRLPVRSLLRFKCVCTSWFFLI-SDPQFAESHFDLNAAPTHRLLLPCL 66
P LP EL+ EIL +P L R C W LI +D +FA H+D NAA L+ +
Sbjct: 4 PNLPKELVEEILSFVPATYLKRLSATCKPWNRLIHNDKRFARKHYD-NAA--KEFLVFMM 60
Query: 67 DKN--------KLESLDLESSSLFVTLNLPPPC--KSRDHNSLYFLGSCRGFMLLAYDYN 116
KN L D S+ + L LP P S D + + C G +L
Sbjct: 61 RKNFRIIRRGVNLHGAD-PSAEVKGELTLPDPYFKNSADEFHIDRVFHCDGLLLCTSKLE 119
Query: 117 RQVIVWNPSTGFYKQILSFSDFMLDSLY-GFGYDNSTDDYFLVLIGLIWVKAIIQAFSVK 175
R+++VWNP TG K I + + D+ + G+ ++ + + ++ + +
Sbjct: 120 RRMVVWNPLTGETKWIQTHEEG--DNFFLGYSQEDKNISCKKSYKIMGFYRSGSKVWEYD 177
Query: 176 TNSCDFKYVNA----QYRDLGYHYRHGVFLNNSLHWLV-TTTDTSFSNDTSFLVVIAYDL 230
NS ++ +N Y D Y V L + + L TD F L + +YD
Sbjct: 178 FNSDSWRVLNGILPNWYFDKSYKC---VSLKGNTYMLAGAVTDMGFD-----LSLQSYDF 229
Query: 231 LEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKGGRRDRAEIWVMKEYKVQ 290
+ + P+S + + GA + VR G L+L Y+ +R +AEIWV +
Sbjct: 230 STEKFA--PVSLPVPSQARSLNGANRLSVVR--GEKLALLYRRDKRSKAEIWVTNKIDDT 285
Query: 291 S----SWTK 295
+ SWTK
Sbjct: 286 TEGAVSWTK 294
>At2g16220 hypothetical protein
Length = 407
Score = 64.7 bits (156), Expect = 1e-10
Identities = 68/245 (27%), Positives = 107/245 (42%), Gaps = 18/245 (7%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
L ++LI E+L RLP++S+ RF CV W + P F E ++A RLL +
Sbjct: 6 LTNDLILEVLSRLPLKSVARFHCVSKRWASMFGSPYFKELFLTRSSAKP-RLLFAIVQNG 64
Query: 70 -----KLESLDLESSSLFVT----LNLPPPCKS--RDHNSLYF-LGSCRGFMLLAYD-YN 116
L+ SS+L T + L P D+ YF +G G + L D Y
Sbjct: 65 VWRFFSSPRLEKSSSTLVATAEFHMKLSPNNLRIYHDNTPRYFSIGYASGLIYLYGDRYE 124
Query: 117 RQVIVWNPSTGFYKQILSFSDFMLDSLYGFGYDNSTDDYFLVLIGLIWVKAIIQAFSVKT 176
++ NP+TG Y IL + FG+D Y + +I+ + +
Sbjct: 125 ATPLICNPNTGRY-TILPKCYTYRKAFSFFGFDPIDKQY--KALSMIYPSGPGHSKILTF 181
Query: 177 NSCDFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTDTSFSND-TSFLVVIAYDLLEKSL 235
D + +YR L Y G+ +N L++L T+D +D TS V++ +DL +S
Sbjct: 182 GDGDMNWKKIKYRVLHDIYSQGICINGVLYYLGDTSDWDNDHDVTSGNVLVCFDLRSESF 241
Query: 236 SEIPL 240
+ I L
Sbjct: 242 TFIGL 246
>At1g80810 unknown protein
Length = 826
Score = 63.9 bits (154), Expect = 2e-10
Identities = 51/151 (33%), Positives = 70/151 (45%), Gaps = 36/151 (23%)
Query: 407 KDEEEEASGLSLHSVKSLTCEFKEATEE----VEAAEDEEEETTEDEDASDDKEDESEEA 462
K E+ G +L S+K L E EE +EA D+ E E ED DK+++S++
Sbjct: 675 KQVEKTREGKNLRSLKELNAETDRTAEEQEVSLEAESDDRSEEQEYEDDCSDKKEQSQDK 734
Query: 463 STE-----------------DGEATEDEEA-----TEDVEATVDEEEEATEVEEATDHEE 500
E +GE +E EE T+D+E DEEEE E++ D E
Sbjct: 735 GVEAETKEEEKQYPNSEGESEGEDSESEEEPKWRETDDMED--DEEEEEEEIDHMED--E 790
Query: 501 VESEKESNKDEEAT------EDEEEESSEDD 525
E EKE D+EA+ E EEEE ED+
Sbjct: 791 AEEEKEEVDDKEASANMSEIEKEEEEEEEDE 821
Score = 51.2 bits (121), Expect = 1e-06
Identities = 41/164 (25%), Positives = 74/164 (45%), Gaps = 13/164 (7%)
Query: 349 KNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPHGFKEANEVEEATKD 408
K + K + RE KN++ E+ E+ + +++ L S ++ E + + K
Sbjct: 671 KRLNKQVEKTREG--KNLRSLKELNAETDRTAEEQEVSLEAESDDRSEEQEYEDDCSDKK 728
Query: 409 EEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETT---EDEDASDDKEDESEEASTE 465
E+ + G+ + K ++ + E E + E EE E +D DD+E+E EE
Sbjct: 729 EQSQDKGVEAET-KEEEKQYPNSEGESEGEDSESEEEPKWRETDDMEDDEEEEEEEIDHM 787
Query: 466 DGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNK 509
+ EA E++E VD++E + + E EE E E E +
Sbjct: 788 EDEAEEEKEE-------VDDKEASANMSEIEKEEEEEEEDEEKR 824
Score = 30.0 bits (66), Expect = 3.3
Identities = 25/112 (22%), Positives = 47/112 (41%), Gaps = 20/112 (17%)
Query: 432 TEEVEAAEDEEEETTEDEDASDDKE-DESEEASTEDGEATEDEEATEDVEATVDEEEEA- 489
+EE+ E+ E +D A +DKE D E D + + +++V +V+ +
Sbjct: 597 SEELNLTEERWELLEDDTSADEDKEIDLPESIPLSDIMQRQKVKKSKNVAVSVEPTSSSG 656
Query: 490 ------------------TEVEEATDHEEVESEKESNKDEEATEDEEEESSE 523
+VE+ + + + S KE N + + T +E+E S E
Sbjct: 657 VRSSSRTLMKKDCGKRLNKQVEKTREGKNLRSLKELNAETDRTAEEQEVSLE 708
>At3g49980 putative protein
Length = 382
Score = 63.2 bits (152), Expect = 4e-10
Identities = 44/154 (28%), Positives = 70/154 (44%), Gaps = 11/154 (7%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLDKN 69
LP EL+ EIL R+P SL + + C W L +D F+ HFD AP L+ ++
Sbjct: 5 LPQELLEEILCRVPATSLKQLRLTCKEWNRLFNDRTFSRKHFD--KAPKQFLITVLEERC 62
Query: 70 KLESLDLESSSLFVTLNLPPPCKSRDHNS------LYFLGSCRGFMLLAYDYNRQVIVWN 123
+L SL + S F + D++S + + C G + + +++VWN
Sbjct: 63 RLSSLSINLHSGFPSEEFTGELSPIDYHSNSSQVIIMKIFHCDGLFVCTILKDTRIVVWN 122
Query: 124 PSTGFYKQIL---SFSDFMLDSLYGFGYDNSTDD 154
P TG K I + + D + G+ DN + D
Sbjct: 123 PCTGQKKWIQTGENLDENGQDFVLGYYQDNKSSD 156
>At2g40910 unknown protein
Length = 449
Score = 62.8 bits (151), Expect = 5e-10
Identities = 78/320 (24%), Positives = 129/320 (39%), Gaps = 35/320 (10%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCL--D 67
+P +L+ EI++RLP +S++RFKC+ W LIS F S F + CL
Sbjct: 69 IPPDLLMEIVMRLPAKSMVRFKCISKQWSSLISCRYFCNSLFTSVTRKKQPHIHMCLVDH 128
Query: 68 KNKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNPST- 126
+ L L S+S T + S +FL RG LL + + ++NP+T
Sbjct: 129 GGQRVLLSLSSTSPDNTCYVVDQDLSLTGMGGFFLNLVRG--LLCFSVREKACIYNPTTR 186
Query: 127 ------GFYKQILSFSDFMLDSLYGFGYDNSTDDYFLVLIGLIWVKAIIQAFSVKTNSCD 180
I++ D D Y G+D D Y LV I AF S
Sbjct: 187 QRLTLPAIKSDIIALKDERKDIRYYIGHDPVNDQYKLVC-----TIGISSAFFTNLKSEH 241
Query: 181 FKYVN------AQYRDLGYHYRH-----GVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYD 229
+ +V + R L ++ H G F+N S+ + + D DT V+++D
Sbjct: 242 WVFVLEAGGSWRKVRTLESYHPHAPSTVGQFINGSVVYYMAWLDM----DT--CAVVSFD 295
Query: 230 LLEKSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKG-GRRDRAEIWVMKEYK 288
+ + L+ I ++ E L A G K ++ G + + ++WV+K+
Sbjct: 296 ITSEELTTIIVTLEAGDVALPA-GRMKAGLIQYHGEIAVFDHTHLKEKFLVDLWVLKDAG 354
Query: 289 VQSSWTKAFVVTDCDIPCIH 308
++ K V+ C +H
Sbjct: 355 MKKWSKKRLVLQPCQKHLVH 374
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 62.0 bits (149), Expect = 8e-10
Identities = 39/129 (30%), Positives = 64/129 (49%), Gaps = 5/129 (3%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+EA + EEA + EEEEA + E +EA + E + EEEE E+ +E
Sbjct: 443 EEARKREEAKRREEEEAKRREEEETERKKREEEEARKREEERKREEEEAKRREEERKKRE 502
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATED 516
+E+E+A + E ++EE A EEE + E + + E ++ ++EEA +
Sbjct: 503 EEAEQARKREEEREKEEEM-----AKKREEERQRKEREEVERKRREEQERKRREEEARKR 557
Query: 517 EEEESSEDD 525
EEE E++
Sbjct: 558 EEERKREEE 566
Score = 58.5 bits (140), Expect = 9e-09
Identities = 42/132 (31%), Positives = 60/132 (44%), Gaps = 11/132 (8%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+EA + EE K EEEEA K E K+ EE E A EEE ++E+ + +E
Sbjct: 475 EEARKREEERKREEEEA--------KRREEERKKREEEAEQARKREEEREKEEEMAKKRE 526
Query: 457 DESEEASTEDGE---ATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEA 513
+E + E+ E E E + EA EEE E E A E+ KE + E
Sbjct: 527 EERQRKEREEVERKRREEQERKRREEEARKREEERKREEEMAKRREQERQRKEREEVERK 586
Query: 514 TEDEEEESSEDD 525
+E+E E++
Sbjct: 587 IREEQERKREEE 598
Score = 58.5 bits (140), Expect = 9e-09
Identities = 44/150 (29%), Positives = 64/150 (42%), Gaps = 12/150 (8%)
Query: 375 ESLLSFPSEQLHLRVTSLPHG--FKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEA- 431
+ LL E + S G K E+EE + EEEE ++ E +EA
Sbjct: 395 KGLLKAQKESVIFECASCAEGELSKLMREIEERKRREEEE--------IERRRKEEEEAR 446
Query: 432 -TEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEEEAT 490
EE + E+EE + E+E+ K +E E E+ E+EEA E EEEA
Sbjct: 447 KREEAKRREEEEAKRREEEETERKKREEEEARKREEERKREEEEAKRREEERKKREEEAE 506
Query: 491 EVEEATDHEEVESEKESNKDEEATEDEEEE 520
+ + + E E E ++EE E EE
Sbjct: 507 QARKREEEREKEEEMAKKREEERQRKEREE 536
Score = 54.7 bits (130), Expect = 1e-07
Identities = 38/131 (29%), Positives = 59/131 (45%), Gaps = 3/131 (2%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
K E E+A K EEE + + + KE EEVE EE+E E+ + +E
Sbjct: 500 KREEEAEQARKREEEREKEEEMAKKREEERQRKER-EEVERKRREEQERKRREEEARKRE 558
Query: 457 DES--EEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEAT 514
+E EE + E + E+VE + EE+E EE E E +K+ ++ E
Sbjct: 559 EERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKEREEMERK 618
Query: 515 EDEEEESSEDD 525
+ EEE ++
Sbjct: 619 KREEEARKREE 629
Score = 52.8 bits (125), Expect = 5e-07
Identities = 40/123 (32%), Positives = 58/123 (46%), Gaps = 6/123 (4%)
Query: 403 EEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKEDESEEA 462
EEA K EEE + + + KE EEVE EE+E +E+ + +E E ++
Sbjct: 552 EEARKREEERKREEEMAKRREQERQRKER-EEVERKIREEQERKREEEMAKRREQERQKK 610
Query: 463 STEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNKDEEATEDEEEESS 522
E+ E + EE EA EEE A EE +E E + ++EEA EEE
Sbjct: 611 EREEMERKKREE-----EARKREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKR 665
Query: 523 EDD 525
E++
Sbjct: 666 EEE 668
Score = 48.9 bits (115), Expect = 7e-06
Identities = 48/172 (27%), Positives = 74/172 (42%), Gaps = 19/172 (11%)
Query: 335 EGKLLEHHKYGQEIKNVRKDLAMYRESEYKNIQIYFEMYRESLLSFPSEQLHLRVTSLPH 394
E + +E + ++ + R++ A RE E K RE ++ EQ R
Sbjct: 533 EREEVERKRREEQERKRREEEARKREEERK---------REEEMAKRREQERQR------ 577
Query: 395 GFKEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDD 454
KE EVE ++E+E + + + KE EE+E + EEE +E+ +
Sbjct: 578 --KEREEVERKIREEQERKREEEMAKRREQERQKKER-EEMERKKREEEARKREEEMAKI 634
Query: 455 KEDESEEASTEDGE-ATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEK 505
+E+E + ED E +EEA E EEE A EE +E E EK
Sbjct: 635 REEERQRKEREDVERKRREEEAMRREEERKREEEAAKRAEEERRKKEEEEEK 686
Score = 46.2 bits (108), Expect = 4e-05
Identities = 35/126 (27%), Positives = 54/126 (42%), Gaps = 2/126 (1%)
Query: 397 KEANEVEEATKDEEEEASGLSLHSVKSLTCEFKEATEEVEAAEDEEEETTEDEDASDDKE 456
+E EE K E+E V+ E +E E E A+ E+E + E +++
Sbjct: 559 EERKREEEMAKRREQERQRKEREEVERKIREEQERKREEEMAKRREQERQKKEREEMERK 618
Query: 457 DESEEASTEDGEATEDEEATEDVEATVDEEEEATEVEEATDHEEVESEKESNK--DEEAT 514
EEA + E + E + D E + E E EE + E+E+ K +EE
Sbjct: 619 KREEEARKREEEMAKIREEERQRKEREDVERKRREEEAMRREEERKREEEAAKRAEEERR 678
Query: 515 EDEEEE 520
+ EEEE
Sbjct: 679 KKEEEE 684
Score = 33.9 bits (76), Expect = 0.23
Identities = 30/100 (30%), Positives = 48/100 (48%), Gaps = 6/100 (6%)
Query: 428 FKEATEEVEAAEDEEEETTEDEDASDDKEDESEEASTEDGEATEDEEATEDVEATVDEEE 487
F + E ++ ++E+ E AS E E + E E EE E++E EEE
Sbjct: 387 FNLSNETIKGLLKAQKESVIFECASC-AEGELSKLMREIEERKRREE--EEIERRRKEEE 443
Query: 488 EATEVEEATDHEEVESEKESNKDEEATEDEEE---ESSED 524
EA + EEA EE E+++ ++ E + EEE + E+
Sbjct: 444 EARKREEAKRREEEEAKRREEEETERKKREEEEARKREEE 483
>At3g17280 hypothetical protein
Length = 386
Score = 61.6 bits (148), Expect = 1e-09
Identities = 41/143 (28%), Positives = 64/143 (44%), Gaps = 10/143 (6%)
Query: 10 LPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPT-----HRLLLP 64
LP++L+ EIL RLP +S+ + K C W+ L DP+F E A T H +
Sbjct: 7 LPYDLLPEILSRLPTKSIPKLKTTCKKWYALFKDPKFVEKKLGKAARETVFLMNHEVNSI 66
Query: 65 CLDKNKLESLDLESSSLFVTLNLPPPCKSRDHNSLYFLGSCRGFMLLAYDYNRQVIVWNP 124
+D + + S TL +P ++ + C G L A N +++VWNP
Sbjct: 67 SVDIHGIPKGYSVSMDFTGTLTIP----EGSDLEIFRIHHCNGLFLCA-TMNCRLVVWNP 121
Query: 125 STGFYKQILSFSDFMLDSLYGFG 147
TG I+ + + D +Y G
Sbjct: 122 CTGQITWIIPRTRYDSDDIYALG 144
>At3g17490 hypothetical protein
Length = 388
Score = 61.2 bits (147), Expect = 1e-09
Identities = 75/309 (24%), Positives = 130/309 (41%), Gaps = 38/309 (12%)
Query: 8 PTLPHELIAEILLRLPVRSLLRFKCVCTSWFFLISDPQFAESHFDLNAAPTHRLLLPCLD 67
P L +L+ EIL R+P SL R + C W L +D +F++ H D AP L L D
Sbjct: 4 PHLSEDLVEEILSRVPAISLKRLRYTCKQWNALFNDQRFSKKHRD--KAPKTYLGLTLKD 61
Query: 68 ------KNKLESLDLESSSLFVTLNLPPPCKS-RDHNSLYF--LGSCRGFMLLAYDYNRQ 118
+ L L L ++++ + + S D N + C G +L + N +
Sbjct: 62 FRIYSMSSNLHGL-LHNNNIDLLMEFKGKLSSLNDLNDFEISQIYPCDGLILCSTKRNTR 120
Query: 119 VIVWNPSTGFYKQILSFSDFMLDSLYGFGYDNS----TDDYFLVLI--GLIWVKAIIQAF 172
++VWNP TG + I + M D+ + FGYDNS ++Y ++ + + + + F
Sbjct: 121 LVVWNPCTGQTRWIKRRNRRMCDT-FAFGYDNSKSSCLNNYKILRVCEKIKGQQFEYEIF 179
Query: 173 SVKTNSCDFKYVNAQYRDLGYHYRHGVFLNNSLHWLVTTTDTSFSNDTSFLVVIAYDLLE 232
+NS VN G V + + +W T T T + + +D
Sbjct: 180 EFSSNSWRVLDVNPNCIIEG----RSVSVKGNSYWFATITKTHY-------FIRRFDFSS 228
Query: 233 KSLSEIPLSPELAKPVLTAEGAPKFYHVRVLGGCLSLCYKGGRRDRAEIWVMKEYKVQS- 291
++ ++PL P + F + LS+ ++ ++ +IWV + +
Sbjct: 229 ETFQKLPL-PFHIFDYNDSRALSAFREEQ-----LSVLHQSFDTEKMDIWVTNKIDETTD 282
Query: 292 -SWTKAFVV 299
SW+K F V
Sbjct: 283 WSWSKFFTV 291
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,011,003
Number of Sequences: 26719
Number of extensions: 636760
Number of successful extensions: 23113
Number of sequences better than 10.0: 1458
Number of HSP's better than 10.0 without gapping: 882
Number of HSP's successfully gapped in prelim test: 599
Number of HSP's that attempted gapping in prelim test: 5546
Number of HSP's gapped (non-prelim): 7456
length of query: 527
length of database: 11,318,596
effective HSP length: 104
effective length of query: 423
effective length of database: 8,539,820
effective search space: 3612343860
effective search space used: 3612343860
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0476b.10


