

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.5
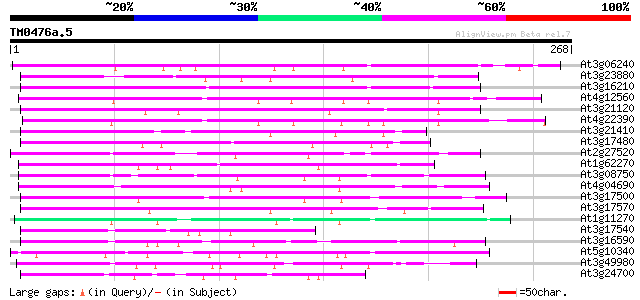
(268 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g06240 unknown protein 106 1e-23
At3g23880 unknown protein 100 1e-21
At3g16210 hypothetical protein 97 9e-21
At4g12560 putative protein 87 7e-18
At3g21120 hypothetical protein 80 1e-15
At4g22390 unknown protein 77 1e-14
At3g21410 hypothetical protein 74 8e-14
At3g17480 hypothetical protein 72 2e-13
At2g27520 hypothetical protein 72 3e-13
At1g62270 hypothetical protein 72 3e-13
At3g08750 hypothetical protein 71 7e-13
At4g04690 69 3e-12
At3g17500 hypothetical protein 69 3e-12
At3g17570 hypothetical protein 69 3e-12
At1g11270 unknown protein (At1g11270) 69 3e-12
At3g17540 hypothetical protein 67 8e-12
At3g16590 hypothetical protein 67 8e-12
At5g10340 putative protein 65 3e-11
At3g49980 putative protein 65 3e-11
At3g24700 hypothetical protein 65 5e-11
>At3g06240 unknown protein
Length = 427
Score = 106 bits (265), Expect = 1e-23
Identities = 98/314 (31%), Positives = 143/314 (45%), Gaps = 61/314 (19%)
Query: 2 EKLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDL-----NAAPTHR 56
E L LP E+I EILLRLP +S+ RF+CV K + L SDP FAK H DL + HR
Sbjct: 32 ESLVLPPEIITEILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLHR 91
Query: 57 LLLRCLFDLDSLGDNS--------AVVTLNLP----PPCMSCI----------------- 87
L+ +L SL NS A V N P P S +
Sbjct: 92 KLIVSSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRRVML 151
Query: 88 --------QNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQILT-----SCDFIIGSL 134
+N + +GS G + ++ V ++NP+TG K++ S ++ +
Sbjct: 152 KLNAKSYRRNWVEIVGSSNGLVCISPGEGAVFLYNPTTGDSKRLPENFRPKSVEYERDNF 211
Query: 135 --YGFGYDNSTDDYFLVLIVLASMNA-KIHAYSVKTNSWDYNHVHVLYMHLGCDYRHGVF 191
YGFG+D TDDY LV +V S + YS+K +SW ++ Y H Y GV
Sbjct: 212 QTYGFGFDGLTDDYKLVKLVATSEDILDASVYSLKADSWR-RICNLNYEHNDGSYTSGVH 270
Query: 192 LNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLLPELAEPVLKAECMPTF--YQVRVLG 249
N ++HW+ + +VV+A+D+ E+P +P+ AE +C F + V L
Sbjct: 271 FNGAIHWVFTESRHNQRVVVAFDIQTEEFREMP-VPDEAE-----DCSHRFSNFVVGSLN 324
Query: 250 GCLSLCYTGSRWDM 263
G LC S +D+
Sbjct: 325 G--RLCVVNSCYDV 336
>At3g23880 unknown protein
Length = 364
Score = 100 bits (248), Expect = 1e-21
Identities = 76/247 (30%), Positives = 112/247 (44%), Gaps = 39/247 (15%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP E++ EILLRLPV+SL RFKCVC SW LIS+ FA H L+L
Sbjct: 14 LPLEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALK---------HALILETSKAT 64
Query: 66 DSLGDNSAVVTLNLPPPCMSCIQNSLY---------------------FLGSCRGFMLLA 104
S V+T + SC +SLY +G+C G +
Sbjct: 65 TSTKSPYGVITTS-RYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQVVGTCHGLVCFH 123
Query: 105 NEYQR-VIVWNPSTGFHKQI----LTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAK 159
+Y + + +WNP+ +++ L + D YGFGYD S DDY +V ++ K
Sbjct: 124 VDYDKSLYLWNPTIKLQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVALLQQRHQVK 183
Query: 160 IHA--YSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSE 217
I YS + W N + + R G+++N +L+W S +S +I+YD+S
Sbjct: 184 IETKIYSTRQKLWRSNTSFPSGVVVADKSRSGIYINGTLNWAATSS-SSSWTIISYDMSR 242
Query: 218 TSLSEIP 224
E+P
Sbjct: 243 DEFKELP 249
>At3g16210 hypothetical protein
Length = 360
Score = 97.1 bits (240), Expect = 9e-21
Identities = 63/220 (28%), Positives = 108/220 (48%), Gaps = 3/220 (1%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP EL EIL+RL ++ L RF+CVCK+W LI+DP F +++ D++ A + + L
Sbjct: 5 LPEELAIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAKFVSFYDKNFYML 64
Query: 66 DSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQILT 125
D G + + P S I S L C G + + + ++VWNP + K +
Sbjct: 65 DVEGKHPVITNKLDFPLDQSMIDESTCVL-HCDGTLCVTLKNHTLMVWNPFSKQFKIVPN 123
Query: 126 SCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHAYSVKTNSWDYNHVHVLYMHLGCD 185
+ ++ GFGYD DDY +V + + H + +T SW + + Y
Sbjct: 124 PGIYQDSNILGFGYDPVHDDYKVVTFIDRLDVSTAHVFEFRTGSWG-ESLRISYPDWHYR 182
Query: 186 YRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPL 225
R G FL+ L+W+ + ++ + ++ ++LS ++PL
Sbjct: 183 DRRGTFLDQYLYWIAY-RSSADRFILCFNLSTHEYRKLPL 221
>At4g12560 putative protein
Length = 408
Score = 87.4 bits (215), Expect = 7e-18
Identities = 77/279 (27%), Positives = 130/279 (45%), Gaps = 36/279 (12%)
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFD--LNAAPTHRLLLRCL 62
T+P +++ +I LRLP ++L+R + + K ++LI+DP F +SH L +LLR
Sbjct: 3 TIPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRGA 62
Query: 63 FDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPST-GFHK 121
L S+ +S ++ P ++ GS G + L+N + V+NPST H+
Sbjct: 63 LRLYSVDLDSLDSVSDVEHPMKRGGPTEVF--GSSNGLIGLSNSPTDLAVFNPSTRQIHR 120
Query: 122 QILTSCDFIIGS------LYGFGYDNSTDDYFLVLIVLASMNA----------KIHAYSV 165
+S D GS YG GYD+ +DDY +V +V +++ ++ +S+
Sbjct: 121 LPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKIDSEDELGCSFPYEVKVFSL 180
Query: 166 KTNSW--------DYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISK--VTSLQVVIAYDL 215
K NSW + Y HL +GV NSLHW++ + + + +++ +DL
Sbjct: 181 KKNSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSLHWVLPRRPGLIAFNLIVRFDL 240
Query: 216 SETSLSEIPLLPELAEPVLKAECMPTFYQVRVLGGCLSL 254
+ EI PE + + + VL GCL L
Sbjct: 241 ALEEF-EIVRFPE----AVANGNVDIQMDIGVLDGCLCL 274
>At3g21120 hypothetical protein
Length = 367
Score = 79.7 bits (195), Expect = 1e-15
Identities = 58/240 (24%), Positives = 105/240 (43%), Gaps = 21/240 (8%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLF-- 63
LP +L+ EIL ++P SL RF+ C+ W L+ D FAK H+ +++ F
Sbjct: 3 LPEDLVLEILSKVPAVSLARFRSTCRRWNALVVDGSFAKKHYAYGPRQYPIVIMLIEFRV 62
Query: 64 -----DLDSLGDNSAVVTLNL-------PPPCMSCIQNSLYFLGSCRGFMLLANEYQRVI 111
DL + +N+ + L P S + + C G +L + +R++
Sbjct: 63 YLVSIDLHGINNNNGAPSAKLTGQFSLKDPLSNSSEEVDIRNAFHCDGLLLCCTKDRRLV 122
Query: 112 VWNPSTGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLI--VLASMNAKIHAYSVKTNS 169
VWNP +G K I + LY GYDN + Y ++ + V + + Y ++S
Sbjct: 123 VWNPCSGETKWIQPRNSYKESDLYALGYDNRSSSYKILRMHPVGNPFHIESEVYDFASHS 182
Query: 170 WDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISK----VTSLQVVIAYDLSETSLSEIPL 225
W V + H+ + +G+ + + +W +SK + ++++D S +PL
Sbjct: 183 WRSVGVTTDF-HIQTNESYGMNVKGTTYWFALSKDWWSSDDRRFLLSFDFSRERFQCLPL 241
>At4g22390 unknown protein
Length = 401
Score = 76.6 bits (187), Expect = 1e-14
Identities = 75/279 (26%), Positives = 124/279 (43%), Gaps = 39/279 (13%)
Query: 7 PHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHF--DLNAAPTHRLLLRCLFD 64
P +LI E+ LRL +L++ + + K F LI P+F SH L +LLR
Sbjct: 5 PTDLINEMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMILLRGPRL 64
Query: 65 LDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPST-GFHKQI 123
L ++ +S ++P P + ++ GS G + L N + ++NPST H+
Sbjct: 65 LRTVELDSPENVSDIPHPLQAGGFTEVF--GSFNGVIGLCNSPVDLAIFNPSTRKIHRLP 122
Query: 124 LTSCDFIIGSL------YGFGYDNSTDDYFLVLIVLASMN---------AKIHAYSVKTN 168
+ DF + YG GYD+ DD+ +V IV + ++ +S+K N
Sbjct: 123 IEPIDFPERDITREYVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFPCPVEVKVFSLKKN 182
Query: 169 SW-------DYNHVHV-LYMHLGCDYRHGVFLNNSLHWLVISK--VTSLQVVIAYDLSET 218
SW ++ + + Y HL +GV +NN LHW++ + V + +I YDL+
Sbjct: 183 SWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNHLHWILPRRQGVIAFNAIIKYDLASD 242
Query: 219 SLSEIPLLPELAEPVLKAECMPTFYQVRVLGGCLSL-CY 256
+ + EL + + VL GC+ L CY
Sbjct: 243 DIGVLSFPQEL--------YIEDNMDIGVLDGCVCLMCY 273
>At3g21410 hypothetical protein
Length = 374
Score = 73.9 bits (180), Expect = 8e-14
Identities = 60/199 (30%), Positives = 90/199 (45%), Gaps = 13/199 (6%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP EL EIL R+P +SLLR K CK W L +D +F H L R
Sbjct: 11 LPFELFEEILCRVPTKSLLRLKLTCKRWLALFNDKRFIYKHLALVREHIIRTNQMVKIIN 70
Query: 66 DSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQI-L 124
+G A + +LP ++ +Y + C G +L E + VWNP + I L
Sbjct: 71 PVVG---ACSSFSLPNKFQ--VKGEIYTMVPCDGLLLCIFETGSMAVWNPCLNQVRWIFL 125
Query: 125 TSCDFIIGSLYGFGYDNSTDDYFLVLIVLA---SMNAKIHAYSVKTNSWDYNHVHV-LYM 180
+ F S YG GYD + D + +L A S ++ Y +K+NSW V + ++
Sbjct: 126 LNPSFRGCSCYGIGYDGLSRDSYKILRFYANTGSYKPEVDIYELKSNSWKTFKVSLDWHV 185
Query: 181 HLGCDYRHGVFLNNSLHWL 199
L C G+ L +++W+
Sbjct: 186 VLRC---KGLSLKGNMYWI 201
>At3g17480 hypothetical protein
Length = 373
Score = 72.4 bits (176), Expect = 2e-13
Identities = 58/211 (27%), Positives = 95/211 (44%), Gaps = 19/211 (9%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCL--- 62
L +L+ +IL R+P SL+R + CK W +++D +F K HFD +LLR L
Sbjct: 11 LTEDLVEDILSRVPATSLVRLRSTCKQWNAILNDRRFIKKHFDTAEKEYLDMLLRSLRVS 70
Query: 63 ---FDLDSLGDN-----SAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWN 114
+L L DN LNL + + + + C G +L N ++VWN
Sbjct: 71 SMSVNLHGLHDNIDPSIELKRELNLKGLLCNSGKVESFEVFHCNGLLLFTNT-STIVVWN 129
Query: 115 PSTGFHKQILTSCDFIIGSLYGFGYDNST--DDYFLVLIVLASMNAKIHAYSVKTNSWD- 171
P TG K I T Y GY+N DY ++ + N ++ Y ++SW
Sbjct: 130 PCTGQTKWIQTESANTRYHKYALGYENKNLCRDYKILRFLDDGTNFELEIYEFNSSSWRV 189
Query: 172 YNHVHVLY-MHLGCDYRHGVFLNNSLHWLVI 201
+ V + + + +G G+ + + +W+VI
Sbjct: 190 LDSVEIDFELDIG---SQGMSVKGNTYWIVI 217
>At2g27520 hypothetical protein
Length = 347
Score = 72.0 bits (175), Expect = 3e-13
Identities = 61/233 (26%), Positives = 102/233 (43%), Gaps = 27/233 (11%)
Query: 1 MEKLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLR 60
M +L LP +L+ EIL RLP SL R + CK W L DP+F F AA +L+
Sbjct: 1 MVRLDLPWDLVDEILSRLPATSLGRLRFTCKRWNALFKDPEFITKQFH-KAAKQDLVLML 59
Query: 61 CLFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANE---YQRVIVWNPST 117
F + S+ N + N+ + + C G +L + E +++V NP T
Sbjct: 60 SNFGVYSMSTNLKEIPNNI----------EIAQVFHCNGLLLCSTEEGNKTKLVVVNPCT 109
Query: 118 GFHKQILTSCDFIIGSLYGFGYDN-----STDDYFLVLIVLASMNAKIHAYSVKTNSWDY 172
G + I D+ GY N S D Y ++ I +I + +K+NSW
Sbjct: 110 GQTRWIEPRTDYNYNHDIALGYGNNSTKKSYDSYKILRITYGCKLVEI--FELKSNSW-- 165
Query: 173 NHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPL 225
+ ++ ++ Y GV + +WL +K ++++D + + +PL
Sbjct: 166 RVLSKVHPNVEKHYYGGVSFKGNTYWLSYTKFN----ILSFDFTTETFRSVPL 214
>At1g62270 hypothetical protein
Length = 383
Score = 72.0 bits (175), Expect = 3e-13
Identities = 62/211 (29%), Positives = 89/211 (41%), Gaps = 14/211 (6%)
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLR---- 60
+LP +L+ +IL R+P SL R + CK W +L +D F K HFD L+LR
Sbjct: 11 SLPWDLVEDILARVPATSLKRLRSTCKQWNFLFNDQIFTKMHFDKAEKQFLVLILRLYTV 70
Query: 61 CLFDLDSLG-----DNSAVV--TLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVW 113
C LD G D S V L+L P S + + + C G +LL ++VW
Sbjct: 71 CSMSLDLRGLHDNIDPSIEVKGELSLIDPHCSSRKTFVSKVFHCNG-LLLCTTMTGLVVW 129
Query: 114 NPSTGFHKQILTSCDFIIGSLYGFGYDNSTDDY-FLVLIVLASMNAKIHAYSVKTNSWDY 172
NP T + I T Y GY N Y + ++ L + + Y V +NSW
Sbjct: 130 NPCTDQTRWIKTEVPHNRNDKYALGYGNYKSCYNYKIMKFLDLESFDLEIYEVNSNSWRV 189
Query: 173 NHVHVLYMHLGCDYRHGVFLNNSLHWLVISK 203
+ D GV L + +W+ K
Sbjct: 190 LGTVTPDFTIPLD-AEGVSLRGNSYWIASHK 219
>At3g08750 hypothetical protein
Length = 369
Score = 70.9 bits (172), Expect = 7e-13
Identities = 62/232 (26%), Positives = 104/232 (44%), Gaps = 17/232 (7%)
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFD 64
+LP ELI EIL ++P SL+RFK CK W+ LI++ +F +H D + +P + +D
Sbjct: 11 SLPFELIEEILYKIPAESLIRFKSTCKKWYNLITEKRFMYNHLD-HYSPERFIR---TYD 66
Query: 65 LDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEY--QRVIVWNPSTGFHKQ 122
+ + +++ L P + +Y + C G ML + VWNP K
Sbjct: 67 QQIIDPVTEILSDALIPDEFRDLY-PIYSMVHCDGLMLCTCRKWDNSLAVWNPVLREIKW 125
Query: 123 ILTSCDFIIGSLYGFGY-DNSTDDYFLVLIVLASM------NAKIHAYSVKTNSWDYNHV 175
I S ++ G GY DN + D + +L +L + + Y K++SW V
Sbjct: 126 IKPSVCYLHTDYVGIGYDDNVSRDNYKILKLLGRLPKDDDSDPNCEIYEFKSDSWK-TLV 184
Query: 176 HVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLLP 227
+ +GV + ++W I+K +I +D S + EI + P
Sbjct: 185 AKFDWDIDIRCNNGVSVKGKMYW--IAKKKEDFTIIRFDFSTETFKEICVCP 234
>At4g04690
Length = 378
Score = 68.9 bits (167), Expect = 3e-12
Identities = 56/236 (23%), Positives = 102/236 (42%), Gaps = 20/236 (8%)
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFD 64
+LP EL+ EIL + P SL RFK CK W+ +I+ +F +H D + R +
Sbjct: 10 SLPFELVEEILKKTPAESLNRFKSTCKQWYGIITSKRFMYNHLDHS---PERFIRIDDHK 66
Query: 65 LDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLA---NEYQR-----VIVWNPS 116
+ D + + P P + +S + C G ML + Y+R + VWNP
Sbjct: 67 TVQIMDPMTGIFSDSPVPDVFRSPHSFASMVHCDGLMLCICSDSSYERTREANLAVWNPV 126
Query: 117 TGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASM---NAKIHAYSVKTNSWDYN 173
T K I + +G GYDN+ + + ++ M + + Y K++SW
Sbjct: 127 TKKIKWIEPLDSYYETDYFGIGYDNTCRENYKIVRFSGPMSFDDTECEIYEFKSDSWRTL 186
Query: 174 HVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLLPEL 229
++ C GV + +++W+ +K + ++ +D S + + + P +
Sbjct: 187 DTKYWDVYTQC---RGVSVKGNMYWIADTKE---KFILRFDFSMETFKNVCVCPPI 236
>At3g17500 hypothetical protein
Length = 438
Score = 68.9 bits (167), Expect = 3e-12
Identities = 61/244 (25%), Positives = 108/244 (44%), Gaps = 24/244 (9%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLR----- 60
LP +L+ EIL R+P SL R + CKSW D +F + H + A L+L+
Sbjct: 4 LPLDLVEEILSRVPATSLKRLRSTCKSWNNCYKDQRFTEKHSVIAAKQFLVLMLKDCRVS 63
Query: 61 -CLFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGF 119
+L+ + +N A ++ L +Q S F C G +L + R+ VWNP TG
Sbjct: 64 SVSVNLNEIHNNIAAPSIELKGVVGPQMQISGIF--HCDGLLLCTTKDDRLEVWNPCTGQ 121
Query: 120 HKQILTSCDFIIGSLYGFGY--DNSTDDYFLVLIVLASMN----AKIHAYSVKTNSWDYN 173
+++ S + S + GY +NS Y ++ M+ ++ Y ++SW +
Sbjct: 122 TRRVQHSIHYKTNSEFVLGYVNNNSRHSYKILRYWNFYMSNYRVSEFEIYDFSSDSWRF- 180
Query: 174 HVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLLPELAEPV 233
+ ++ C V L + +WL + + +++ +D S I L P+
Sbjct: 181 ---IDEVNPYCLTEGEVSLKGNTYWLASDEKRDIDLILRFDFS------IERYQRLNLPI 231
Query: 234 LKAE 237
LK++
Sbjct: 232 LKSD 235
>At3g17570 hypothetical protein
Length = 381
Score = 68.6 bits (166), Expect = 3e-12
Identities = 54/231 (23%), Positives = 95/231 (40%), Gaps = 15/231 (6%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP +L EIL R+P SL + K CK W+ L DP+F K H L+ ++ L
Sbjct: 5 LPRDLETEILSRVPATSLQKLKPTCKRWYTLFKDPEFLKKHVGRAEREVISLMSLRVYSL 64
Query: 66 --DSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQI 123
+ G +S+V + + + C G +L + R++VWNP TG + I
Sbjct: 65 SVNLSGIHSSVEMTGMLNSLKDSEDVKISDITECNGLLLCTTDDSRLVVWNPYTGETRWI 124
Query: 124 --LTSCDFIIGSLYGFGYDNSTDDYFLVLIV-----LASMNAKIHAYSVKTNSWDYNHVH 176
++ + + + GYDN+ + I+ L + Y ++SW + +
Sbjct: 125 PYKSNSPYEMYQKFVLGYDNTNKSRYSYKILRCYHGLIDFGYEFEIYEFNSHSWRRFYDN 184
Query: 177 VLYMHLGCDYR-HGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLSEIPLL 226
C + GV L + +W S +++ +D + + LL
Sbjct: 185 ----SPNCSFESKGVTLKGNTYWFA-SDTEGRHIILRFDFATERFGRLSLL 230
>At1g11270 unknown protein (At1g11270)
Length = 312
Score = 68.6 bits (166), Expect = 3e-12
Identities = 73/263 (27%), Positives = 105/263 (39%), Gaps = 36/263 (13%)
Query: 3 KLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHF--DLNAAPTHRLLLR 60
KL LPH+++ IL RLPV SLLRFKCV W I F + + + L++
Sbjct: 32 KLLLPHDVVGLILERLPVESLLRFKCVSNQWKSTIESQCFQERQLIRRMESRGPDVLVVS 91
Query: 61 CLFDLDSLG-----DNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNP 115
D D G +S V T P ++L GSC G + + Y IV NP
Sbjct: 92 FADDEDKYGRKAVFGSSIVSTFRFP------TLHTLICYGSCEGLICIYCVYSPNIVVNP 145
Query: 116 STGFHKQILTS--------------CDFIIGSLYGFGYDNSTDDYFLVLIVLASM----- 156
+T +H+ S DF L FG D Y V + +S
Sbjct: 146 ATKWHRSCPLSNLQQFLDDKFEKKEYDFPTPKL-AFGKDKLNGTYKQVWLYNSSEFRLDD 204
Query: 157 NAKIHAYSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLS 216
+ N+W Y VH + DY+ V+ + S+HWL K + + + +
Sbjct: 205 VTTCEVFDFSNNAWRY--VHPASPYRINDYQDPVYSDGSVHWLTEGKESKILSFHLHTET 262
Query: 217 ETSLSEIPLLPELAEPVLKAECM 239
L E P L E +PV + C+
Sbjct: 263 FQVLCEAPFLRE-RDPVGDSMCI 284
>At3g17540 hypothetical protein
Length = 396
Score = 67.4 bits (163), Expect = 8e-12
Identities = 49/152 (32%), Positives = 72/152 (47%), Gaps = 15/152 (9%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LPHE+ EIL R+P +SL + CK W+ L DP+F K +F RL+L F +
Sbjct: 10 LPHEIESEILSRVPTKSLAKLHTTCKRWYALFRDPRFVKKNF---GKSERRLMLHSNFGV 66
Query: 66 DSLGDNSAVVTLNLPPPCM---SCIQN-------SLYFLGSCRGFMLLA-NEYQRVIVWN 114
+ D+ + LN P + S + N ++ + C G +L + E R++VWN
Sbjct: 67 YKITDDLHGI-LNSGDPSLEFTSKLSNLKISEDLTITKIFHCDGLILCSTKENTRLVVWN 125
Query: 115 PSTGFHKQILTSCDFIIGSLYGFGYDNSTDDY 146
P TG + I S + Y GY NS Y
Sbjct: 126 PCTGQTRWIKPSKRYRSDDSYCLGYVNSKSSY 157
>At3g16590 hypothetical protein
Length = 374
Score = 67.4 bits (163), Expect = 8e-12
Identities = 69/252 (27%), Positives = 105/252 (41%), Gaps = 46/252 (18%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFD- 64
LP EL EILLR+P SL RF+ VCK W L +D +F +H A + +LR D
Sbjct: 5 LPLELEDEILLRVPPLSLTRFRTVCKRWNTLFNDQRFINNHL---ACVRPQFILRTEKDS 61
Query: 65 -LDSLG----DNSAVVTLNL----PPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNP 115
+ S+G D+ V LNL P + +N Y C GF+L V VWNP
Sbjct: 62 KIYSIGINIDDSLEVRELNLETQGPNKKLKVYRNLFY----CDGFLLCPALLDEVAVWNP 117
Query: 116 -------------------STGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASM 156
G+ + C I+G +G+GY + + + +
Sbjct: 118 WLRKQTKWIEPKRSRFNLYGLGYDNRRPEKCYKILG--FGYGYSSEINGSY------NRI 169
Query: 157 NAKIHAYSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVI-AYDL 215
N ++ + +TN+W + HL R + LN +L+W+ + + I ++D
Sbjct: 170 NPRVSVFEFETNAWKDLKFGLFDWHLRSP-RTVLSLNGTLYWIAVRCESGGDGFIQSFDF 228
Query: 216 SETSLSEIPLLP 227
S LLP
Sbjct: 229 SREMFEPFCLLP 240
>At5g10340 putative protein
Length = 445
Score = 65.5 bits (158), Expect = 3e-11
Identities = 73/272 (26%), Positives = 120/272 (43%), Gaps = 51/272 (18%)
Query: 1 MEKLTLPHELI-FEILLRLPVRSLLRFKCVCKSWFYLISDPQFAK----SHFDLNAAPTH 55
ME+L LPH++I + I++RL V++LL+FK V K W I P F + H ++ H
Sbjct: 65 MEEL-LPHDVIEYHIMVRLDVKTLLKFKSVSKQWMSTIQSPSFQERQLIHHLSQSSGDPH 123
Query: 56 RLLLRCLFD---------------LDSLGDNSAVVTLNLPPPCMSCIQNSLYFL--GSCR 98
+LL L+D L + S+ ++ +P P ++ LYF+ SC
Sbjct: 124 -VLLVSLYDPCARQQDPSISSFEALRTFLVESSAASVQIPTPW----EDKLYFVCNTSCD 178
Query: 99 GFMLLANEYQ-RVIVWNPSTGFHK-------QILTS-------CDFIIGSLYGFGYDNST 143
G + L + Y+ IV NP+T +H+ Q++ + C + GFG D +
Sbjct: 179 GLICLFSFYELPSIVVNPTTRWHRTFPKCNYQLVAADKGERHECFKVACPTPGFGKDKIS 238
Query: 144 DDYFLVLIVLAS---MNAK---IHAYSVKTNSWDYNHVHVLYMHLGCDYRHGVFLNNSLH 197
Y V + ++ +N K + TN+W Y V HL + V+++ SLH
Sbjct: 239 GTYKPVWLYNSAELDLNDKPTTCEVFDFATNAWRY--VFPASPHLILHTQDPVYVDGSLH 296
Query: 198 WLVISKVTSLQVVIAYDLSETSLSEIPLLPEL 229
W +V++ DL + I P L
Sbjct: 297 WFTALSHEGETMVLSLDLHSETFQVISKAPFL 328
>At3g49980 putative protein
Length = 382
Score = 65.5 bits (158), Expect = 3e-11
Identities = 70/244 (28%), Positives = 100/244 (40%), Gaps = 45/244 (18%)
Query: 4 LTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLL---- 59
L LP EL+ EIL R+P SL + + CK W L +D F++ HFD AP L+
Sbjct: 3 LNLPQELLEEILCRVPATSLKQLRLTCKEWNRLFNDRTFSRKHFD--KAPKQFLITVLEE 60
Query: 60 RCLFDLDSL----GDNSAVVTLNLPPPCMSCIQNSLYFLG--SCRG-FMLLANEYQRVIV 112
RC S+ G S T L P + + + C G F+ + R++V
Sbjct: 61 RCRLSSLSINLHSGFPSEEFTGELSPIDYHSNSSQVIIMKIFHCDGLFVCTILKDTRIVV 120
Query: 113 WNPSTGFHKQILT-------SCDFIIGSLYGFGYDNSTDDYFLVLIVLASMN---AKIHA 162
WNP TG K I T DF++ G+ DN + D ++ N +
Sbjct: 121 WNPCTGQKKWIQTGENLDENGQDFVL----GYYQDNKSSDKSYKILSYKGYNYGDQEFKI 176
Query: 163 YSVKTNSW---DYNHVHVLYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETS 219
Y +K+N+W D + Y DYR V L + +W AYDL +
Sbjct: 177 YDIKSNTWRNLDVTPIPGNYFTCS-DYR--VSLKGNTYW------------FAYDLKDEQ 221
Query: 220 LSEI 223
L I
Sbjct: 222 LGLI 225
>At3g24700 hypothetical protein
Length = 249
Score = 64.7 bits (156), Expect = 5e-11
Identities = 60/196 (30%), Positives = 88/196 (44%), Gaps = 42/196 (21%)
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLL-LRCLFD 64
LP +L EIL R+P SL R K CK W+ L DP+F K +L A TH + R +
Sbjct: 5 LPLDLESEILSRVPATSLQRLKTTCKRWYALFRDPRFVKK--NLGKAATHVIFDNRSGYS 62
Query: 65 LDSLGDNSAVVTLNLPPPCMSCIQNSL----------------------YFLGSCRGFML 102
+ + NS + ++NL IQNS + + C G +L
Sbjct: 63 MTDI--NSLIHSINL-----RGIQNSFDPSIGVDVKLNVLKDPRHDKISHIISHCDGLLL 115
Query: 103 LANE-YQRVIVWNPSTGFHKQILTSCDFIIGSLYGFGYDN---STDDY----FLVLIVLA 154
E Y R++VWNP TG K I + ++ +Y GY N S + Y F +L + +
Sbjct: 116 CKTEDYGRLVVWNPCTGQIKWI--QANNMLMDVYVLGYVNNNKSCNSYKILNFGILPLNS 173
Query: 155 SMNAKIHAYSVKTNSW 170
S + K Y ++SW
Sbjct: 174 SHDNKSKIYEFNSDSW 189
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,389,945
Number of Sequences: 26719
Number of extensions: 265484
Number of successful extensions: 1302
Number of sequences better than 10.0: 280
Number of HSP's better than 10.0 without gapping: 253
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 901
Number of HSP's gapped (non-prelim): 332
length of query: 268
length of database: 11,318,596
effective HSP length: 98
effective length of query: 170
effective length of database: 8,700,134
effective search space: 1479022780
effective search space used: 1479022780
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0476a.5


