

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0437b.10
(101 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
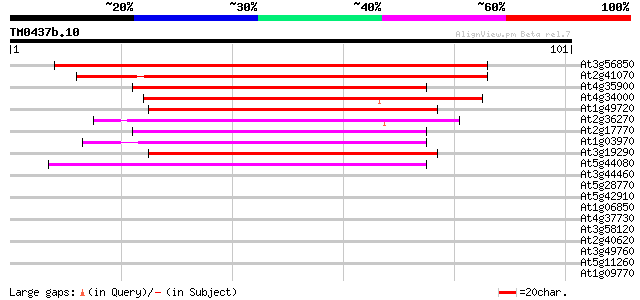
Score E
Sequences producing significant alignments: (bits) Value
At3g56850 ABA-responsive element binding bZip transcription fact... 72 4e-14
At2g41070 bZIP transcription factor AtbZIP12 / DPBF4 60 2e-10
At4g35900 bZIP transcription factor AtbZIP14 47 1e-06
At4g34000 abscisic acid responsive elements-binding factor (ABRE... 46 3e-06
At1g49720 abscisic acid responsive elements-binding factor (ABRE... 46 3e-06
At2g36270 bZip transcription factor AtbZip39 45 8e-06
At2g17770 bZIP transcription factor atbzip27 45 8e-06
At1g03970 G-box binding bZip transcription factor GBF4 / AtbZip40 44 2e-05
At3g19290 abscisic acid responsive elements-binding factor (ABRE... 43 2e-05
At5g44080 bZIP protein AtbZIP13 39 3e-04
At3g44460 bZIP transcription factor DPBF2 / AtbZip67 37 0.002
At5g28770 bZip transcription factor BZO2H3 / AtbZip63 35 0.005
At5g42910 bzip protein AtbZip15 35 0.008
At1g06850 bZip transcription factor AtbZip52 34 0.011
At4g37730 bZip transcription factor AtbZip7 33 0.018
At3g58120 bZip transcription factor AtbZip61 33 0.018
At2g40620 bZIP transcription factor AtbZip18 33 0.018
At3g49760 bZIP transcription factor AtbZip5 33 0.031
At5g11260 bZip transcription factor HY5 / AtbZip56 32 0.070
At1g09770 putative DNA-binding protein, Myb 31 0.12
>At3g56850 ABA-responsive element binding bZip transcription factor
AREB3 / AtbZip66
Length = 297
Score = 72.4 bits (176), Expect = 4e-14
Identities = 41/78 (52%), Positives = 52/78 (66%)
Query: 9 VSNIQKHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVE 68
+S+ Q GRK +A ++VE TV+RRQKRMIKN S A SRA KQAY + LE V LE E
Sbjct: 204 LSDTQTPGRKRVASGEVVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEE 263
Query: 69 IERLMKQTAIQNLLTQIP 86
ERL KQ ++ +L +P
Sbjct: 264 NERLRKQKEVEKILPSVP 281
>At2g41070 bZIP transcription factor AtbZIP12 / DPBF4
Length = 262
Score = 59.7 bits (143), Expect = 2e-10
Identities = 38/74 (51%), Positives = 47/74 (63%), Gaps = 1/74 (1%)
Query: 13 QKHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERL 72
Q GRK +A E +VE TV+RRQKRMIKN S A SRA KQAY + LE V LE E E+L
Sbjct: 174 QAPGRKRVAGE-IVEKTVERRQKRMIKNRESAARSRARKQAYTHELEIKVSRLEEENEKL 232
Query: 73 MKQTAIQNLLTQIP 86
+ ++ +L P
Sbjct: 233 RRLKEVEKILPSEP 246
>At4g35900 bZIP transcription factor AtbZIP14
Length = 285
Score = 47.4 bits (111), Expect = 1e-06
Identities = 28/53 (52%), Positives = 33/53 (61%)
Query: 23 EDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQ 75
+D E + +RR KRMIKN S A SRA KQAY N LE V L+ E RL +Q
Sbjct: 207 QDSNEGSGNRRHKRMIKNRESAARSRARKQAYTNELELEVAHLQAENARLKRQ 259
>At4g34000 abscisic acid responsive elements-binding factor
(ABRE/ABF3) / bZip transcription factor AtbZip37
Length = 449
Score = 46.2 bits (108), Expect = 3e-06
Identities = 28/62 (45%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query: 25 MVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SL-EVEIERLMKQTAIQNLLT 83
++E ++RRQKRMIKN S A SRA KQAY LE + L E+ E KQ + + L+
Sbjct: 367 VLEKVIERRQKRMIKNRESAARSRARKQAYTMELEAEIAQLKELNEELQKKQVCLASSLS 426
Query: 84 QI 85
Q+
Sbjct: 427 QL 428
>At1g49720 abscisic acid responsive elements-binding factor
(ABRE/ABF1)/ bZip transcription factor AtbZip35
Length = 392
Score = 46.2 bits (108), Expect = 3e-06
Identities = 27/52 (51%), Positives = 34/52 (64%)
Query: 26 VENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQTA 77
+E V+RRQKRMIKN S A SRA KQAY LE + SL++ + L K+ A
Sbjct: 307 LEKVVERRQKRMIKNRESAARSRARKQAYTLELEAEIESLKLVNQDLQKKQA 358
>At2g36270 bZip transcription factor AtbZip39
Length = 442
Score = 44.7 bits (104), Expect = 8e-06
Identities = 32/76 (42%), Positives = 42/76 (55%), Gaps = 11/76 (14%)
Query: 16 GRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLE--------- 66
GRK + ++ VE V+RRQ+RMIKN S A SRA KQAY LE + L+
Sbjct: 342 GRKRV-VDGPVEKVVERRQRRMIKNRESAARSRARKQAYTVELEAELNQLKEENAQLKHA 400
Query: 67 -VEIERLMKQTAIQNL 81
E+ER KQ ++L
Sbjct: 401 LAELERKRKQQYFESL 416
>At2g17770 bZIP transcription factor atbzip27
Length = 234
Score = 44.7 bits (104), Expect = 8e-06
Identities = 27/53 (50%), Positives = 31/53 (57%)
Query: 23 EDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQ 75
+D + DRR KRMIKN S A SRA KQAY N LE + L+ E RL Q
Sbjct: 156 QDSDDTRGDRRYKRMIKNRESAARSRARKQAYTNELELEIAHLQTENARLKIQ 208
>At1g03970 G-box binding bZip transcription factor GBF4 / AtbZip40
Length = 270
Score = 43.5 bits (101), Expect = 2e-05
Identities = 29/62 (46%), Positives = 36/62 (57%), Gaps = 3/62 (4%)
Query: 14 KHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLM 73
K GR MM + ++ +RQKRMIKN S A SR KQAY LE LE E E+L+
Sbjct: 174 KRGRVMM---EAMDKAAAQRQKRMIKNRESAARSRERKQAYQVELETLAAKLEEENEQLL 230
Query: 74 KQ 75
K+
Sbjct: 231 KE 232
>At3g19290 abscisic acid responsive elements-binding factor
(ABRE/ABF4) / bZip transcription factor AtbZip38
Length = 431
Score = 43.1 bits (100), Expect = 2e-05
Identities = 24/52 (46%), Positives = 32/52 (61%)
Query: 26 VENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQTA 77
+E ++RRQ+RMIKN S A SRA KQAY LE + L+ + L K+ A
Sbjct: 347 LEKVIERRQRRMIKNRESAARSRARKQAYTLELEAEIEKLKKTNQELQKKQA 398
>At5g44080 bZIP protein AtbZIP13
Length = 315
Score = 39.3 bits (90), Expect = 3e-04
Identities = 26/68 (38%), Positives = 35/68 (51%)
Query: 8 DVSNIQKHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEV 67
DV G++ + + ++ +RQ+RMIKN S A SR KQAY LE LE
Sbjct: 209 DVYGGGARGKRARVMVEPLDKAAAQRQRRMIKNRESAARSRERKQAYQVELEALAAKLEE 268
Query: 68 EIERLMKQ 75
E E L K+
Sbjct: 269 ENELLSKE 276
>At3g44460 bZIP transcription factor DPBF2 / AtbZip67
Length = 331
Score = 36.6 bits (83), Expect = 0.002
Identities = 23/55 (41%), Positives = 32/55 (57%)
Query: 18 KMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERL 72
K ++ E ++RRQ+RMIKN S A SRA +QAY LE + +L E +L
Sbjct: 235 KKRIIDGPPEILMERRQRRMIKNRESAARSRARRQAYTVELELELNNLTEENTKL 289
>At5g28770 bZip transcription factor BZO2H3 / AtbZip63
Length = 307
Score = 35.4 bits (80), Expect = 0.005
Identities = 20/43 (46%), Positives = 26/43 (59%)
Query: 32 RRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMK 74
+R KRM+ N S SR KQA+++ LE V L VE +LMK
Sbjct: 146 KRVKRMLSNRESARRSRRRKQAHLSELETQVSQLRVENSKLMK 188
>At5g42910 bzip protein AtbZip15
Length = 370
Score = 34.7 bits (78), Expect = 0.008
Identities = 20/46 (43%), Positives = 29/46 (62%)
Query: 30 VDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQ 75
VD++ +R IKN S A SRA KQA +E + +L+ + E L+KQ
Sbjct: 293 VDKKLRRKIKNRESAARSRARKQAQTMEVEVELENLKKDYEELLKQ 338
>At1g06850 bZip transcription factor AtbZip52
Length = 337
Score = 34.3 bits (77), Expect = 0.011
Identities = 24/64 (37%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Query: 17 RKMMALEDMVE--NTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMK 74
+K M E + E N +R KR++ N S A S+ K YI LER V SL+ E L
Sbjct: 133 KKAMPPEKLSELWNIDPKRAKRILANRQSAARSKERKARYIQELERKVQSLQTEATTLSA 192
Query: 75 QTAI 78
Q +
Sbjct: 193 QLTL 196
>At4g37730 bZip transcription factor AtbZip7
Length = 305
Score = 33.5 bits (75), Expect = 0.018
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Query: 3 TSSTYDVSNIQKHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV 62
T+ T S H RK M +M + +R++KRM N S SR KQ++I+ L V
Sbjct: 171 TAITNFGSGENNHNRKKMIQPEMTD---ERKRKRMESNRESAKRSRMRKQSHIDNLREQV 227
Query: 63 *SLEVEIERL 72
L++E L
Sbjct: 228 NRLDLENREL 237
>At3g58120 bZip transcription factor AtbZip61
Length = 329
Score = 33.5 bits (75), Expect = 0.018
Identities = 22/57 (38%), Positives = 31/57 (53%), Gaps = 3/57 (5%)
Query: 32 RRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQTAI---QNLLTQI 85
+R KR++ N S SR K YI+ LER+V SL+ E+ L + A Q LL +
Sbjct: 204 KRVKRILANRQSAQRSRVRKLQYISELERSVTSLQTEVSVLSPRVAFLDHQRLLLNV 260
>At2g40620 bZIP transcription factor AtbZip18
Length = 367
Score = 33.5 bits (75), Expect = 0.018
Identities = 24/64 (37%), Positives = 34/64 (52%), Gaps = 2/64 (3%)
Query: 17 RKMMALEDMVENTV--DRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMK 74
+K MA + + E V +R KR+I N S A S+ K YI LER V +L+ E L
Sbjct: 133 KKAMAPDKLAELWVVDPKRAKRIIANRQSAARSKERKARYILELERKVQTLQTEATTLSA 192
Query: 75 QTAI 78
Q ++
Sbjct: 193 QLSL 196
>At3g49760 bZIP transcription factor AtbZip5
Length = 156
Score = 32.7 bits (73), Expect = 0.031
Identities = 19/71 (26%), Positives = 36/71 (49%), Gaps = 4/71 (5%)
Query: 5 STYDVSNIQKHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV*S 64
ST+D ++ + G M +NT +R++KR + N S SR KQ ++ + +
Sbjct: 49 STHDYGSVNQIGSDMSP----TDNTDERKKKRKLSNRESAKRSREKKQKHLEEMSIQLNQ 104
Query: 65 LEVEIERLMKQ 75
L+++ + L Q
Sbjct: 105 LKIQNQELKNQ 115
>At5g11260 bZip transcription factor HY5 / AtbZip56
Length = 168
Score = 31.6 bits (70), Expect = 0.070
Identities = 18/51 (35%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Query: 31 DRRQKRMIKN*VSGASSRAVKQAYINVLERNV*SLEVEIERLMKQ-TAIQN 80
++R KR+++N VS +R K+AY++ LE V LE + L ++ + +QN
Sbjct: 89 NKRLKRLLRNRVSAQQARERKKAYLSELENRVKDLENKNSELEERLSTLQN 139
>At1g09770 putative DNA-binding protein, Myb
Length = 844
Score = 30.8 bits (68), Expect = 0.12
Identities = 21/85 (24%), Positives = 42/85 (48%), Gaps = 6/85 (7%)
Query: 3 TSSTYDVSNIQKHGRKMMALEDMVENTVDRRQKRMIKN*VSGASSRAVKQAYINVLERNV 62
T S Y++S++ + K+ A ++ +EN +K+M ++ +A + Y ER
Sbjct: 674 TRSAYELSSVAGNADKVAAFQEEMENV----RKKMEEDEKKAEHMKAKYKTYTKGHERRA 729
Query: 63 *SLEVEIERLMKQTAIQNLLTQIPC 87
++ +IE +KQ I T++ C
Sbjct: 730 ETVWTQIEATLKQAEIGG--TEVEC 752
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,656,652
Number of Sequences: 26719
Number of extensions: 43761
Number of successful extensions: 233
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 196
Number of HSP's gapped (non-prelim): 42
length of query: 101
length of database: 11,318,596
effective HSP length: 77
effective length of query: 24
effective length of database: 9,261,233
effective search space: 222269592
effective search space used: 222269592
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0437b.10


