

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.11
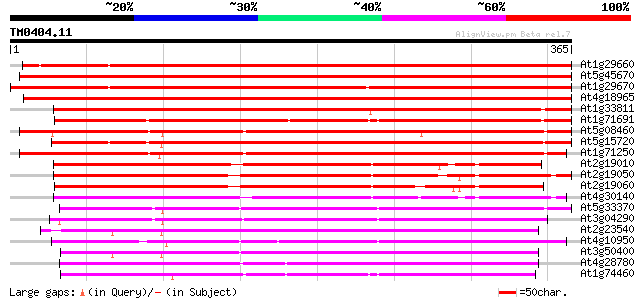
(365 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g29660 lipase/hydrolase like protein 518 e-147
At5g45670 GDSL-motif lipase/hydrolase-like protein 511 e-145
At1g29670 lipase/hydrolase-like protein 507 e-144
At4g18965 unknown protein 504 e-143
At1g33811 unknown protein 376 e-104
At1g71691 unknown protein 353 1e-97
At5g08460 GDSL-motif lipase/acylhydrolase-like protein 327 5e-90
At5g15720 unknown protein 327 6e-90
At1g71250 putative GDSL-motif lipase/acylhydrolase 322 2e-88
At2g19010 putative GDSL-motif lipase/hydrolase 284 6e-77
At2g19050 putative GDSL-motif lipase/hydrolase 280 9e-76
At2g19060 putative GDSL-motif lipase/hydrolase 268 3e-72
At4g30140 unknown protein 257 8e-69
At5g33370 unknown protein 238 3e-63
At3g04290 GDSL-motif lipase/acylhydrolase like protein 235 2e-62
At2g23540 putative GDSL-motif lipase/hydrolase 232 3e-61
At4g10950 unknown protein 228 5e-60
At3g50400 unknown protein 226 2e-59
At4g28780 Proline-rich APG - like protein 223 2e-58
At1g74460 putative lipase/acylhydrolase 214 5e-56
>At1g29660 lipase/hydrolase like protein
Length = 364
Score = 518 bits (1334), Expect = e-147
Identities = 251/357 (70%), Positives = 294/357 (82%), Gaps = 2/357 (0%)
Query: 9 VVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGP 68
+V++ V++LGL G V A PQVPCYFIFGDSLVDNGNNN LRS+ARADY PYGIDF GGP
Sbjct: 10 LVSVWVLLLGL-GFKVKAEPQVPCYFIFGDSLVDNGNNNRLRSIARADYFPYGIDF-GGP 67
Query: 69 SGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRI 128
+GRFSNG+TTVDV+ ELLGFD++IP Y + SG +IL+GVN+ASAAAGIREETG QLG RI
Sbjct: 68 TGRFSNGRTTVDVLTELLGFDNYIPAYSTVSGQEILQGVNYASAAAGIREETGAQLGQRI 127
Query: 129 SFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQY 188
+FSGQV+NY++TV+QVV ILG+E AA+YL +CIYS+G+GSNDYLNNYFMPQFYSTSRQY
Sbjct: 128 TFSGQVENYKNTVAQVVEILGDEYTAADYLKRCIYSVGMGSNDYLNNYFMPQFYSTSRQY 187
Query: 189 TTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDA 248
T +QYAD LI Y +QL LYN+GARK L GIG IGCSPN LAQ SQDG TCV IN A
Sbjct: 188 TPEQYADDLISRYRDQLNALYNYGARKFALVGIGAIGCSPNALAQGSQDGTTCVERINSA 247
Query: 249 NQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNG 308
N+IFNN+L S+V Q NN DA YINAYG FQDIIA+P+ YGF+NTN CCG+GRN G
Sbjct: 248 NRIFNNRLISMVQQLNNAHSDASFTYINAYGAFQDIIANPSAYGFTNTNTACCGIGRNGG 307
Query: 309 QITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
Q+TCLP + PC NR EY+FWDAFHPS A N IA+R+Y+AQ SD YPIDI +LAQ+
Sbjct: 308 QLTCLPGEPPCLNRDEYVFWDAFHPSAAANTAIAKRSYNAQRSSDVYPIDISQLAQL 364
>At5g45670 GDSL-motif lipase/hydrolase-like protein
Length = 362
Score = 511 bits (1317), Expect = e-145
Identities = 243/359 (67%), Positives = 289/359 (79%)
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
M ++ M++MV + P PCYFIFGDSLVDNGNNN L+SLARA+Y PYGIDF
Sbjct: 4 MSLMIMMIMVAVTMINIAKSDPIAPCYFIFGDSLVDNGNNNQLQSLARANYFPYGIDFAA 63
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGG 126
GP+GRFSNG TTVDVIA+LLGF+D+I PY S G DILRGVN+ASAAAGIR+ETG+QLGG
Sbjct: 64 GPTGRFSNGLTTVDVIAQLLGFEDYITPYASARGQDILRGVNYASAAAGIRDETGRQLGG 123
Query: 127 RISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSR 186
RI+F+GQV N+ +TVSQVVNILG++++A+NYLSKCIYSIGLGSNDYLNNYFMP FYST
Sbjct: 124 RIAFAGQVANHVNTVSQVVNILGDQNEASNYLSKCIYSIGLGSNDYLNNYFMPTFYSTGN 183
Query: 187 QYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDIN 246
Q++ + YAD L+ YTEQL+ LY GARK L G+G IGCSPNELAQ S+DGRTC IN
Sbjct: 184 QFSPESYADDLVARYTEQLRVLYTNGARKFALIGVGAIGCSPNELAQNSRDGRTCDERIN 243
Query: 247 DANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRN 306
AN+IFN+KL S+VD FN PDA+ YINAYGIFQDII +PA YGF TN GCCGVGRN
Sbjct: 244 SANRIFNSKLISIVDAFNQNTPDAKFTYINAYGIFQDIITNPARYGFRVTNAGCCGVGRN 303
Query: 307 NGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
NGQITCLP Q PC NR EY+FWDAFHP EA N+VI +R++ ++ SDA+P DI++LA +
Sbjct: 304 NGQITCLPGQAPCLNRNEYVFWDAFHPGEAANIVIGRRSFKREAASDAHPYDIQQLASL 362
>At1g29670 lipase/hydrolase-like protein
Length = 363
Score = 507 bits (1306), Expect = e-144
Identities = 247/365 (67%), Positives = 295/365 (80%), Gaps = 2/365 (0%)
Query: 1 MDLNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPY 60
M+ + V ++++ G A QVPC+F+FGDSLVDNGNNN L S+AR++Y PY
Sbjct: 1 MESYLTKWCVVLVLLCFGFSVVKAQAQAQVPCFFVFGDSLVDNGNNNGLISIARSNYFPY 60
Query: 61 GIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREET 120
GIDF GGP+GRFSNGKTTVDVIAELLGF+ +IP Y + SG IL GVN+ASAAAGIREET
Sbjct: 61 GIDF-GGPTGRFSNGKTTVDVIAELLGFNGYIPAYNTVSGRQILSGVNYASAAAGIREET 119
Query: 121 GQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQ 180
G+QLG RISFSGQV+NYQ+TVSQVV +LG+E +AA+YL +CIYS+GLGSNDYLNNYFMP
Sbjct: 120 GRQLGQRISFSGQVRNYQTTVSQVVQLLGDETRAADYLKRCIYSVGLGSNDYLNNYFMPT 179
Query: 181 FYSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRT 240
FYS+SRQ+T +QYA+ LI Y+ QL LYN+GARK L GIG +GCSPN LA S DGRT
Sbjct: 180 FYSSSRQFTPEQYANDLISRYSTQLNALYNYGARKFALSGIGAVGCSPNALA-GSPDGRT 238
Query: 241 CVSDINDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGC 300
CV IN ANQIFNNKL+S+VDQ NN PDA+ IYINAYGIFQD+I +PA +GF TN GC
Sbjct: 239 CVDRINSANQIFNNKLRSLVDQLNNNHPDAKFIYINAYGIFQDMITNPARFGFRVTNAGC 298
Query: 301 CGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIK 360
CG+GRN GQITCLP Q PC +R Y+FWDAFHP+EA NV+IA+R+Y+AQS SDAYP+DI
Sbjct: 299 CGIGRNAGQITCLPGQRPCRDRNAYVFWDAFHPTEAANVIIARRSYNAQSASDAYPMDIS 358
Query: 361 RLAQI 365
RLAQ+
Sbjct: 359 RLAQL 363
>At4g18965 unknown protein
Length = 361
Score = 504 bits (1299), Expect = e-143
Identities = 244/356 (68%), Positives = 284/356 (79%)
Query: 10 VAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPS 69
V M+ M + + P PCYFIFGDSLVD+GNNN L SLARA+Y PYGIDF GP+
Sbjct: 6 VMMMAMAIAMAMNIAMGDPIAPCYFIFGDSLVDSGNNNRLTSLARANYFPYGIDFQYGPT 65
Query: 70 GRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRIS 129
GRFSNGKTTVDVI ELLGFDD+I PY G+DILRGVN+ASAAAGIREETG+QLG RI+
Sbjct: 66 GRFSNGKTTVDVITELLGFDDYITPYSEARGEDILRGVNYASAAAGIREETGRQLGARIT 125
Query: 130 FSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYT 189
F+GQV N+ +TVSQVVNILG+E++AANYLSKCIYSIGLGSNDYLNNYFMP +YST QY+
Sbjct: 126 FAGQVANHVNTVSQVVNILGDENEAANYLSKCIYSIGLGSNDYLNNYFMPVYYSTGSQYS 185
Query: 190 TDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDAN 249
D YA+ LI YTEQL+ +YN GARK L GIG IGCSPNELAQ S+DG TC IN AN
Sbjct: 186 PDAYANDLINRYTEQLRIMYNNGARKFALVGIGAIGCSPNELAQNSRDGVTCDERINSAN 245
Query: 250 QIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQ 309
+IFN+KL S+VD FN P A+ YINAYGIFQD++A+P+ YGF TN GCCGVGRNNGQ
Sbjct: 246 RIFNSKLVSLVDHFNQNTPGAKFTYINAYGIFQDMVANPSRYGFRVTNAGCCGVGRNNGQ 305
Query: 310 ITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
ITCLP Q PC NR EY+FWDAFHP EA NVVI R++ +S SDA+P DI++LA++
Sbjct: 306 ITCLPGQAPCLNRDEYVFWDAFHPGEAANVVIGSRSFQRESASDAHPYDIQQLARL 361
>At1g33811 unknown protein
Length = 370
Score = 376 bits (965), Expect = e-104
Identities = 183/342 (53%), Positives = 243/342 (70%), Gaps = 7/342 (2%)
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
QVPC FIFGDSLVDNGNNN L SLARA+Y PYGIDFP G +GRF+NG+T VD +A++LGF
Sbjct: 31 QVPCLFIFGDSLVDNGNNNRLLSLARANYRPYGIDFPQGTTGRFTNGRTYVDALAQILGF 90
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
++IPPY G ILRG NFAS AAGIR+ETG LG S + QV+ Y + V Q++
Sbjct: 91 RNYIPPYSRIRGQAILRGANFASGAAGIRDETGDNLGAHTSMNQQVELYTTAVQQMLRYF 150
Query: 149 -GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQT 207
G+ ++ YLS+CI+ G+GSNDYLNNYFMP FYSTS Y +A+ LI+ YT+QL
Sbjct: 151 RGDTNELQRYLSRCIFYSGMGSNDYLNNYFMPDFYSTSTNYNDKTFAESLIKNYTQQLTR 210
Query: 208 LYNFGARKMVLFGIGQIGCSPNELAQ---RSQDGRTCVSDINDANQIFNNKLKSVVDQFN 264
LY FGARK+++ G+GQIGC P +LA+ R+ C IN+A +FN ++K +VD+ N
Sbjct: 211 LYQFGARKVIVTGVGQIGCIPYQLARYNNRNNSTGRCNEKINNAIVVFNTQVKKLVDRLN 270
Query: 265 -NQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRR 323
QL A+ +Y+++Y D+ + A YGF ++GCCGVGRNNGQITCLP+QTPC +R
Sbjct: 271 KGQLKGAKFVYLDSYKSTYDLAVNGAAYGFEVVDKGCCGVGRNNGQITCLPLQTPCPDRT 330
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
+YLFWDAFHP+E N+++A+ + S + YPI+I+ LA +
Sbjct: 331 KYLFWDAFHPTETANILLAKSNF--YSRAYTYPINIQELANL 370
>At1g71691 unknown protein
Length = 384
Score = 353 bits (905), Expect = 1e-97
Identities = 169/336 (50%), Positives = 237/336 (70%), Gaps = 5/336 (1%)
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFD 89
VP F+FGDSL+DNGNNN + S A+A+Y PYGIDF GGP+GRF NG T VD IA+LLG
Sbjct: 53 VPALFVFGDSLIDNGNNNNIPSFAKANYFPYGIDFNGGPTGRFCNGLTMVDGIAQLLGLP 112
Query: 90 DFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILG 149
IP Y +GD +LRGVN+ASAAAGI +TG GRI F Q+ N+++T+ QV + G
Sbjct: 113 -LIPAYSEATGDQVLRGVNYASAAAGILPDTGGNFVGRIPFDQQIHNFETTLDQVASKSG 171
Query: 150 NEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLY 209
A+ +++ ++ IG+GSNDYLNNY MP F T QY + Q+ D+L+Q YT+QL LY
Sbjct: 172 GAVAIADSVTRSLFFIGMGSNDYLNNYLMPNF-PTRNQYNSQQFGDLLVQHYTDQLTRLY 230
Query: 210 NFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPD 269
N G RK V+ G+G++GC P+ LAQ DG+ C ++N FN +K+++ N LPD
Sbjct: 231 NLGGRKFVVAGLGRMGCIPSILAQ-GNDGK-CSEEVNQLVLPFNTNVKTMISNLNQNLPD 288
Query: 270 ARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWD 329
A+ IY++ +F+DI+A+ A YG + ++GCCG+G+N GQITCLP +TPC NR +Y+FWD
Sbjct: 289 AKFIYLDIAHMFEDIVANQAAYGLTTMDKGCCGIGKNRGQITCLPFETPCPNRDQYVFWD 348
Query: 330 AFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
AFHP+E N+++A++A+ A + AYPI+I++LA +
Sbjct: 349 AFHPTEKVNLIMAKKAF-AGDRTVAYPINIQQLASL 383
>At5g08460 GDSL-motif lipase/acylhydrolase-like protein
Length = 385
Score = 327 bits (839), Expect = 5e-90
Identities = 168/367 (45%), Positives = 240/367 (64%), Gaps = 11/367 (2%)
Query: 7 MMVVAMLVMVLGLWGGWVGA--APQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDF 64
+++V ++V L GG + P F+FGDSLVDNGNNN L SLAR++Y+PYGIDF
Sbjct: 22 LVLVPWFLVVFVLAGGEDSSETTAMFPAMFVFGDSLVDNGNNNHLNSLARSNYLPYGIDF 81
Query: 65 PGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVST--SGDDILRGVNFASAAAGIREETG 121
G P+GRFSNGKT VD I ELLG + IP ++ T G DIL GVN+ASAA GI EETG
Sbjct: 82 AGNQPTGRFSNGKTIVDFIGELLGLPE-IPAFMDTVDGGVDILHGVNYASAAGGILEETG 140
Query: 122 QQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQF 181
+ LG R S QV+N++ T+ ++ + E Y++K + + LG+NDY+NNY P+
Sbjct: 141 RHLGERFSMGRQVENFEKTLMEISRSMRKES-VKEYMAKSLVVVSLGNNDYINNYLKPRL 199
Query: 182 YSTSRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTC 241
+ +S Y +AD+L+ +T L LY G RK V+ G+G +GC P++LA ++ C
Sbjct: 200 FLSSSIYDPTSFADLLLSNFTTHLLELYGKGFRKFVIAGVGPLGCIPDQLAAQAALPGEC 259
Query: 242 VSDINDANQIFNNKLKSVVDQFNNQ---LPDARVIYINAYGIFQDIIASPATYGFSNTNE 298
V +N+ ++FNN+L S+VD+ N+ +A +Y N YG DI+ +P YGF T+
Sbjct: 260 VEAVNEMAELFNNRLVSLVDRLNSDNKTASEAIFVYGNTYGAAVDILTNPFNYGFEVTDR 319
Query: 299 GCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPID 358
GCCGVGRN G+ITCLP+ PC R ++FWDAFHP++A N++IA RA++ S SD YPI+
Sbjct: 320 GCCGVGRNRGEITCLPLAVPCAFRDRHVFWDAFHPTQAFNLIIALRAFNG-SKSDCYPIN 378
Query: 359 IKRLAQI 365
+ +L+++
Sbjct: 379 LSQLSRL 385
>At5g15720 unknown protein
Length = 364
Score = 327 bits (838), Expect = 6e-90
Identities = 160/342 (46%), Positives = 231/342 (66%), Gaps = 7/342 (2%)
Query: 28 PQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLG 87
P P +F+FGDSLVD+GNNN + +LARA+Y PYGIDF G P+GRF NG+T VD A LG
Sbjct: 26 PLAPAFFVFGDSLVDSGNNNYIPTLARANYFPYGIDF-GFPTGRFCNGRTVVDYGATYLG 84
Query: 88 FDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVS-QV 144
+PPY+S + G + LRGVN+ASAAAGI +ETG+ G R +F+GQ+ ++ T+ ++
Sbjct: 85 LP-LVPPYLSPLSIGQNALRGVNYASAAAGILDETGRHYGARTTFNGQISQFEITIELRL 143
Query: 145 VNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQ 204
N YL+K I I +GSNDY+NNY MP+ YSTS+ Y+ + YAD+LI+ + Q
Sbjct: 144 RRFFQNPADLRKYLAKSIIGINIGSNDYINNYLMPERYSTSQTYSGEDYADLLIKTLSAQ 203
Query: 205 LQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRT-CVSDINDANQIFNNKLKSVVDQF 263
+ LYN GARKMVL G G +GC P++L+ + + + CV+ IN+ +FN++LK + +
Sbjct: 204 ISRLYNLGARKMVLAGSGPLGCIPSQLSMVTGNNTSGCVTKINNMVSMFNSRLKDLANTL 263
Query: 264 NNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRR 323
N LP + +Y N + +F D++ +P+ YG +NE CCG GR G +TCLP+Q PC +R
Sbjct: 264 NTTLPGSFFVYQNVFDLFHDMVVNPSRYGLVVSNEACCGNGRYGGALTCLPLQQPCLDRN 323
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
+Y+FWDAFHP+E N +IA +S +S + +YPI + LA++
Sbjct: 324 QYVFWDAFHPTETANKIIAHNTFS-KSANYSYPISVYELAKL 364
>At1g71250 putative GDSL-motif lipase/acylhydrolase
Length = 374
Score = 322 bits (826), Expect = 2e-88
Identities = 158/358 (44%), Positives = 236/358 (65%), Gaps = 5/358 (1%)
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
++++A+ V V+ V +VP F+ GDSLVD GNNN L+++ARA+++PYGID
Sbjct: 16 VLILALTVSVILQQPELVTGQARVPAMFVLGDSLVDAGNNNFLQTVARANFLPYGIDMNY 75
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYV--STSGDDILRGVNFASAAAGIREETGQQL 124
P+GRFSNG T +D++A LL PP+ +TSG+ IL+GVN+ASAAAGI + +G
Sbjct: 76 QPTGRFSNGLTFIDLLARLLEIPS-PPPFADPTTSGNRILQGVNYASAAAGILDVSGYNY 134
Query: 125 GGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYST 184
GGR S + Q+ N ++T+SQ+ ++ ++ +YL++ + + GSNDY+NNY MP Y +
Sbjct: 135 GGRFSLNQQMVNLETTLSQLRTMMSPQN-FTDYLARSLVVLVFGSNDYINNYLMPNLYDS 193
Query: 185 SRQYTTDQYADVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSD 244
S ++ +A++L+ Y QL TLY+ G RK+ + G+ +GC PN+ A+ CV
Sbjct: 194 SIRFRPPDFANLLLSQYARQLLTLYSLGLRKIFIPGVAPLGCIPNQRARGISPPDRCVDS 253
Query: 245 INDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG 304
+N FN LKS+VDQ N + P A +Y N Y DI+ +PA YGFS + CCG+G
Sbjct: 254 VNQILGTFNQGLKSLVDQLNQRSPGAIYVYGNTYSAIGDILNNPAAYGFSVVDRACCGIG 313
Query: 305 RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRL 362
RN GQITCLP+QTPC NR +Y+FWDAFHP++ N ++A+RA+ PSDAYP++++++
Sbjct: 314 RNQGQITCLPLQTPCPNRNQYVFWDAFHPTQTANSILARRAFYG-PPSDAYPVNVQQM 370
>At2g19010 putative GDSL-motif lipase/hydrolase
Length = 344
Score = 284 bits (726), Expect = 6e-77
Identities = 142/320 (44%), Positives = 204/320 (63%), Gaps = 16/320 (5%)
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
Q PC+F+FGDS+ DNGNNN L+S A+ ++ PYG DFP GP+GRFSNG+T D+I EL GF
Sbjct: 23 QAPCFFVFGDSMSDNGNNNNLKSEAKVNFSPYGNDFPKGPTGRFSNGRTIPDIIGELSGF 82
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
DFIPP+ S + G+N+AS +G+REET + LG RIS Q+QN+++++++
Sbjct: 83 KDFIPPFAEASPEQAHTGMNYASGGSGLREETSEHLGDRISIRKQLQNHKTSITKA---- 138
Query: 149 GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTL 208
+ A L +C+Y I +GSNDY+NNYFM + Y+T R+YT QYA LI Y L+ L
Sbjct: 139 ---NVPAERLQQCLYMINIGSNDYINNYFMSKPYNTKRRYTPKQYAYSLIIIYRSHLKNL 195
Query: 209 YNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLP 268
+ GARK+ +FG+ QIGC+P + S DG+ C ++N+A +IFN L +V FN ++
Sbjct: 196 HRLGARKVAVFGLSQIGCTPKIMKSHS-DGKICSREVNEAVKIFNKNLDDLVMDFNKKVR 254
Query: 269 DARVIYINAY--GIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYL 326
A+ Y++ + G Q I GF + CC V N G+ C+P Q C NR EY+
Sbjct: 255 GAKFTYVDLFSGGDPQAFI----FLGFKVGGKSCCTV--NPGEELCVPNQPVCANRTEYV 308
Query: 327 FWDAFHPSEAGNVVIAQRAY 346
FWD H +EA N+V+A+ ++
Sbjct: 309 FWDDLHSTEATNMVVAKGSF 328
>At2g19050 putative GDSL-motif lipase/hydrolase
Length = 349
Score = 280 bits (716), Expect = 9e-76
Identities = 141/340 (41%), Positives = 210/340 (61%), Gaps = 21/340 (6%)
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
+VPCYF+FGDS+ DNGNNN L + A+ +Y PYGIDF GP+GRFSNG+ D+IAEL+ F
Sbjct: 28 RVPCYFVFGDSVFDNGNNNVLNTSAKVNYSPYGIDFARGPTGRFSNGRNIPDIIAELMRF 87
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
D+IPP+ S + G+N+AS GIREET Q LG ISF Q++N++S +
Sbjct: 88 SDYIPPFTGASPEQAHIGINYASGGGGIREETSQHLGEIISFKKQIKNHRSMIM------ 141
Query: 149 GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTL 208
L+KC+Y+I +GSNDYLNNYFMP Y T+++++ D+YAD LI++Y L++L
Sbjct: 142 -TAKVPEEKLNKCLYTINIGSNDYLNNYFMPAPYMTNKKFSFDEYADSLIRSYRSYLKSL 200
Query: 209 YNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLP 268
Y GARK+ +FG+ ++GC+P +A G C +++N A + FN LK++V +FN
Sbjct: 201 YVLGARKVAVFGVSKLGCTPRMIASHG-GGNGCAAEVNKAVEPFNKNLKALVYEFNRDFA 259
Query: 269 DARVIYINAYGIFQDIIASPATY---GFSNTNEGCCGVGRNNGQITCLPMQTPCENRREY 325
DA+ +++ + SP + GF T++ CC V G+ C + C +R Y
Sbjct: 260 DAKFTFVDIFS-----GQSPFAFFMLGFRVTDKSCCTV--KPGEELCATNEPVCPVQRRY 312
Query: 326 LFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
++WD H +EA N+V+A+ AY+ S P + LA++
Sbjct: 313 VYWDNVHSTEAANMVVAKAAYAGLITS---PYSLSWLARL 349
>At2g19060 putative GDSL-motif lipase/hydrolase
Length = 349
Score = 268 bits (685), Expect = 3e-72
Identities = 139/324 (42%), Positives = 198/324 (60%), Gaps = 22/324 (6%)
Query: 30 VPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFD 89
VPCYF+FGDS+ DNGNNN L +LA+ +Y PYGIDF GP+GRFSNG+ D IAE L
Sbjct: 28 VPCYFVFGDSVFDNGNNNELDTLAKVNYSPYGIDFARGPTGRFSNGRNIPDFIAEELRIS 87
Query: 90 DFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILG 149
IPP+ S + G+N+AS AG+ EET Q LG RISF Q+ N++ +
Sbjct: 88 YDIPPFTRASTEQAHTGINYASGGAGLLEETSQHLGERISFEKQITNHRKMIM------- 140
Query: 150 NEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLY 209
L KC+Y+I +GSNDYLNNYFMP Y+T+ ++ D+YAD LIQ+Y L++LY
Sbjct: 141 TAGVPPEKLKKCLYTINIGSNDYLNNYFMPAPYTTNENFSFDEYADFLIQSYRSYLKSLY 200
Query: 210 NFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPD 269
GARK+ +FG+ ++GC+P +A G+ C +++N A + FN KLK ++ +FN
Sbjct: 201 VLGARKVAVFGVSKLGCTPRMIASHG-GGKGCATEVNKAVEPFNKKLKDLISEFN----- 254
Query: 270 ARVIYI-NAYGIFQDIIAS--PATY---GFSNTNEGCCGVGRNNGQITCLPMQTPCENRR 323
R+ + +A F D+ +S P Y GF+ T++ CC V +GQ C + C NR
Sbjct: 255 -RISVVDHAKFTFVDLFSSQNPIEYFILGFTVTDKSCCTV--ESGQELCAANKPVCPNRE 311
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYS 347
Y++WD H +EA N V+ + A++
Sbjct: 312 RYVYWDNVHSTEAANKVVVKAAFA 335
>At4g30140 unknown protein
Length = 348
Score = 257 bits (656), Expect = 8e-69
Identities = 145/335 (43%), Positives = 194/335 (57%), Gaps = 19/335 (5%)
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
Q PCYF+FGDS+ DNGNNNAL + A+ +Y+PYGID+ GP+GRFSNG+ DVIAEL GF
Sbjct: 30 QTPCYFVFGDSVFDNGNNNALNTKAKVNYLPYGIDYFQGPTGRFSNGRNIPDVIAELAGF 89
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
++ IPP+ S G+N+AS A GIREET + +G RIS QV N+ S + L
Sbjct: 90 NNPIPPFAGASQAQANIGLNYASGAGGIREETSENMGERISLRQQVNNHFSAIITAAVPL 149
Query: 149 GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTL 208
L +C+Y+I +GSNDYLNNYF+ R + DQYA LI Y L L
Sbjct: 150 SR-------LRQCLYTINIGSNDYLNNYFLSPPTLARRLFNPDQYARSLISLYRIYLTQL 202
Query: 209 YNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLP 268
Y GAR + LFGIG+IGC+P +A G C ++N A IFN KLK++V FNN+ P
Sbjct: 203 YVLGARNVALFGIGKIGCTPRIVATLG-GGTGCAEEVNQAVIIFNTKLKALVTDFNNK-P 260
Query: 269 DARVIYINAY-GIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLF 327
A Y++ + G +D A T G + CC V N G+ C C +R +++F
Sbjct: 261 GAMFTYVDLFSGNAEDFAALGITVG----DRSCCTV--NPGEELCAANGPVCPDRNKFIF 314
Query: 328 WDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRL 362
WD H +E N V+A A++ S P +I +L
Sbjct: 315 WDNVHTTEVINTVVANAAFNGPIAS---PFNISQL 346
>At5g33370 unknown protein
Length = 366
Score = 238 bits (608), Expect = 3e-63
Identities = 129/336 (38%), Positives = 198/336 (58%), Gaps = 7/336 (2%)
Query: 33 YFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDF 91
+ +FGDSLVDNGNN+ L + ARAD PYGIDFP P+GRFSNG D+I+E LG +
Sbjct: 31 FLVFGDSLVDNGNNDFLATTARADNYPYGIDFPTHRPTGRFSNGLNIPDLISEHLGQESP 90
Query: 92 IPPYVST--SGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILG 149
+P Y+S D +LRG NFASA GI +TG Q I + Q++ ++ +V ++G
Sbjct: 91 MP-YLSPMLKKDKLLRGANFASAGIGILNDTGIQFLNIIRITKQLEYFEQYKVRVSGLVG 149
Query: 150 NEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLY 209
E++ ++ + I LG ND++NNY++ F + SRQ++ Y +I Y + L+ +Y
Sbjct: 150 -EEEMNRLVNGALVLITLGGNDFVNNYYLVPFSARSRQFSLPDYVVFVISEYRKVLRKMY 208
Query: 210 NFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPD 269
+ GAR++++ G G +GC P ELAQRS++G C +++ A +FN +L ++ NN++
Sbjct: 209 DLGARRVLVTGTGPMGCVPAELAQRSRNGE-CATELQRAASLFNPQLIQMITDLNNEVGS 267
Query: 270 ARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWD 329
+ I N + D I+ P YGF + CCG G NG C P+ C NR + FWD
Sbjct: 268 SAFIAANTQQMHMDFISDPQAYGFVTSKVACCGQGPYNGIGLCTPLSNLCPNRDLFAFWD 327
Query: 330 AFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
FHPSE + +IAQ+ + SP +P+++ + +
Sbjct: 328 PFHPSEKASRIIAQQILNG-SPEYMHPMNLSTILTV 362
>At3g04290 GDSL-motif lipase/acylhydrolase like protein
Length = 366
Score = 235 bits (600), Expect = 2e-62
Identities = 130/329 (39%), Positives = 191/329 (57%), Gaps = 8/329 (2%)
Query: 27 APQVP--CYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIA 83
APQV +F+FGDSLVDNGNN+ L + ARAD PYGID+P P+GRFSNG D+I+
Sbjct: 22 APQVKSRAFFVFGDSLVDNGNNDYLVTTARADNYPYGIDYPTRRPTGRFSNGLNIPDIIS 81
Query: 84 ELLGFDDFIPPYVST--SGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTV 141
E +G +P Y+S +G+++L G NFASA GI +TG Q I S Q++ ++
Sbjct: 82 EAIGMPSTLP-YLSPHLTGENLLVGANFASAGIGILNDTGIQFVNIIRISKQMEYFEQYQ 140
Query: 142 SQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAY 201
+V ++G E +++ + I LG ND++NNY++ F + SRQY Y LI Y
Sbjct: 141 LRVSALIGPE-ATQQLVNQALVLITLGGNDFVNNYYLIPFSARSRQYALPDYVVYLISEY 199
Query: 202 TEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVD 261
+ L+ LY GAR++++ G G +GC+P ELAQ S++G C + A +FN +L ++
Sbjct: 200 GKILRKLYELGARRVLVTGTGAMGCAPAELAQHSRNGE-CYGALQTAAALFNPQLVDLIA 258
Query: 262 QFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCEN 321
N ++ + NAY + D +++P +GF + CCG G NG C P+ C N
Sbjct: 259 SVNAEIGQDVFVAANAYQMNMDYLSNPEQFGFVTSKVACCGQGPYNGIGLCTPVSNLCPN 318
Query: 322 RREYLFWDAFHPSEAGNVVIAQRAYSAQS 350
R Y FWDAFHP+E N +I + + S
Sbjct: 319 RDLYAFWDAFHPTEKANRIIVNQILTGSS 347
>At2g23540 putative GDSL-motif lipase/hydrolase
Length = 387
Score = 232 bits (591), Expect = 3e-61
Identities = 128/331 (38%), Positives = 187/331 (55%), Gaps = 13/331 (3%)
Query: 21 GGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFP---GGPSGRFSNGKT 77
GG +GA+ FIFGDSLVD GNNN L +L+RA+ P GIDF G P+GRF+NG+T
Sbjct: 43 GGGLGAS------FIFGDSLVDAGNNNYLSTLSRANMKPNGIDFKASGGTPTGRFTNGRT 96
Query: 78 TVDVIAELLGFDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQ 135
D++ E LG ++ P+++ G +L GVN+AS GI TG+ R+ QV
Sbjct: 97 IGDIVGEELGSANYAIPFLAPDAKGKALLAGVNYASGGGGIMNATGRIFVNRLGMDVQVD 156
Query: 136 NYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSR-QYTTDQYA 194
+ +T Q ++LG E K I+SI +G+ND+LNNY P +R T D +
Sbjct: 157 FFNTTRKQFDDLLGKEKAKDYIAKKSIFSITIGANDFLNNYLFPLLSVGTRFTQTPDDFI 216
Query: 195 DVLIQAYTEQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNN 254
+++ +QL LY ARK V+ +G IGC P + D CV N +N
Sbjct: 217 GDMLEHLRDQLTRLYQLDARKFVIGNVGPIGCIPYQKTINQLDENECVDLANKLANQYNV 276
Query: 255 KLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCG-VGRNNGQITCL 313
+LKS++++ N +LP A ++ N Y + ++I + YGF + + CCG G+ G I C
Sbjct: 277 RLKSLLEELNKKLPGAMFVHANVYDLVMELITNYDKYGFKSATKACCGNGGQYAGIIPCG 336
Query: 314 PMQTPCENRREYLFWDAFHPSEAGNVVIAQR 344
P + CE R +Y+FWD +HPSEA NV+IA++
Sbjct: 337 PTSSLCEERDKYVFWDPYHPSEAANVIIAKQ 367
>At4g10950 unknown protein
Length = 400
Score = 228 bits (580), Expect = 5e-60
Identities = 120/339 (35%), Positives = 196/339 (57%), Gaps = 12/339 (3%)
Query: 28 PQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELL 86
P VP F+FGDS VD+G NN L +LARAD +PYG DF P+GRF NG+ VD +
Sbjct: 67 PFVPALFVFGDSSVDSGTNNFLGTLARADRLPYGRDFDTHQPTGRFCNGRIPVDYLGL-- 124
Query: 87 GFDDFIPPYVSTSG--DDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQV 144
F+P Y+ +G +D+ +GVN+ASA AGI +G +LG R+SF+ QV+ + T Q+
Sbjct: 125 ---PFVPSYLGQTGTVEDMFQGVNYASAGAGIILSSGSELGQRVSFAMQVEQFVDTFQQM 181
Query: 145 VNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQ 204
+ +G E + +S ++ I +G NDY++ +++ + YT + L ++
Sbjct: 182 ILSIG-EKASERLVSNSVFYISIGVNDYIH-FYIRNISNVQNLYTPWNFNQFLASNMRQE 239
Query: 205 LQTLYNFGARKMVLFGIGQIGCSPNELAQ-RSQDGRTCVSDINDANQIFNNKLKSVVDQF 263
L+TLYN R+MV+ G+ IGC+P + + RSQ+G C ++N N ++ VD+
Sbjct: 240 LKTLYNVKVRRMVVMGLPPIGCAPYYMWKYRSQNGE-CAEEVNSMIMESNFVMRYTVDKL 298
Query: 264 NNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRR 323
N +LP A +IY + + DI+ + YGF+ T + CCG+GR G + C+ + C +
Sbjct: 299 NRELPGASIIYCDVFQSAMDILRNHQHYGFNETTDACCGLGRYKGWLPCISPEMACSDAS 358
Query: 324 EYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRL 362
+L+WD FHP++A N ++A ++ + YP +++ +
Sbjct: 359 GHLWWDQFHPTDAVNAILADNVWNGRHVDMCYPTNLETM 397
>At3g50400 unknown protein
Length = 374
Score = 226 bits (576), Expect = 2e-59
Identities = 123/320 (38%), Positives = 188/320 (58%), Gaps = 10/320 (3%)
Query: 34 FIFGDSLVDNGNNNALRSLARADYMPYGIDFP---GGPSGRFSNGKTTVDVIAELLGFDD 90
F+FGDSLVD GNNN L++L+RA+ P GIDF G P+GRF+NG+T D++ E LG
Sbjct: 36 FVFGDSLVDAGNNNYLQTLSRANSPPNGIDFKPSRGNPTGRFTNGRTIADIVGEKLGQQS 95
Query: 91 FIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
+ PY++ SG+ +L GVN+AS GI TG R+ QV + +T Q +L
Sbjct: 96 YAVPYLAPNASGEALLNGVNYASGGGGILNATGSVFVNRLGMDIQVDYFTNTRKQFDKLL 155
Query: 149 GNEDQAANYLSK-CIYSIGLGSNDYLNNYFMPQFYSTSR-QYTTDQYADVLIQAYTEQLQ 206
G +D+A +Y+ K ++S+ +GSND+LNNY +P + +R T + + D +I QL+
Sbjct: 156 G-QDKARDYIRKRSLFSVVIGSNDFLNNYLVPFVAAQARLTQTPETFVDDMISHLRNQLK 214
Query: 207 TLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVD-QFNN 265
LY+ ARK V+ + IGC P + + + + CV N +N +LK ++ + +
Sbjct: 215 RLYDMDARKFVVGNVAPIGCIPYQKSINQLNDKQCVDLANKLAIQYNARLKDLLTVELKD 274
Query: 266 QLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGV-GRNNGQITCLPMQTPCENRRE 324
L DA +Y N Y +F D+I + YGF +E CC GR G + C P + C +R +
Sbjct: 275 SLKDAHFVYANVYDLFMDLIVNFKDYGFRTASEACCETRGRLAGILPCGPTSSLCTDRSK 334
Query: 325 YLFWDAFHPSEAGNVVIAQR 344
++FWDA+HP+EA N++IA +
Sbjct: 335 HVFWDAYHPTEAANLLIADK 354
>At4g28780 Proline-rich APG - like protein
Length = 367
Score = 223 bits (567), Expect = 2e-58
Identities = 120/314 (38%), Positives = 175/314 (55%), Gaps = 4/314 (1%)
Query: 33 YFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDF 91
+F+FGDSLVD+GNNN L + ARAD PYGID+P G P+GRFSNG D+I+E +G +
Sbjct: 32 FFVFGDSLVDSGNNNYLVTTARADSPPYGIDYPTGRPTGRFSNGLNLPDIISEQIGSEPT 91
Query: 92 IPPYV-STSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGN 150
+P +G+ +L G NFASA GI +TG Q + Q + +Q +V I+G+
Sbjct: 92 LPILSPELTGEKLLIGANFASAGIGILNDTGVQFLNILRIGRQFELFQEYQERVSEIIGS 151
Query: 151 EDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLYN 210
D+ ++ + + LG ND++NNYF P + RQ + +++ +LI Y + L +LY
Sbjct: 152 -DKTQQLVNGALVLMTLGGNDFVNNYFFP-ISTRRRQSSLGEFSQLLISEYKKILTSLYE 209
Query: 211 FGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPDA 270
GAR++++ G G +GC P ELA C + A IFN L ++ N ++
Sbjct: 210 LGARRVMVTGTGPLGCVPAELASSGSVNGECAPEAQQAAAIFNPLLVQMLQGLNREIGSD 269
Query: 271 RVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWDA 330
I NA+ D I +P +GF + CCG G NGQ C P+ T C +R Y FWD
Sbjct: 270 VFIGANAFNTNADFINNPQRFGFVTSKVACCGQGAYNGQGVCTPLSTLCSDRNAYAFWDP 329
Query: 331 FHPSEAGNVVIAQR 344
FHP+E +I Q+
Sbjct: 330 FHPTEKATRLIVQQ 343
>At1g74460 putative lipase/acylhydrolase
Length = 366
Score = 214 bits (546), Expect = 5e-56
Identities = 119/313 (38%), Positives = 181/313 (57%), Gaps = 8/313 (2%)
Query: 34 FIFGDSLVDNGNNNAL-RSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDF 91
FIFGDSL D GNN L RSLA A+ YGIDF G P+GRF+NG+T D+I + +G
Sbjct: 25 FIFGDSLSDVGNNKNLPRSLATANLPFYGIDFGNGLPNGRFTNGRTVSDIIGDKIGLPRP 84
Query: 92 IPPYVSTSGDDIL--RGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILG 149
+ + +D++ GVN+AS GI ETG R S Q++ +Q T VV +G
Sbjct: 85 VAFLDPSMNEDVILENGVNYASGGGGILNETGGYFIQRFSLWKQIELFQGTQDVVVAKIG 144
Query: 150 NEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTLY 209
++ A + Y + LGSND++NNY MP YS S +Y + D L++ QL+ L+
Sbjct: 145 KKE-ADKFFQDARYVVALGSNDFINNYLMP-VYSDSWKYNDQTFVDYLMETLESQLKVLH 202
Query: 210 NFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQLPD 269
+ GARK+++FG+G +GC P + A S DG C + ++ + FN +++ +LP+
Sbjct: 203 SLGARKLMVFGLGPMGCIPLQRAL-SLDGN-CQNKASNLAKRFNKAATTMLLDLETKLPN 260
Query: 270 ARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFWD 329
A + AY + D+I +P YGF N++ CC R +TC+P T C++R +Y+FWD
Sbjct: 261 ASYRFGEAYDLVNDVITNPKKYGFDNSDSPCCSFYRIRPALTCIPASTLCKDRSKYVFWD 320
Query: 330 AFHPSEAGNVVIA 342
+HP++ N ++A
Sbjct: 321 EYHPTDKANELVA 333
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,244,057
Number of Sequences: 26719
Number of extensions: 362847
Number of successful extensions: 1256
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 712
Number of HSP's gapped (non-prelim): 125
length of query: 365
length of database: 11,318,596
effective HSP length: 101
effective length of query: 264
effective length of database: 8,619,977
effective search space: 2275673928
effective search space used: 2275673928
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0404.11


