

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.9
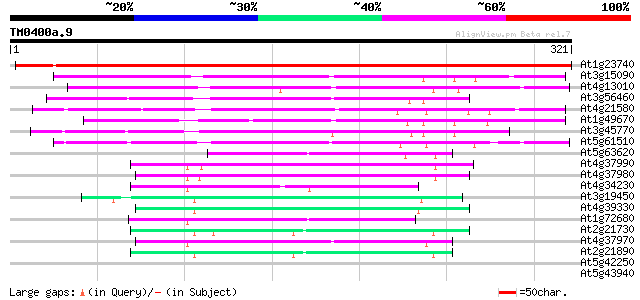
(321 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g23740 auxin-induced protein like 415 e-116
At3g15090 zinc-binding dehydrogenase, putative 135 2e-32
At4g13010 unknown protein 133 1e-31
At3g56460 quinone reductase-like protein 108 5e-24
At4g21580 putative NADPH quinone oxidoreductase 103 1e-22
At1g49670 ARP protein (At1g49670; F14J22.10) 92 4e-19
At3g45770 nuclear receptor binding factor-like protein 78 7e-15
At5g61510 quinone oxidoreductase - like protein 67 2e-11
At5g63620 alcohol dehydrogenase-like protein 65 6e-11
At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2 65 6e-11
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 64 8e-11
At4g34230 cinnamyl alcohol dehydrogenase like protein 63 2e-10
At3g19450 putative cinnamyl alcohol dehydrogenase 2 63 2e-10
At4g39330 cinnamyl-alcohol dehydrogenase CAD1 61 7e-10
At1g72680 putative cinnamyl-alcohol dehydrogenase 57 1e-08
At2g21730 putative cinnamyl-alcohol dehydrogenase 52 6e-07
At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa 51 7e-07
At2g21890 putative cinnamyl-alcohol dehydrogenase 51 7e-07
At5g42250 alcohol dehydrogenase 37 0.019
At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III (pir||S71... 33 0.21
>At1g23740 auxin-induced protein like
Length = 386
Score = 415 bits (1067), Expect = e-116
Identities = 210/318 (66%), Positives = 253/318 (79%), Gaps = 1/318 (0%)
Query: 4 TPYSNIPSQTKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALG 63
T ++IP + KAWVYS YG ++ +LK + N+ P +K DQVLIKV AAALNPVD+KR G
Sbjct: 69 TADASIPKEMKAWVYSDYGGVD-VLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQG 127
Query: 64 HFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTA 123
FK DSPLPTVPGYDVAGVVVKVGS VK LK GDEVY +V+E L+ PK GSLAEYTA
Sbjct: 128 KFKATDSPLPTVPGYDVAGVVVKVGSAVKDLKEGDEVYANVSEKALEGPKQFGSLAEYTA 187
Query: 124 AEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLA 183
EEK+LA KP N+ F +AA LPLAI TA +GL +FSAGKSILVL GAGGVGSLVIQLA
Sbjct: 188 VEEKLLALKPKNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLA 247
Query: 184 KHVFEASKIAATASTGKLEFLRKLGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRALKA 243
KHV+ ASK+AATAST KLE +R LGADL IDYTKEN E++ +K+DVV+DA+G D+A+K
Sbjct: 248 KHVYGASKVAATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKV 307
Query: 244 VKEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEA 303
+KEGGKVV + TPP F++TS+G VL+KL PY+ESGKVKP++DPK PFPFS+ +A
Sbjct: 308 IKEGGKVVALTGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADA 367
Query: 304 FSYLNTSRATGEVVIYPI 321
FSYL T+ ATG+VV+YPI
Sbjct: 368 FSYLETNHATGKVVVYPI 385
>At3g15090 zinc-binding dehydrogenase, putative
Length = 366
Score = 135 bits (341), Expect = 2e-32
Identities = 99/331 (29%), Positives = 167/331 (49%), Gaps = 48/331 (14%)
Query: 26 EILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSP-LPTVPGYDVAGVV 84
E+ + NVP P+L ++VL+K +A ++NP+D + G+ + + P LP + G DV+G V
Sbjct: 44 EVFELRENVPVPNLNPNEVLVKAKAVSVNPLDCRIRAGYGRSVFQPHLPIIVGRDVSGEV 103
Query: 85 VKVGSQVKKLKVGDEVYGDVNEITLDHPKTV-GSLAEYTAAEEKVLAHKPSNLSFIEAAS 143
+G+ VK LKVG EV+G + HP + G+ +Y E L KPS++S +EA++
Sbjct: 104 AAIGTSVKSLKVGQEVFGAL------HPTALRGTYTDYGILSEDELTEKPSSISHVEASA 157
Query: 144 LPLAIITAYQGLET-AQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLE 202
+P A +TA++ L++ A+ + G+ +LV GG G VG IQLA V + A+ +
Sbjct: 158 IPFAALTAWRALKSNARITEGQRLLVFGGGGAVGFSAIQLA--VASGCHVTASCVGQTKD 215
Query: 203 FLRKLGADLPIDYTKENFE-EVSEKFDVVYDAVG--QSDR-ALKAVKEGGKVVTI----- 253
+ GA+ +DYT E+ E V KFD V D +G +++R + +++GG +T+
Sbjct: 216 RILAAGAEQAVDYTTEDIELAVKGKFDAVLDTIGGPETERIGINFLRKGGNYMTLQGEAA 275
Query: 254 -------LPPGTPPAIPFL-------------------LTSDGAVLEKLQPYLESGKVKP 287
G P A L + +D L ++Q + +GK+K
Sbjct: 276 SLTDKYGFVVGLPLATSLLMKKKIQYQYSHGIDYWWTYMRADPEGLAEIQRLVGAGKLK- 334
Query: 288 ILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
+ + FP + V A + G+VV+
Sbjct: 335 -IPVEKTFPITDVVAAHEAKEKKQIPGKVVL 364
>At4g13010 unknown protein
Length = 329
Score = 133 bits (334), Expect = 1e-31
Identities = 100/314 (31%), Positives = 148/314 (46%), Gaps = 37/314 (11%)
Query: 34 VPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKD-IDSPLPTVPGYDVAGVVVKVGSQVK 92
VP P K ++V +K+EA +LNPVD K G + + P +P DVAG VV+VGS VK
Sbjct: 26 VPVPTPKSNEVCLKLEATSLNPVDWKIQKGMIRPFLPRKFPCIPATDVAGEVVEVGSGVK 85
Query: 93 KLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAY 152
K GD+V ++ + G LAE+ A EK+ +P + EAA+LP+A +TA
Sbjct: 86 NFKAGDKVVAVLSHLG------GGGLAEFAVATEKLTVKRPQEVGAAEAAALPVAGLTAL 139
Query: 153 Q------GLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRK 206
Q GL+ +ILV +GGVG +QLAK + + AT +EF++
Sbjct: 140 QALTNPAGLKLDGTGKKANILVTAASGGVGHYAVQLAK--LANAHVTATCGARNIEFVKS 197
Query: 207 LGADLPIDYTKENFEEVSEKFDVVYDAVGQSDRAL------KAVKEGGKVVTILP----- 255
LGAD +DY + YDAV + + E GKV+ I P
Sbjct: 198 LGADEVLDYKTPEGAALKSPSGKKYDAVVHCANGIPFSVFEPNLSENGKVIDITPGPNAM 257
Query: 256 ---------PGTPPAIPFLLTSDGAVLEKLQPYLESGKVKPILDPKSPFPFSQTVEAFSY 306
+P LL LE + ++ GKVK ++D K P S+ +A++
Sbjct: 258 WTYAVKKITMSKKQLVPLLLIPKAENLEFMVNLVKEGKVKTVIDSK--HPLSKAEDAWAK 315
Query: 307 LNTSRATGEVVIYP 320
ATG++++ P
Sbjct: 316 SIDGHATGKIIVEP 329
>At3g56460 quinone reductase-like protein
Length = 348
Score = 108 bits (269), Expect = 5e-24
Identities = 81/256 (31%), Positives = 129/256 (49%), Gaps = 25/256 (9%)
Query: 22 GDIEEILKFDPNVPTPHLK*D-QVLIKVEAAALNPVDSKRALGHFKDIDSPLPTVPGYDV 80
G E ++ P P L D V ++V A +LN + + LG +++ PLP +PG D
Sbjct: 18 GSPESPVEVSKTHPIPSLNSDTSVRVRVIATSLNYANYLQILGKYQE-KPPLPFIPGSDY 76
Query: 81 AGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIE 140
+G+V +G V K +VGD V + +GS A++ A++ L P +
Sbjct: 77 SGIVDAIGPAVTKFRVGDRVCSFAD---------LGSFAQFIVADQSRLFLVPERCDMVA 127
Query: 141 AASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTG 199
AA+LP+A T++ L A+ ++G+ +LVLG AGGVG +Q+ K V A IA T
Sbjct: 128 AAALPVAFGTSHVALVHRARLTSGQVLLVLGAAGGVGLAAVQIGK-VCGAIVIAVARGTE 186
Query: 200 KLEFLRKLGADLPIDYTKENFEEVSEKF---------DVVYDAVG--QSDRALKAVKEGG 248
K++ L+ +G D +D EN ++F DV+YD VG + ++K +K G
Sbjct: 187 KIQLLKSMGVDHVVDLGTENVISSVKEFIKTRKLKGVDVLYDPVGGKLTKESMKVLKWGA 246
Query: 249 KVVTI-LPPGTPPAIP 263
+++ I G P IP
Sbjct: 247 QILVIGFASGEIPVIP 262
>At4g21580 putative NADPH quinone oxidoreductase
Length = 325
Score = 103 bits (257), Expect = 1e-22
Identities = 98/338 (28%), Positives = 158/338 (45%), Gaps = 49/338 (14%)
Query: 14 KAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLP 73
KA V S+ G E + D V P +K D+VLI+V A ALN D+ + LG + P
Sbjct: 2 KAIVISEPGKPEVLQLRD--VADPEVKDDEVLIRVLATALNRADTLQRLGLYNPPPGSSP 59
Query: 74 TVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKP 133
+ G + +G + VG V + KVGD+V ++ G AE + + P
Sbjct: 60 YL-GLECSGTIESVGKGVSRWKVGDQVCALLSG---------GGYAEKVSVPAGQIFPIP 109
Query: 134 SNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKI 192
+ +S +AA+ P T + + + S G+S L+ GG+ G+G+ IQ+AKH+ ++
Sbjct: 110 AGISLKDAAAFPEVACTVWSTVFMMGRLSVGESFLIHGGSSGIGTFAIQIAKHL--GVRV 167
Query: 193 AATA-STGKLEFLRKLGADLPIDYTKENF------EEVSEKFDVVYDAVGQS--DRALKA 243
TA S KL ++LGAD+ I+Y E+F E + DV+ D +G + L +
Sbjct: 168 FVTAGSDEKLAACKELGADVCINYKTEDFVAKVKAETDGKGVDVILDCIGAPYLQKNLDS 227
Query: 244 VKEGGKVVTILPPGTPPA-------IPFLLTSDGAVL----------------EKLQPYL 280
+ G++ I G A +P LT GA L + + P +
Sbjct: 228 LNFDGRLCIIGLMGGANAEIKLSSLLPKRLTVLGAALRPRSPENKAVVVREVEKNVWPAI 287
Query: 281 ESGKVKPILDPKSPFPFSQTVEAFSYLNTSRATGEVVI 318
E+GKVKP++ P SQ E S + +S G++++
Sbjct: 288 EAGKVKPVI--YKYLPLSQAAEGHSLMESSNHIGKILL 323
>At1g49670 ARP protein (At1g49670; F14J22.10)
Length = 629
Score = 92.0 bits (227), Expect = 4e-19
Identities = 88/313 (28%), Positives = 139/313 (44%), Gaps = 51/313 (16%)
Query: 43 QVLIKVEAAALNPVDSKRALGHFKDIDSP-LPTVPGYDVAGVVVKVGSQVKKLKVGDEVY 101
QVL+K+ A +N D + G + SP LP G++ G++ VG VK L+VG
Sbjct: 318 QVLLKIIYAGVNASDVNFSSGRYFTGGSPKLPFDAGFEGVGLIAAVGESVKNLEVG---- 373
Query: 102 GDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETA-QF 160
T T G+ +EY K + P E ++ + +TA LE A Q
Sbjct: 374 ------TPAAVMTFGAYSEYMIVSSKHVLPVPRPDP--EVVAMLTSGLTALIALEKAGQM 425
Query: 161 SAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAAT-ASTGKLEFLRKLGADLPIDYTKEN 219
+G+++LV AGG G +QLAK +K+ AT + K + L++LG D IDY EN
Sbjct: 426 KSGETVLVTAAAGGTGQFAVQLAK--LSGNKVIATCGGSEKAKLLKELGVDRVIDYKSEN 483
Query: 220 FEEVSEK-----FDVVYDAVG--QSDRALKAVKEGGKVVTI------------LPPGTPP 260
+ V +K +++Y++VG D L A+ G+++ I P P
Sbjct: 484 IKTVLKKEFPKGVNIIYESVGGQMFDMCLNALAVYGRLIVIGMISQYQGEKGWEPAKYPG 543
Query: 261 AIPFLLTSDGAV---------------LEKLQPYLESGKVKPILDPKSPFPFSQTVEAFS 305
+L V L+KL GK+K +D K + +A
Sbjct: 544 LCEKILAKSQTVAGFFLVQYSQLWKQNLDKLFNLYALGKLKVGIDQKKFIGLNAVADAVE 603
Query: 306 YLNTSRATGEVVI 318
YL++ ++TG+VV+
Sbjct: 604 YLHSGKSTGKVVV 616
>At3g45770 nuclear receptor binding factor-like protein
Length = 375
Score = 77.8 bits (190), Expect = 7e-15
Identities = 82/290 (28%), Positives = 128/290 (43%), Gaps = 26/290 (8%)
Query: 13 TKAWVYSQYGDIEEILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPL 72
+KA VY ++G + + + N+P +K + V +K+ AA +NP D R G + + P+
Sbjct: 45 SKAIVYEEHGSPDSVTRL-VNLPPVEVKENDVCVKMIAAPINPSDINRIEGVYP-VRPPV 102
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHK 132
P V GY+ G V VGS V GD V + P + G+ Y EE V
Sbjct: 103 PAVGGYEGVGEVYAVGSNVNGFSPGDWV--------IPSPPSSGTWQTYVVKEESVWHKI 154
Query: 133 PSNLSFIEAASLPLAIITAYQGLET-AQFSAGKSILVLGGAGGVGSLVIQLA--KHVFEA 189
AA++ + +TA + LE ++G S++ G VG VIQLA + +
Sbjct: 155 DKECPMEYAATITVNPLTALRMLEDFVNLNSGDSVVQNGATSIVGQCVIQLARLRGISTI 214
Query: 190 SKIAATASTGKL-EFLRKLGADLPIDYTKENFEEVSEKFD------VVYDAVG--QSDRA 240
+ I A + + E L+ LGAD ++ N + V + ++ VG +
Sbjct: 215 NLIRDRAGSDEAREQLKALGADEVFSESQLNVKNVKSLLGNLPEPALGFNCVGGNAASLV 274
Query: 241 LKAVKEGGKVVTI----LPPGTPPAIPFLLTSDGAVLEKLQPYLESGKVK 286
LK ++EGG +VT P T F+ LQ +L GKVK
Sbjct: 275 LKYLREGGTMVTYGGMSKKPITVSTTSFIFKDLALRGFWLQSWLSMGKVK 324
>At5g61510 quinone oxidoreductase - like protein
Length = 406
Score = 66.6 bits (161), Expect = 2e-11
Identities = 89/329 (27%), Positives = 140/329 (42%), Gaps = 52/329 (15%)
Query: 26 EILKFDPNVPTPHLK*DQVLIKVEAAALNPVDSKRALGHFKDIDSPLPTVPGYDVAGVVV 85
E+LK++ +V K ++ +K +A LN +D G +K + +P PG + G VV
Sbjct: 96 EVLKWE-DVEVGEPKEGEIRVKNKAIGLNFIDVYFRKGVYKP--ASMPFTPGMEAVGEVV 152
Query: 86 KVGSQVKKLKVGDEVYGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLP 145
VGS + +GD V N + G+ AE + PS++ I AAS+
Sbjct: 153 AVGSGLTGRMIGDLVAYAGNPM--------GAYAEEQILPADKVVPVPSSIDPIVAASIM 204
Query: 146 LAIITAYQGLETA-QFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFL 204
L +TA L + G +ILV AGGVGSL+ Q A + A+ I ++ K
Sbjct: 205 LKGMTAQFLLRRCFKVEPGHTILVHAAAGGVGSLLCQWA-NALGATVIGTVSTNEKAAQA 263
Query: 205 RKLGADLPIDYTKENFEE------VSEKFDVVYDAVGQS--DRALKAVKEGGKVVTI-LP 255
++ G I Y E+F + +VVYD+VG+ +L +K G +V+
Sbjct: 264 KEDGCHHVIMYKNEDFVSRVNDITSGKGVNVVYDSVGKDTFKGSLACLKSRGYMVSFGQS 323
Query: 256 PGTPPAIPF------------------------LLTSDGAVLEKLQPYLESGKVKPILDP 291
G+P IP LL G V + SG +K ++
Sbjct: 324 SGSPDPIPLSDLAPKSLFLTRPSMMHYNETRDELLECAGEVFAN----VTSGILKARVNH 379
Query: 292 KSPFPFSQTVEAFSYLNTSRATGEVVIYP 320
K +P S+ +A + L +G VV+ P
Sbjct: 380 K--YPLSRVADAHADLENRITSGSVVLLP 406
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 64.7 bits (156), Expect = 6e-11
Identities = 50/150 (33%), Positives = 77/150 (51%), Gaps = 11/150 (7%)
Query: 114 TVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGL-ETAQFSAGKSILVLGGA 172
++G +AEY LA P +L + E+A L A+ TAY + A+ G SI V+G
Sbjct: 196 SMGGMAEYCVTPAHGLAPLPESLPYSESAILGCAVFTAYGAMAHAAEIRPGDSIAVIG-I 254
Query: 173 GGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGADLPIDYTKEN-FEEVSE-----K 226
GGVGS +Q+A+ + IA KL+ + LGA ++ KE+ E + E
Sbjct: 255 GGVGSSCLQIARAFGASDIIAVDVQDDKLQKAKTLGATHIVNAAKEDAVERIREITGGMG 314
Query: 227 FDVVYDAVGQSDRALK---AVKEGGKVVTI 253
DV +A+G+ ++ +VK+GGK V I
Sbjct: 315 VDVAVEALGKPQTFMQCTLSVKDGGKAVMI 344
>At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2
Length = 359
Score = 64.7 bits (156), Expect = 6e-11
Identities = 55/226 (24%), Positives = 95/226 (41%), Gaps = 30/226 (13%)
Query: 70 SPLPTVPGYDVAGVVVKVGSQVKKLKVGDEV--------YGDVNEIT------------- 108
S P VPG+++ GVV +VG++V K K G++V G + T
Sbjct: 60 STYPLVPGHEIVGVVTEVGAKVTKFKTGEKVGVGCLVSSCGSCDSCTEGMENYCPKSIQT 119
Query: 109 -----LDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAG 163
D+ T G +++ EE + P NL AA L A IT Y ++
Sbjct: 120 YGFPYYDNTITYGGYSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGITVYSPMKYHGLDKP 179
Query: 164 KSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENFEE 222
+ + G GG+G + ++ AK + + +T+ + E + +LGAD + + ++
Sbjct: 180 GMHIGVVGLGGLGHVGVKFAKAMGTKVTVISTSEKKRDEAINRLGADAFLVSRDPKQIKD 239
Query: 223 VSEKFDVVYDAVGQSDRALK---AVKEGGKVVTILPPGTPPAIPFL 265
D + D V + L +K GK+V + P P +P +
Sbjct: 240 AMGTMDGIIDTVSATHSLLPLLGLLKHKGKLVMVGAPEKPLELPVM 285
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 64.3 bits (155), Expect = 8e-11
Identities = 54/221 (24%), Positives = 91/221 (40%), Gaps = 30/221 (13%)
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEV----------------YGDVNEI--------- 107
P VPG+++ GVV +VG++VKK GD+V GD N
Sbjct: 63 PLVPGHEIVGVVTEVGAKVKKFNAGDKVGVGYMAGSCRSCDSCNDGDENYCPKMILTSGA 122
Query: 108 -TLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSI 166
D T G +++ E + P NL AA L A +T Y ++
Sbjct: 123 KNFDDTMTHGGYSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVTVYSPMKYHGLDKPGMH 182
Query: 167 LVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENFEEVSE 225
+ + G GG+G + ++ AK + + +T+ + E + +LGAD + + ++
Sbjct: 183 IGVVGLGGLGHVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFLVSRDPKQMKDAMG 242
Query: 226 KFDVVYDAVGQSDRALK---AVKEGGKVVTILPPGTPPAIP 263
D + D V + L +K GK+V + P P +P
Sbjct: 243 TMDGIIDTVSATHPLLPLLGLLKNKGKLVMVGAPAEPLELP 283
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 63.2 bits (152), Expect = 2e-10
Identities = 54/194 (27%), Positives = 86/194 (43%), Gaps = 31/194 (15%)
Query: 70 SPLPTVPGYDVAGVVVKVGSQVKKLKVGDEV--------------------------YGD 103
S P VPG++V G VV+VGS V K VGD V
Sbjct: 61 SNYPMVPGHEVVGEVVEVGSDVSKFTVGDIVGVGCLVGCCGGCSPCERDLEQYCPKKIWS 120
Query: 104 VNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAG 163
N++ ++ T G A+ T +K + P ++ +AA L A +T Y L + F
Sbjct: 121 YNDVYINGQPTQGGFAKATVVHQKFVVKIPEGMAVEQAAPLLCAGVTVYSPL--SHFGLK 178
Query: 164 KSILVLG--GAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGA-DLPIDYTKENF 220
+ L G G GGVG + +++AK + + ++++ + E L+ LGA D I +
Sbjct: 179 QPGLRGGILGLGGVGHMGVKIAKAMGHHVTVISSSNKKREEALQDLGADDYVIGSDQAKM 238
Query: 221 EEVSEKFDVVYDAV 234
E+++ D V D V
Sbjct: 239 SELADSLDYVIDTV 252
>At3g19450 putative cinnamyl alcohol dehydrogenase 2
Length = 365
Score = 62.8 bits (151), Expect = 2e-10
Identities = 65/251 (25%), Positives = 100/251 (38%), Gaps = 38/251 (15%)
Query: 42 DQVLIKVEAAALNPVDS---KRALGHFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGD 98
D V IKV + D K LG S P VPG++V G V++VGS V K VGD
Sbjct: 36 DDVYIKVICCGICHTDIHQIKNDLGM-----SNYPMVPGHEVVGEVLEVGSDVSKFTVGD 90
Query: 99 EVYGDV--------------------------NEITLDHPKTVGSLAEYTAAEEKVLAHK 132
V V N++ D T G A+ +K +
Sbjct: 91 VVGVGVVVGCCGSCKPCSSELEQYCNKRIWSYNDVYTDGKPTQGGFADTMIVNQKFVVKI 150
Query: 133 PSNLSFIEAASLPLAIITAYQGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKI 192
P ++ +AA L A +T Y L A + G GGVG + +++AK + +
Sbjct: 151 PEGMAVEQAAPLLCAGVTVYSPLSHFGLMASGLKGGILGLGGVGHMGVKIAKAMGHHVTV 210
Query: 193 AATASTGKLEFLRKLGA-DLPIDYTKENFEEVSEKFDVVYDAV---GQSDRALKAVKEGG 248
+++ K E + LGA D + + +++ D + D V D L +K G
Sbjct: 211 ISSSDKKKEEAIEHLGADDYVVSSDPAEMQRLADSLDYIIDTVPVFHPLDPYLACLKLDG 270
Query: 249 KVVTILPPGTP 259
K++ + TP
Sbjct: 271 KLILMGVINTP 281
>At4g39330 cinnamyl-alcohol dehydrogenase CAD1
Length = 360
Score = 61.2 bits (147), Expect = 7e-10
Identities = 54/221 (24%), Positives = 86/221 (38%), Gaps = 30/221 (13%)
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDV--------------------------NE 106
P VPG+++ G+ KVG V K K GD V V N
Sbjct: 67 PVVPGHEIVGIATKVGKNVTKFKEGDRVGVGVISGSCQSCESCDQDLENYCPQMSFTYNA 126
Query: 107 ITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSI 166
I D K G +E +++ + P NL A L A IT Y ++ +
Sbjct: 127 IGSDGTKNYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGMTEAGKH 186
Query: 167 LVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENFEEVSE 225
L + G GG+G + +++ K + +++ST E + LGAD + + +
Sbjct: 187 LGVAGLGGLGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQKMKAAIG 246
Query: 226 KFDVVYD---AVGQSDRALKAVKEGGKVVTILPPGTPPAIP 263
D + D AV L +K GK++ + P P +P
Sbjct: 247 TMDYIIDTISAVHALYPLLGLLKVNGKLIALGLPEKPLELP 287
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 57.4 bits (137), Expect = 1e-08
Identities = 51/192 (26%), Positives = 80/192 (41%), Gaps = 29/192 (15%)
Query: 69 DSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEV--------------------------YG 102
DS P VPG+++AG+V KVG V++ KVGD V
Sbjct: 61 DSKYPLVPGHEIAGIVTKVGPNVQRFKVGDHVGVGTYVNSCRECEYCNEGQEVNCAKGVF 120
Query: 103 DVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSA 162
N I D T G + + E+ P + AA L A IT Y + +
Sbjct: 121 TFNGIDHDGSVTKGGYSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNMNQ 180
Query: 163 -GKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENF 220
GKS+ V+G GG+G + ++ K + + +T+ + K E L LGA+ I +
Sbjct: 181 PGKSLGVIG-LGGLGHMAVKFGKAFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQM 239
Query: 221 EEVSEKFDVVYD 232
+ + + D + D
Sbjct: 240 KALEKSLDFLVD 251
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 51.6 bits (122), Expect = 6e-07
Identities = 53/227 (23%), Positives = 89/227 (38%), Gaps = 34/227 (14%)
Query: 70 SPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDV------------NEITLDHPKTV-- 115
S P +PG+++ G+ KVG V K K GD V V ++ PK V
Sbjct: 58 SRYPIIPGHEIVGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFT 117
Query: 116 -------------GSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFS- 161
G ++ + + + P L A L A IT Y ++ +
Sbjct: 118 YNSRSSDGTSRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTK 177
Query: 162 -AGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKEN 219
+GK + V G GG+G + +++ K + + +S + E + +LGAD + +
Sbjct: 178 ESGKRLGV-NGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQK 236
Query: 220 FEEVSEKFDVVYDAVGQSDRAL---KAVKEGGKVVTILPPGTPPAIP 263
+E D + D V L +K GK+V + P P +P
Sbjct: 237 MKEAVGTMDFIIDTVSAEHALLPLFSLLKVNGKLVALGLPEKPLDLP 283
>At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa
Length = 363
Score = 51.2 bits (121), Expect = 7e-07
Identities = 47/212 (22%), Positives = 86/212 (40%), Gaps = 32/212 (15%)
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEV--------------------------YGDVNE 106
P VPG+++ G V ++G++V K +GD+V N
Sbjct: 68 PLVPGHEIIGEVSEIGNKVSKFNLGDKVGVGCIVDSCRTCESCREDQENYCTKAIATYNG 127
Query: 107 ITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFSAGKSI 166
+ D G +++ +E+ P L + AA L A I+ Y ++ +
Sbjct: 128 VHHDGTINYGGYSDHIVVDERYAVKIPHTLPLVSAAPLLCAGISMYSPMKYFGLTGPDKH 187
Query: 167 LVLGGAGGVGSLVIQLAKHVFEASKIAATASTGK-LEFLRKLGAD-LPIDYTKENFEEVS 224
+ + G GG+G + ++ AK F +++TGK + L LGAD + ++ +
Sbjct: 188 VGIVGLGGLGHIGVRFAK-AFGTKVTVVSSTTGKSKDALDTLGADGFLVSTDEDQMKAAM 246
Query: 225 EKFDVVYDAVGQS---DRALKAVKEGGKVVTI 253
D + D V S + +K GK+V +
Sbjct: 247 GTMDGIIDTVSASHSISPLIGLLKSNGKLVLL 278
>At2g21890 putative cinnamyl-alcohol dehydrogenase
Length = 375
Score = 51.2 bits (121), Expect = 7e-07
Identities = 49/216 (22%), Positives = 84/216 (38%), Gaps = 33/216 (15%)
Query: 70 SPLPTVPGYDVAGVVVKVGSQVKKLKVGDEVYGDV------------------------- 104
S P +PG+++ G+ KVG V K K GD V V
Sbjct: 58 SRYPIIPGHEIVGIATKVGKNVTKFKEGDRVGVGVIIGSCQSCESCNQDLENYCPKVVFT 117
Query: 105 -NEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITAYQGLETAQFS-- 161
N + D + G ++ + + + P L A L A IT Y ++ +
Sbjct: 118 YNSRSSDGTRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGMTKE 177
Query: 162 AGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD-LPIDYTKENF 220
+GK + V G GG+G + +++ K + + +S + E + +LGAD + +
Sbjct: 178 SGKRLGV-NGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQKM 236
Query: 221 EEVSEKFDVVYDAVGQSDRAL---KAVKEGGKVVTI 253
+E D + D V L +K GK+V +
Sbjct: 237 KEAVGTMDFIIDTVSAEHALLPLFSLLKVSGKLVAL 272
>At5g42250 alcohol dehydrogenase
Length = 390
Score = 36.6 bits (83), Expect = 0.019
Identities = 51/235 (21%), Positives = 84/235 (35%), Gaps = 55/235 (23%)
Query: 73 PTVPGYDVAGVVVKVGSQVKKLKVGDEV-------------------------------- 100
P + G++ GVV VG VK++ GD V
Sbjct: 73 PRILGHEAIGVVESVGENVKEVVEGDTVLPTFMPDCGDCVDCKSHKSNLCSKFPFKVSPW 132
Query: 101 ---------YGDVNEITLDHPKTVGSLAEYTAAEEKVLAHKPSNLSFIEAASLPLAIITA 151
+ D+N TL H V S +EYT + + S++ A L + T
Sbjct: 133 MPRYDNSSRFTDLNGETLFHFLNVSSFSEYTVLDVANVVKIDSSIPPSRACLLSCGVSTG 192
Query: 152 Y-QGLETAQFSAGKSILVLGGAGGVGSLVIQLAKHVFEASKIAATASTGKLEFLRKLGAD 210
ETA+ G ++++ G G +G V + A+ + I + K + +K G
Sbjct: 193 VGAAWETAKVEKGSTVVIF-GLGSIGLAVAEGARLCGASRIIGVDINPTKFQVGQKFGVT 251
Query: 211 LPIDYTKENFEEVSEKF--------DVVYDAVGQS---DRALKAVKEG-GKVVTI 253
++ VSE D ++ VG S A ++G GK +T+
Sbjct: 252 EFVNSMTCEKNRVSEVINEMTDGGADYCFECVGSSSLVQEAYACCRQGWGKTITL 306
>At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III
(pir||S71244)
Length = 379
Score = 33.1 bits (74), Expect = 0.21
Identities = 20/58 (34%), Positives = 31/58 (52%), Gaps = 2/58 (3%)
Query: 43 QVLIKVEAAALNPVDSKRALGHFKDIDSPLPTVPGYDVAGVVVKVGSQVKKLKVGDEV 100
+V IK+ AL D+ G KD + P + G++ AG+V VG V +++ GD V
Sbjct: 36 EVRIKILYTALCHTDAYTWSG--KDPEGLFPCILGHEAAGIVESVGEGVTEVQAGDHV 91
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,198,863
Number of Sequences: 26719
Number of extensions: 313485
Number of successful extensions: 837
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 777
Number of HSP's gapped (non-prelim): 52
length of query: 321
length of database: 11,318,596
effective HSP length: 99
effective length of query: 222
effective length of database: 8,673,415
effective search space: 1925498130
effective search space used: 1925498130
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0400a.9


