

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.8
(721 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
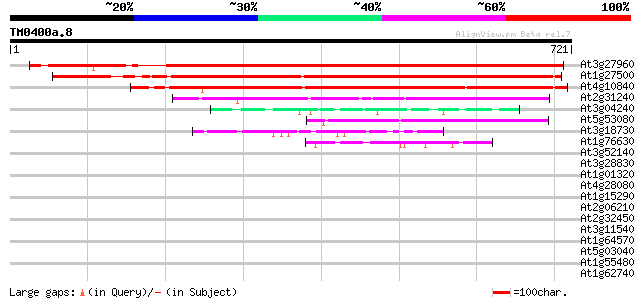
Score E
Sequences producing significant alignments: (bits) Value
At3g27960 hypothetical protein 846 0.0
At1g27500 unknown protein 691 0.0
At4g10840 unknown protein 645 0.0
At2g31240 putative kinesin light chain protein (At2g31240) 170 3e-42
At3g04240 O-linked GlcNAc transferase like protein 60 3e-09
At5g53080 unknown protein 46 8e-05
At3g18730 hypothetical protein 45 2e-04
At1g76630 hypothetical protein, 3' partial 44 3e-04
At3g52140 putative protein 40 0.005
At3g28830 hypothetical protein 39 0.008
At1g01320 unknown protein 39 0.008
At4g28080 putative protein 39 0.010
At1g15290 unknown protein 38 0.023
At2g06210 putative TPR repeat nuclear phosphoprotein 37 0.039
At2g32450 putative O-GlcNAc transferase 36 0.066
At3g11540 spindly (gibberellin signal transduction protein) 36 0.087
At1g64570 hypothetical protein 35 0.15
At5g03040 unknown protein 35 0.19
At1g55480 unknown protein 34 0.25
At1g62740 TPR-repeat protein 34 0.33
>At3g27960 hypothetical protein
Length = 663
Score = 846 bits (2186), Expect = 0.0
Identities = 448/695 (64%), Positives = 539/695 (77%), Gaps = 58/695 (8%)
Query: 26 KDNFNQQASPRSTLSPRSIQSSDSIDLAIDGVVDTSIEQLYYNVCEMRSSD-QSPSRASF 84
KD+ QASPRS LS SIDLAIDG ++ SIEQLY+NVCEM SSD QSPSRASF
Sbjct: 11 KDDSALQASPRSPLS--------SIDLAIDGAMNASIEQLYHNVCEMESSDDQSPSRASF 62
Query: 85 YSYGEESRIDSELGHLVGGILE---ITKEVVTENKEESNGNAA--EKDIVSCGKEATKKD 139
SYG ESRID EL HLVG + E KE++ E KEESNG + +K +S GK+ K
Sbjct: 63 ISYGAESRIDLELRHLVGDVGEEGESKKEIILEKKEESNGEGSLSQKKPLSNGKKVAKTS 122
Query: 140 NNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIE 199
N P SRI S+ KS +L
Sbjct: 123 PNNPKMPGSRI------SSRKSPDLG---------------------------------- 142
Query: 200 ERIATGLDNPDLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADGKPSL--ELVM 257
+++ ++P+LG LLKQ R+++SSGEN KALDLALRA+K FE C +G+ L LVM
Sbjct: 143 -KVSVDEESPELGVVLLKQARELVSSGENLNKALDLALRAVKVFEKCGEGEKQLGLNLVM 201
Query: 258 CLHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIEN 317
LH+LA IY LG+YN+A+P+LERSI+IP++EDG+DHALAKFAGCMQLGD Y +MG +EN
Sbjct: 202 SLHILAAIYAGLGRYNDAVPVLERSIEIPMIEDGEDHALAKFAGCMQLGDMYGLMGQVEN 261
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNG 377
SI+ YTAGLEIQ QVLGE+D R GETCRY+AEAHVQA+QF+EA RLCQMAL+IH+ NG
Sbjct: 262 SIMLYTAGLEIQRQVLGESDARVGETCRYLAEAHVQAMQFEEASRLCQMALDIHKENGAA 321
Query: 378 SSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLA 437
++AS+EE+ADR+LMGLI D+KGDYE ALEHYVLASMAMS+ +DVA+VD SIGDAY++
Sbjct: 322 ATASIEEAADRKLMGLICDAKGDYEVALEHYVLASMAMSSQNHREDVAAVDCSIGDAYMS 381
Query: 438 LARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF 497
LAR+DEA+F+YQKAL VFK KGE H +VA VYVRLADLYNKIGK +DSKSYCENAL+I+
Sbjct: 382 LARFDEAIFAYQKALAVFKQGKGETHSSVALVYVRLADLYNKIGKTRDSKSYCENALKIY 441
Query: 498 GKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQM 557
K PG P EE+A G I+++AIYQSMN+L++ LKLL++ALKIY +APGQQ+TIAGIEAQM
Sbjct: 442 LKPTPGTPMEEVATGFIEISAIYQSMNELDQALKLLRRALKIYANAPGQQNTIAGIEAQM 501
Query: 558 GVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFE 617
GV+ YM+GNYS+SY+IFKSA++KFR SGEKK+ALFGIALNQMGLACVQRYAINEAADLFE
Sbjct: 502 GVVTYMMGNYSESYDIFKSAISKFRNSGEKKTALFGIALNQMGLACVQRYAINEAADLFE 561
Query: 618 EARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVD 677
EA+TILEKE G YHPDTL VYSNLAGTYDAMGR+DDAIEILE+VVG REEKLGTANP+V+
Sbjct: 562 EAKTILEKECGPYHPDTLAVYSNLAGTYDAMGRLDDAIEILEYVVGTREEKLGTANPEVE 621
Query: 678 DEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRL 712
DEK+RLA LLKEAGRGR++++ R+L TLLD N +
Sbjct: 622 DEKQRLAALLKEAGRGRSKRN-RALLTLLDNNPEI 655
>At1g27500 unknown protein
Length = 650
Score = 691 bits (1784), Expect = 0.0
Identities = 370/658 (56%), Positives = 485/658 (73%), Gaps = 32/658 (4%)
Query: 55 DGVVDTSIEQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHLVGGILEITKEVVTE 114
D + DT+IE+L N+CE++SS+QSPSR SF SYG+ES+IDS+L HL G + + E
Sbjct: 14 DQMFDTTIEELCKNLCELQSSNQSPSRQSFGSYGDESKIDSDLQHLALGEMRDIDILEDE 73
Query: 115 NKEESNGNAAEKDIVSCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRS 174
E+ E D+ S + S+ +E + K T K++
Sbjct: 74 GDEDEVAKPEEFDVKS--------------NSSNLDLEVMPRDMEKQ--------TGKKN 111
Query: 175 ERSSRKGSLYPMRKHR--SLALRGIIEERIATGLDNPDLGPFLLKQTRDMISSGENPQKA 232
S G + MRK + L+ EE + +N +L FLL Q R+++SSG++ KA
Sbjct: 112 VTKSNVG-VGGMRKKKVGGTKLQNGNEEPSS---ENVELARFLLNQARNLVSSGDSTHKA 167
Query: 233 LDLALRALKSFEMCAD-GKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLEDG 291
L+L RA K FE A+ GKP LE +MCLHV A ++C L +YNEAIP+L+RS++IPV+E+G
Sbjct: 168 LELTHRAAKLFEASAENGKPCLEWIMCLHVTAAVHCKLKEYNEAIPVLQRSVEIPVVEEG 227
Query: 292 QDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAH 351
++HALAKFAG MQLGDTYAM+G +E+SI YT GL IQ +VLGE DPR GETCRY+AEA
Sbjct: 228 EEHALAKFAGLMQLGDTYAMVGQLESSISCYTEGLNIQKKVLGENDPRVGETCRYLAEAL 287
Query: 352 VQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLA 411
VQAL+FDEA+++C+ AL IHR +G S+ E+ADRRLMGLI ++KGD+E ALEH VLA
Sbjct: 288 VQALRFDEAQQVCETALSIHRESG--LPGSIAEAADRRLMGLICETKGDHENALEHLVLA 345
Query: 412 SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYV 471
SMAM+ANGQE +VA VD+SIGD+YL+L+R+DEA+ +YQK+LT K+ KGENHP V SVY+
Sbjct: 346 SMAMAANGQESEVAFVDTSIGDSYLSLSRFDEAICAYQKSLTALKTAKGENHPAVGSVYI 405
Query: 472 RLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLK 531
RLADLYN+ GK +++KSYCENALRI+ I PEEIA+GL D++ I +SMN++E+ +
Sbjct: 406 RLADLYNRTGKVREAKSYCENALRIYESHNLEISPEEIASGLTDISVICESMNEVEQAIT 465
Query: 532 LLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSAL 591
LL+KALKIY +PGQ+ IAGIEAQMGV+YYM+G Y +SYN FKSA++K RA+G+K+S
Sbjct: 466 LLQKALKIYADSPGQKIMIAGIEAQMGVLYYMMGKYMESYNTFKSAISKLRATGKKQSTF 525
Query: 592 FGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRV 651
FGIALNQMGLAC+Q AI EA +LFEEA+ ILE+E G YHP+TLG+YSNLAG YDA+GR+
Sbjct: 526 FGIALNQMGLACIQLDAIEEAVELFEEAKCILEQECGPYHPETLGLYSNLAGAYDAIGRL 585
Query: 652 DDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDAN 709
DDAI++L VVG+REEKLGTANP +DEKRRLA+LLKEAG RK+K SL+TL+D++
Sbjct: 586 DDAIKLLGHVVGVREEKLGTANPVTEDEKRRLAQLLKEAGNVTGRKAK-SLKTLIDSD 642
>At4g10840 unknown protein
Length = 609
Score = 645 bits (1664), Expect = 0.0
Identities = 341/573 (59%), Positives = 429/573 (74%), Gaps = 24/573 (4%)
Query: 156 KSTPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERIATGLDNPDLGPFL 215
K TP S R +PS ++ + + P + A+ + + LDNPDLGPFL
Sbjct: 38 KKTPSSTPSRSKPSPNRSTGKKDS-----PTVSSSTAAVIDVDDP----SLDNPDLGPFL 88
Query: 216 LKQTRDMISSGENPQKALDLALRALKSFEMC-----------ADGKPSLELVMCLHVLAT 264
LK RD I+SGE P KALD A+RA KSFE C +DG P L+L M LHVLA
Sbjct: 89 LKLARDAIASGEGPNKALDYAIRATKSFERCCAAVAPPIPGGSDGGPVLDLAMSLHVLAA 148
Query: 265 IYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTA 324
IYC+LG+++EA+P LER+I +P G DH+LA F+G MQLGDT +M+G I+ SI Y
Sbjct: 149 IYCSLGRFDEAVPPLERAIQVPDPTRGPDHSLAAFSGHMQLGDTLSMLGQIDRSIACYEE 208
Query: 325 GLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEE 384
GL+IQ Q LG+TDPR GETCRY+AEA+VQA+QF++AE LC+ LEIHR + AS+EE
Sbjct: 209 GLKIQIQTLGDTDPRVGETCRYLAEAYVQAMQFNKAEELCKKTLEIHRAHSE--PASLEE 266
Query: 385 SADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEA 444
+ADRRLM +I ++KGDYE ALEH VLASMAM A+GQE +VAS+D SIG+ Y++L R+DEA
Sbjct: 267 AADRRLMAIICEAKGDYENALEHLVLASMAMIASGQESEVASIDVSIGNIYMSLCRFDEA 326
Query: 445 VFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGI 504
VFSYQKALTVFK++KGE HP VASV+VRLA+LY++ GK ++SKSYCENALRI+ K PG
Sbjct: 327 VFSYQKALTVFKASKGETHPTVASVFVRLAELYHRTGKLRESKSYCENALRIYNKPVPGT 386
Query: 505 PPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYML 564
EEIA GL +++AIY+S+++ E+ LKLL+K++K+ PGQQS IAG+EA+MGVMYY +
Sbjct: 387 TVEEIAGGLTEISAIYESVDEPEEALKLLQKSMKLLEDKPGQQSAIAGLEARMGVMYYTV 446
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
G Y D+ N F+SAV K RA+GE KSA FG+ LNQMGLACVQ + I+EA +LFEEAR ILE
Sbjct: 447 GRYEDARNAFESAVTKLRAAGE-KSAFFGVVLNQMGLACVQLFKIDEAGELFEEARGILE 505
Query: 625 KEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLA 684
+E G DTLGVYSNLA TYDAMGR++DAIEILE V+ +REEKLGTANPD +DEK+RLA
Sbjct: 506 QERGPCDQDTLGVYSNLAATYDAMGRIEDAIEILEQVLKLREEKLGTANPDFEDEKKRLA 565
Query: 685 ELLKEAGRGRNRKSKRSLETLLDANSRLIKNNS 717
ELLKEAGR RN K+K SL+ L+D N+R K S
Sbjct: 566 ELLKEAGRSRNYKAK-SLQNLIDPNARPPKKES 597
>At2g31240 putative kinesin light chain protein (At2g31240)
Length = 617
Score = 170 bits (430), Expect = 3e-42
Identities = 136/491 (27%), Positives = 232/491 (46%), Gaps = 16/491 (3%)
Query: 210 DLGPFLLKQTRDMISSGENPQKALDLALRALKSFEMCADG-KPSLELVMCLHVLATIYCN 268
+LG LK + GE+P+K L A +ALKSF+ DG KP+L + M ++ +
Sbjct: 116 ELGLSALKLGLHLDREGEDPEKVLSYADKALKSFD--GDGNKPNLLVAMASQLMGSANYG 173
Query: 269 LGQYNEAIPILERS--IDIPVLEDG----QDHALAKFAGCMQLGDTYAMMGNIENSILFY 322
L ++++++ L R+ I + + DG +D A ++L + MG E +I
Sbjct: 174 LKRFSDSLGYLNRANRILVKLEADGDCVVEDVRPVLHAVQLELANVKNAMGRREEAIENL 233
Query: 323 TAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASV 382
LEI+ E G R +A+A+V L F+EA ALEIH+ +SA V
Sbjct: 234 KKSLEIKEMTFDEDSKEMGVANRSLADAYVAVLNFNEALPYALKALEIHKKELGNNSAEV 293
Query: 383 EESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYD 442
+ DRRL+G+IY ++ ALE L+ + G + ++ + + +AL +Y+
Sbjct: 294 AQ--DRRLLGVIYSGLEQHDKALEQNRLSQRVLKNWGMKLELIRAEIDAANMKVALGKYE 351
Query: 443 EAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKP 502
EA+ + +V + T ++ A V++ ++ KF +SK E A I K +
Sbjct: 352 EAIDILK---SVVQQTDKDSEMR-AMVFISMSKALVNQQKFAESKRCLEFACEILEKKET 407
Query: 503 GIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYY 562
+P E +A +VA Y+SMN+ E + LL+K L I P +Q + + A++G +
Sbjct: 408 ALPVE-VAEAYSEVAMQYESMNEFETAISLLQKTLGILEKLPQEQHSEGSVSARIGWLLL 466
Query: 563 MLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTI 622
G S + +SA + + S K G N +G A ++ AA +F A+ I
Sbjct: 467 FSGRVSQAVPYLESAAERLKESFGAKHFGVGYVYNNLGAAYLELGRPQSAAQMFAVAKDI 526
Query: 623 LEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRR 682
++ G H D++ NL+ Y MG A+E + V+ + +A ++ + KR
Sbjct: 527 MDVSLGPNHVDSIDACQNLSKAYAGMGNYSLAVEFQQRVINAWDNHGDSAKDEMKEAKRL 586
Query: 683 LAELLKEAGRG 693
L +L +A G
Sbjct: 587 LEDLRLKARGG 597
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 60.5 bits (145), Expect = 3e-09
Identities = 97/448 (21%), Positives = 162/448 (35%), Gaps = 107/448 (23%)
Query: 259 LHVLATIYCNLGQYNEAIPILERSIDIPVLEDGQDHALAKFAGCM-QLGDTYAMMGNIEN 317
L ++ IY L +Y+ I E ++ I +FA C + + + G+ +
Sbjct: 90 LLLIGAIYYQLQEYDMCIARNEEALRIQ----------PQFAECYGNMANAWKEKGDTDR 139
Query: 318 SILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEI------- 370
+I +Y +E++ P F + +A A+++ + EA + CQ AL +
Sbjct: 140 AIRYYLIAIELR--------PNFADAWSNLASAYMRKGRLSEATQCCQQALSLNPLLVDA 191
Query: 371 HRGNGNGSSAS--VEES----------------ADRRLMGLIYDSKGDYEAALEHYVLAS 412
H GN A + E+ A L GL +S GD AL++Y A
Sbjct: 192 HSNLGNLMKAQGLIHEAYSCYLEAVRIQPTFAIAWSNLAGLFMES-GDLNRALQYYKEAV 250
Query: 413 MAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYV- 471
A ++G+ Y AL R EA+ YQ AL + + N+AS+Y
Sbjct: 251 KLKPA------FPDAYLNLGNVYKALGRPTEAIMCYQHALQM-RPNSAMAFGNIASIYYE 303
Query: 472 -------------------RLADLYNKIGKFKDSKSYCENALRIFGKSKPGIP--PEEIA 510
R + YN +G + A+R + + P P+ +A
Sbjct: 304 QGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEAVRCYNQCLALQPNHPQAMA 363
Query: 511 NGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEA---QMGVMYYMLGNY 567
N + IY N + L K L + G+ A + ++Y GNY
Sbjct: 364 N----LGNIYMEWNMMGPASSLFKATLAV----------TTGLSAPFNNLAIIYKQQGNY 409
Query: 568 SDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY 627
SD+ + + + + L AL G + + EA + A
Sbjct: 410 SDAISCYNEVL--------RIDPLAADALVNRGNTYKEIGRVTEAIQDYMHAI------- 454
Query: 628 GQYHPDTLGVYSNLAGTYDAMGRVDDAI 655
+ P ++NLA Y G V+ AI
Sbjct: 455 -NFRPTMAEAHANLASAYKDSGHVEAAI 481
Score = 41.6 bits (96), Expect = 0.002
Identities = 55/267 (20%), Positives = 103/267 (37%), Gaps = 46/267 (17%)
Query: 399 GDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVFKST 458
GD++ ALEH ++M N D + IG Y L YD + ++AL +
Sbjct: 67 GDFKQALEH---SNMVYQRNPLRTDNLLL---IGAIYYQLQEYDMCIARNEEALRI---- 116
Query: 459 KGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAA 518
P A Y +A+ + + G + Y A+ + +P A+ ++A+
Sbjct: 117 ----QPQFAECYGNMANAWKEKGDTDRAIRYYLIAIEL----RPNF-----ADAWSNLAS 163
Query: 519 IYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAV 578
Y L + + ++AL + S + + G+++ ++Y+ + AV
Sbjct: 164 AYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIH-------EAYSCYLEAV 216
Query: 579 AKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVY 638
+ F IA + + ++ +N A ++EA + P Y
Sbjct: 217 --------RIQPTFAIAWSNLAGLFMESGDLNRALQYYKEAVKL--------KPAFPDAY 260
Query: 639 SNLAGTYDAMGRVDDAIEILEFVVGMR 665
NL Y A+GR +AI + + MR
Sbjct: 261 LNLGNVYKALGRPTEAIMCYQHALQMR 287
>At5g53080 unknown protein
Length = 564
Score = 45.8 bits (107), Expect = 8e-05
Identities = 57/317 (17%), Positives = 131/317 (40%), Gaps = 9/317 (2%)
Query: 382 VEESADRRLMGLIYDSKGDYE---AALEHYVLASMAMSANGQEQDVASVDSSIGDAYLAL 438
+E++A ++ L Y + GD + A L+ ++ + + E + SV +G Y +
Sbjct: 142 IEQAAVLDIIALGYMAVGDLKPVPALLD--MINKIVDNLKDSEPLLDSVLMHVGSMYSVI 199
Query: 439 ARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFG 498
+++ A+ +Q+A+ + ++ G+ + + + + +A + GK + E L I
Sbjct: 200 GKFENAILVHQRAIRILENRYGKCNTLLVTPLLGMAKSFASDGKATKAIGVYERTLTILE 259
Query: 499 KSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIE-AQM 557
+++ G E++ L + + + + IY G++ G+ +
Sbjct: 260 RNR-GSESEDLVVPLFSLGKLLLKEGKAAEAEIPFTSIVNIYKKIYGERDGRVGMAMCSL 318
Query: 558 GVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAI--NEAADL 615
G+ +++ +I+++A+ + S + ++ LA + + +E +L
Sbjct: 319 ANAKCSKGDANEAVDIYRNALRIIKDSNYMTIDNSILENMRIDLAELLHFVGRGDEGREL 378
Query: 616 FEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPD 675
EE I E+ G+ HP NLA +Y +A +L + + E +G+
Sbjct: 379 LEECLLINERFKGKNHPSMATHLINLAASYSRSKNYVEAERLLRTCLNIMEVSVGSEGQS 438
Query: 676 VDDEKRRLAELLKEAGR 692
+ LA L + R
Sbjct: 439 ITFPMLNLAVTLSQLNR 455
Score = 38.5 bits (88), Expect = 0.013
Identities = 97/481 (20%), Positives = 165/481 (34%), Gaps = 90/481 (18%)
Query: 215 LLKQTRDMISSGENPQKALDLALRA--LKSFEMCADGKPSLELVMCLHVLATIYCNLGQY 272
L + + M+ G+ A+DL LRA + E G +E L ++A Y +G
Sbjct: 104 LFNEVKSMVKIGKESD-AMDL-LRANYVAVKEELDSGLKGIEQAAVLDIIALGYMAVGDL 161
Query: 273 NEAIPILERSIDIPVLEDGQDHALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQV 332
+ +P L I+ ++++ +D + M +G Y+++G EN+IL + + I
Sbjct: 162 -KPVPALLDMIN-KIVDNLKDSEPLLDSVLMHVGSMYSVIGKFENAILVHQRAIRI---- 215
Query: 333 LGETDPRFGETCRYVAEAHVQALQFDEAERLCQMALEIHRGNGNGSSASVEESADRRLMG 392
+ R+G+ C L S AS
Sbjct: 216 ---LENRYGK---------------------CNTLLVTPLLGMAKSFAS----------- 240
Query: 393 LIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKAL 452
D K + L + + + +D+ S+G L + EA + +
Sbjct: 241 ---DGKATKAIGVYERTLTILERNRGSESEDLVVPLFSLGKLLLKEGKAAEAEIPFTSIV 297
Query: 453 TVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSK-PGIPPEEIAN 511
++K GE V LA+ G ++ NALRI S I + N
Sbjct: 298 NIYKKIYGERDGRVGMAMCSLANAKCSKGDANEAVDIYRNALRIIKDSNYMTIDNSILEN 357
Query: 512 GLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSY 571
ID+A + + ++G +LL++ L I G+ M L N + SY
Sbjct: 358 MRIDLAELLHFVGRGDEGRELLEECLLINERFKGKNHPS---------MATHLINLAASY 408
Query: 572 NIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYH 631
+ K+ V EA L I+E G
Sbjct: 409 SRSKNYV--------------------------------EAERLLRTCLNIMEVSVGSEG 436
Query: 632 PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLAELLKEAG 691
NLA T + R ++A +I V+ +RE+ G + V + L + G
Sbjct: 437 QSITFPMLNLAVTLSQLNRDEEAEQIALKVLRIREKAFGEDSLPVGEALDCLVSIQARLG 496
Query: 692 R 692
R
Sbjct: 497 R 497
>At3g18730 hypothetical protein
Length = 1220
Score = 44.7 bits (104), Expect = 2e-04
Identities = 75/347 (21%), Positives = 144/347 (40%), Gaps = 59/347 (17%)
Query: 236 ALRALKSFEMCADGKPSLELVMCLHVLATIYCNLGQYNEAIPILERSIDIPVLE-DGQDH 294
A RA + E D + E + + I N G+Y +A+ DI V G+D
Sbjct: 9 AKRAYRKAEEVGDRR---EQARWANNVGDILKNHGEYVDALKWFRIDYDISVKYLPGKD- 64
Query: 295 ALAKFAGCMQLGDTYAMMGNIENSILFYTAGLEIQGQVLGETD-----PRFGETCRYV-- 347
C LG+ Y + N E ++++ L++ + + + G T +
Sbjct: 65 ---LLPTCQSLGEIYLRLENFEEALIYQKKHLQLAEEANDTVEKQRACTQLGRTYHEMFL 121
Query: 348 -AEAHVQALQ-----FDEAERLCQMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDY 401
+E +A+Q F +A L Q+ E + SS +EE + +++ G
Sbjct: 122 KSEDDCEAIQSAKKYFKKAMELAQILKE--KPPPGESSGFLEEYIN------AHNNIGML 173
Query: 402 EAALEHYVLASMAMSANGQ---EQDVASVDS-------SIGDAYLALARYDEAVFSYQKA 451
+ L++ A + Q E++V D+ ++G+ ++AL +DEA +
Sbjct: 174 DLDLDNPEAARTILKKGLQICDEEEVREYDAARSRLHHNLGNVFMALRSWDEAKKHIEMD 233
Query: 452 LTVFKSTKGENH-PNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIA 510
+ + NH A Y+ LA+L+NK K+ D AL +GK+ +A
Sbjct: 234 INICHKI---NHVQGEAKGYINLAELHNKTQKYID-------ALLCYGKA------SSLA 277
Query: 511 NGLIDVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQM 557
+ D +A+ + +E K++KK++K+ ++ + + A+M
Sbjct: 278 KSMQDESAL---VEQIEHNTKIVKKSMKVMEELREEELMLKKLSAEM 321
Score = 32.0 bits (71), Expect = 1.3
Identities = 43/206 (20%), Positives = 82/206 (38%), Gaps = 29/206 (14%)
Query: 383 EESADRRL-------MGLIYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAY 435
EE DRR +G I + G+Y AL+ + + +D+ S+G+ Y
Sbjct: 17 EEVGDRREQARWANNVGDILKNHGEYVDALKWFRIDYDISVKYLPGKDLLPTCQSLGEIY 76
Query: 436 LALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKI--------GKFKDSK 487
L L ++EA+ +K L + + A +L Y+++ + +K
Sbjct: 77 LRLENFEEALIYQKKHLQLAEEANDTVEKQRAC--TQLGRTYHEMFLKSEDDCEAIQSAK 134
Query: 488 SYCENALRIFGKSKPGIPP-------EEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIY 540
Y + A+ + K PP EE N ++ + +++ E +LKK L+I
Sbjct: 135 KYFKKAMELAQILKEKPPPGESSGFLEEYINAHNNIGMLDLDLDNPEAARTILKKGLQIC 194
Query: 541 GHAPGQQSTIAGIEAQMGVMYYMLGN 566
+ + +A +++ LGN
Sbjct: 195 -----DEEEVREYDAARSRLHHNLGN 215
>At1g76630 hypothetical protein, 3' partial
Length = 971
Score = 43.9 bits (102), Expect = 3e-04
Identities = 69/269 (25%), Positives = 111/269 (40%), Gaps = 46/269 (17%)
Query: 381 SVEESADRRLM----GL-IYDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAY 435
SVEE+ D + GL ++D+ GD E A EH+VL++ + N A +G Y
Sbjct: 9 SVEENPDDSSLQFELGLYLWDNGGDSEKAAEHFVLSAKSDPNN------AVAFKYLGHYY 62
Query: 436 LALA-RYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENAL 494
+ + A YQ+A+ + +PN + L DL+++ GK + C +A
Sbjct: 63 SRVTLDLNRAAKCYQRAVLI--------NPNDSDSGEALCDLFDRQGKEILEIAVCRDAS 114
Query: 495 RIFGKS-----KPGIPP------EEIANGLIDVAAIYQSMNDLEKGLKL-------LKKA 536
K+ + G E L Y +M+DL + L L A
Sbjct: 115 EKSPKAFWAFCRLGYIQLHQKKWSEAVQSLQHAIRGYPTMSDLWEALGLAYQRLGMFTAA 174
Query: 537 LKIYGHAPGQQSTIAGIEAQMGVMYYMLGNY---SDSYNIFKSAVAKFRASGEKKSALFG 593
+K YG A T + ++ MLG+Y + +F+ A+ + S + S L+G
Sbjct: 175 IKAYGRAIELDETKIFALVESANIFLMLGSYRKVTFGVELFEQAL---KISPQNISVLYG 231
Query: 594 IA--LNQMGLACVQRYAINEAADLFEEAR 620
+A L C+ A AA L E+AR
Sbjct: 232 LASGLLSWSKECINLGAFGWAASLLEDAR 260
>At3g52140 putative protein
Length = 1403
Score = 40.0 bits (92), Expect = 0.005
Identities = 57/260 (21%), Positives = 101/260 (37%), Gaps = 32/260 (12%)
Query: 389 RLMGLIYDSKGDYEAAL--EHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVF 446
R + ++ GD A+ +H L D A ++ Y L + + A+
Sbjct: 1054 RYLAMVLYHAGDMAGAIMQQHKELIINERCLGLDHPDTAHSYGNMALFYHGLNQTELALQ 1113
Query: 447 SYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKSKPGIPP 506
+ +AL + + G +HP+VA+ ++ +A +Y +GK + Y + AL+ K++ + P
Sbjct: 1114 NMGRALLLLGLSSGPDHPDVAATFINVAMMYQDMGKMDTALRYLQEALK---KNERLLGP 1170
Query: 507 EEIANGLI--DVAAIYQSMNDLEKGLKLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYML 564
E I + +A + M + + KK I G +
Sbjct: 1171 EHIQTAVCYHALAIAFNCMGAFKLSHQHEKKTYDILVKQLGDDDS--------------- 1215
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
DS N K+ + G K + G LNQM + A N A ++A +L+
Sbjct: 1216 -RTRDSLNWMKTFKMLYILFGFLKFS--GWVLNQMTAQKQKGQAANAANT--QKAIDLLK 1270
Query: 625 KEYGQYHPDTLGVYSNLAGT 644
HPD + + N A T
Sbjct: 1271 A-----HPDLIHAFQNAAAT 1285
Score = 33.9 bits (76), Expect = 0.33
Identities = 25/81 (30%), Positives = 33/81 (39%), Gaps = 10/81 (12%)
Query: 590 ALFGIALNQMGLACVQRYAINEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMG 649
ALF LNQ LA A +L G HPD + N+A Y MG
Sbjct: 1099 ALFYHGLNQTELALQN----------MGRALLLLGLSSGPDHPDVAATFINVAMMYQDMG 1148
Query: 650 RVDDAIEILEFVVGMREEKLG 670
++D A+ L+ + E LG
Sbjct: 1149 KMDTALRYLQEALKKNERLLG 1169
Score = 32.7 bits (73), Expect = 0.73
Identities = 28/115 (24%), Positives = 55/115 (47%), Gaps = 4/115 (3%)
Query: 601 LACVQRYAINEAADLFEEARTIL--EKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEIL 658
LA V +A + A + ++ + ++ E+ G HPDT Y N+A Y + + + A++ +
Sbjct: 1056 LAMVLYHAGDMAGAIMQQHKELIINERCLGLDHPDTAHSYGNMALFYHGLNQTELALQNM 1115
Query: 659 EFVVGMREEKLGTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLI 713
+ + G +PDV +A + ++ G+ + R L+ L N RL+
Sbjct: 1116 GRALLLLGLSSGPDHPDVAATFINVAMMYQD--MGKMDTALRYLQEALKKNERLL 1168
>At3g28830 hypothetical protein
Length = 536
Score = 39.3 bits (90), Expect = 0.008
Identities = 49/196 (25%), Positives = 80/196 (40%), Gaps = 18/196 (9%)
Query: 43 SIQSSDSIDLAIDGV-VDTSIEQLYYNVCEMRSSD-QSPSRASFYSYGEESRIDSELGHL 100
S Q SI++A G+ VD + + SS +S S +S S +S+ S G
Sbjct: 169 SEQKGKSINIASYGLDVDVNDSSIVGGAASSESSSTKSGSVSSSGSVSTKSKESSSSGSS 228
Query: 101 VGGILEITKEVVTENKEESNGNAAEKDIVSC-GKEATKKDNNQSHSPSSRIIEGSAKSTP 159
G V T++KE S G+AA K S G ATK + S ++ GS +P
Sbjct: 229 ASG------SVATKSKESSGGSAATKSKESSGGSAATKSKESSGGSATTGKTSGSPSGSP 282
Query: 160 KSRNLRERPSTDKRSERSSRKGSLYPM---RKHRSLALRGIIEERIATGLDNPDLGPFLL 216
K+ + S +SS KGS S + +G + + + G +
Sbjct: 283 KA------SPSGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRRESGAMAM 336
Query: 217 KQTRDMISSGENPQKA 232
++R+ +S + K+
Sbjct: 337 SKSRETKTSSQRQSKS 352
>At1g01320 unknown protein
Length = 1047
Score = 39.3 bits (90), Expect = 0.008
Identities = 36/155 (23%), Positives = 65/155 (41%), Gaps = 2/155 (1%)
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
G D+ A+AK A + A + + + N+A ++A I E
Sbjct: 146 GKLEDAVTYGTKALAKLVAVCGPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKALDINE 205
Query: 625 KEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLA 684
+E G HPDT+ Y +LA Y + + A++ ++ + + G ++P+ +A
Sbjct: 206 RELGLDHPDTMKSYGDLAVFYYRLQHTELALKYVKRALYLLHLTCGPSHPNTAATYINVA 265
Query: 685 ELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIK 719
+ E G G + R L L N RL+ + I+
Sbjct: 266 --MMEEGLGNVHVALRYLHKALKCNQRLLGPDHIQ 298
Score = 36.6 bits (83), Expect = 0.051
Identities = 28/102 (27%), Positives = 48/102 (46%), Gaps = 9/102 (8%)
Query: 441 YDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF--- 497
+++A QKAL + + G +HP+ Y LA Y ++ + + Y + AL +
Sbjct: 190 FNQATIYQQKALDINERELGLDHPDTMKSYGDLAVFYYRLQHTELALKYVKRALYLLHLT 249
Query: 498 -GKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALK 538
G S P A I+VA + + + ++ L+ L KALK
Sbjct: 250 CGPSHP-----NTAATYINVAMMEEGLGNVHVALRYLHKALK 286
Score = 35.4 bits (80), Expect = 0.11
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 1/98 (1%)
Query: 440 RYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGK 499
+ ++AV KAL + G H A Y LA + G F + Y + AL I +
Sbjct: 147 KLEDAVTYGTKALAKLVAVCGPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKALDI-NE 205
Query: 500 SKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKAL 537
+ G+ + D+A Y + E LK +K+AL
Sbjct: 206 RELGLDHPDTMKSYGDLAVFYYRLQHTELALKYVKRAL 243
>At4g28080 putative protein
Length = 1791
Score = 38.9 bits (89), Expect = 0.010
Identities = 36/155 (23%), Positives = 64/155 (41%), Gaps = 2/155 (1%)
Query: 565 GNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLACVQRYAINEAADLFEEARTILE 624
G D+ N A+AK A + A + + + N+A ++A I E
Sbjct: 878 GKLEDAVNYGTKALAKLVAVCGPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKALDINE 937
Query: 625 KEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKLGTANPDVDDEKRRLA 684
+E G HPDT+ Y +LA Y + + A++ + + + G ++P+ +A
Sbjct: 938 RELGLDHPDTMKSYGDLAVFYYRLQHTELALKYVNRALYLLHLTCGPSHPNTAATYINVA 997
Query: 685 ELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIK 719
+ E G + R L L N RL+ + I+
Sbjct: 998 --MMEEGMKNAHVALRYLHEALKCNQRLLGADHIQ 1030
Score = 35.4 bits (80), Expect = 0.11
Identities = 36/147 (24%), Positives = 59/147 (39%), Gaps = 11/147 (7%)
Query: 398 KGDYEAALEH--YVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTVF 455
KG E A+ + LA + + A S + +++A QKAL +
Sbjct: 877 KGKLEDAVNYGTKALAKLVAVCGPYHRMTAGAYSLLAVVLYHTGDFNQATIYQQKALDIN 936
Query: 456 KSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIF----GKSKPGIPPEEIAN 511
+ G +HP+ Y LA Y ++ + + Y AL + G S P A
Sbjct: 937 ERELGLDHPDTMKSYGDLAVFYYRLQHTELALKYVNRALYLLHLTCGPSHP-----NTAA 991
Query: 512 GLIDVAAIYQSMNDLEKGLKLLKKALK 538
I+VA + + M + L+ L +ALK
Sbjct: 992 TYINVAMMEEGMKNAHVALRYLHEALK 1018
>At1g15290 unknown protein
Length = 1558
Score = 37.7 bits (86), Expect = 0.023
Identities = 28/110 (25%), Positives = 53/110 (47%), Gaps = 2/110 (1%)
Query: 610 NEAADLFEEARTILEKEYGQYHPDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMREEKL 669
N+A ++A I E+E G HPDT+ Y +L+ Y + + A++ + + +
Sbjct: 857 NQATIYQQKALDINERELGLDHPDTMKSYGDLSVFYYRLQHFELALKYVNRALFLLHFTC 916
Query: 670 GTANPDVDDEKRRLAELLKEAGRGRNRKSKRSLETLLDANSRLIKNNSIK 719
G ++P+ +A + KE G + + R L L +N RL+ + I+
Sbjct: 917 GLSHPNTAATYINVAMMEKEV--GNDHLALRYLHEALKSNKRLLGADHIQ 964
Score = 36.6 bits (83), Expect = 0.051
Identities = 27/98 (27%), Positives = 42/98 (42%), Gaps = 1/98 (1%)
Query: 440 RYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGK 499
+ D+AV KAL + G H N A Y LA + G F + Y + AL I +
Sbjct: 813 KLDDAVSYGTKALVKMIAVCGPYHRNTACAYSLLAVVLYHTGDFNQATIYQQKALDI-NE 871
Query: 500 SKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKAL 537
+ G+ + D++ Y + E LK + +AL
Sbjct: 872 RELGLDHPDTMKSYGDLSVFYYRLQHFELALKYVNRAL 909
Score = 33.9 bits (76), Expect = 0.33
Identities = 25/98 (25%), Positives = 46/98 (46%), Gaps = 1/98 (1%)
Query: 441 YDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRIFGKS 500
+++A QKAL + + G +HP+ Y L+ Y ++ F+ + Y AL +
Sbjct: 856 FNQATIYQQKALDINERELGLDHPDTMKSYGDLSVFYYRLQHFELALKYVNRALFLL-HF 914
Query: 501 KPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALK 538
G+ A I+VA + + + + L+ L +ALK
Sbjct: 915 TCGLSHPNTAATYINVAMMEKEVGNDHLALRYLHEALK 952
>At2g06210 putative TPR repeat nuclear phosphoprotein
Length = 501
Score = 37.0 bits (84), Expect = 0.039
Identities = 25/102 (24%), Positives = 43/102 (41%), Gaps = 10/102 (9%)
Query: 395 YDSKGDYEAALEHYVLASMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKALTV 454
Y SKGD+E A +Y+ A N + +G L L +VF+++K L V
Sbjct: 76 YHSKGDFEKAGMYYMAA--IKETNNNPHEFVFPYFGLGQVQLKLGELKGSVFNFEKVLEV 133
Query: 455 FKSTKGENHPNVASVYVRLADLYNKIGKFKDSKSYCENALRI 496
+ P+ L LY ++G+ + + Y A ++
Sbjct: 134 Y--------PDNCETLKALGHLYTQLGQNEKALEYMRKATKL 167
>At2g32450 putative O-GlcNAc transferase
Length = 802
Score = 36.2 bits (82), Expect = 0.066
Identities = 34/161 (21%), Positives = 62/161 (38%), Gaps = 26/161 (16%)
Query: 531 KLLKKALKIYGHAPGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSA 590
+L K+AL + A Q T + G Y+LG Y +S + F A+ + G + +
Sbjct: 243 QLFKEALVSFKRACELQPTDVRPHFKAGNCLYVLGKYKESKDEFLLALEAAESGGNQWAY 302
Query: 591 LFGIALNQMGLACVQRYAINEAADLFEEARTILEKEY-------------GQYH------ 631
L +G++ + A + + EA + Y G+Y
Sbjct: 303 LLPQIYVNLGISLEGEGMVLSACEYYREAAILCPTHYRALKLLGSALFGVGEYRAAVKAL 362
Query: 632 -------PDTLGVYSNLAGTYDAMGRVDDAIEILEFVVGMR 665
PD + +LA + AMG + AIE+ + + ++
Sbjct: 363 EEAIYLKPDYADAHCDLASSLHAMGEDERAIEVFQRAIDLK 403
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 35.8 bits (81), Expect = 0.087
Identities = 63/320 (19%), Positives = 120/320 (36%), Gaps = 53/320 (16%)
Query: 305 LGDTYAMMGNIENSILFYTAGLEIQGQVLGETDPRFGETCRYVAEAHVQALQFDEAERLC 364
LG + + GN + I Y L+I DP + + + + +Q+D A C
Sbjct: 158 LGTSLKLAGNTQEGIQKYYEALKI--------DPHYAPAYYNLGVVYSEMMQYDNALS-C 208
Query: 365 QMALEIHRGNGNGSSASVEESADRRLMGLIYDSKGDYEAALEHYVLASMAMSANGQ--EQ 422
+ R + + MG+IY ++GD E A+ Y +A+S N + +
Sbjct: 209 YEKAALERPMYAEAYCN---------MGVIYKNRGDLEMAITCYE-RCLAVSPNFEIAKN 258
Query: 423 DVASVDSSIGDAYLALARYDEAVFSYQKALTVFKSTKGENHPNVASVYVRLADLYNKIGK 482
++A + +G + V Y+KAL + + A L Y ++ K
Sbjct: 259 NMAIALTDLGTKVKLEGDVTQGVAYYKKALYY--------NWHYADAMYNLGVAYGEMLK 310
Query: 483 FKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKALKIYGH 542
F + + E A A ++ +Y+ ++L+K ++ + AL I
Sbjct: 311 FDMAIVFYELAFHF---------NPHCAEACNNLGVLYKDRDNLDKAVECYQMALSI--- 358
Query: 543 APGQQSTIAGIEAQMGVMYYMLGNYSDSYNIFKSAVAKFRASGEKKSALFGIALNQMGLA 602
+ A +GV+Y + G + ++ + A+ + + A N +G+
Sbjct: 359 ----KPNFAQSLNNLGVVYTVQGKMDAAASMIEKAIL--------ANPTYAEAFNNLGVL 406
Query: 603 CVQRYAINEAADLFEEARTI 622
I A D +EE I
Sbjct: 407 YRDAGNITMAIDAYEECLKI 426
>At1g64570 hypothetical protein
Length = 1239
Score = 35.0 bits (79), Expect = 0.15
Identities = 42/165 (25%), Positives = 59/165 (35%), Gaps = 6/165 (3%)
Query: 41 PRSIQSSDSIDLAIDGVVDTSIEQLYYNVCEMRSSDQSPSRASFYSYGEESRIDSELGHL 100
P S SD+I I+ + D S + E+ SD + E DSE
Sbjct: 1051 PGSAPPSDNISRCIEEMADQSNLGIVMEQEELSDSDDEMMEEEHVEFECEEMADSEGEE- 1109
Query: 101 VGGILEITKEVVTENKEESNGNAAEKDIVSCGKEATKKDNNQ---SHSPSSRIIEGSAKS 157
G E E+ ++ S D+ S GKE K N S PSSR + + K
Sbjct: 1110 -GSECEENIEMQDKDNRNSVVEITSTDVDS-GKELGKDSPNSPWLSLDPSSRRLSSNVKD 1167
Query: 158 TPKSRNLRERPSTDKRSERSSRKGSLYPMRKHRSLALRGIIEERI 202
K E + RS +KG+ R S G+ R+
Sbjct: 1168 REKIEATNETTISQFGPSRSRKKGTPVKSRDAESKEEAGVARLRL 1212
>At5g03040 unknown protein
Length = 446
Score = 34.7 bits (78), Expect = 0.19
Identities = 33/126 (26%), Positives = 57/126 (45%), Gaps = 7/126 (5%)
Query: 61 SIEQLYYNVCEMRSSDQSPSRASFYSYGEES--RIDSELGHLVGGILEITKEVVTENKEE 118
S E++ N+ + RA YSY + + +S+ G+ + V +NK
Sbjct: 214 SKEKVEANLLSKYEATMRRERALAYSYSHQQNWKNNSKSGNPMFMDPSNPTWVPRKNKSN 273
Query: 119 SNGN--AAEKDIVSCGKEATKKDNNQSHSPSSRIIEGSAKSTPKSRNLRERPSTDKRSER 176
SN + A+ K ++ + A N S P++ SA+ TP+++N P T R +
Sbjct: 274 SNNDNAASVKGSINRNEAAKSLTRNGSTQPNT---PSSARGTPRNKNSFFSPPTPSRLNQ 330
Query: 177 SSRKGS 182
SSRK +
Sbjct: 331 SSRKSN 336
>At1g55480 unknown protein
Length = 335
Score = 34.3 bits (77), Expect = 0.25
Identities = 21/62 (33%), Positives = 35/62 (55%), Gaps = 6/62 (9%)
Query: 477 YNKIGKFKDSKSYCENALRIFGKSKPGIPPEEIANGLIDVAAIYQSMNDLEKGLKLLKKA 536
++K GK++++ E+ L SKP PEE + +VA Y +N ++ GL L++A
Sbjct: 227 FSKNGKYEEALERFESVLG----SKP--TPEEASVASYNVACCYSKLNQVQAGLSALEEA 280
Query: 537 LK 538
LK
Sbjct: 281 LK 282
>At1g62740 TPR-repeat protein
Length = 571
Score = 33.9 bits (76), Expect = 0.33
Identities = 22/69 (31%), Positives = 36/69 (51%), Gaps = 1/69 (1%)
Query: 393 LIYDSKGDYEAALEHYVLA-SMAMSANGQEQDVASVDSSIGDAYLALARYDEAVFSYQKA 451
+++ ++ A+L HY A S A + D S +G A+L L ++DEAV +Y K
Sbjct: 37 VLFSNRSAAHASLNHYDEALSDAKKTVELKPDWGKGYSRLGAAHLGLNQFDEAVEAYSKG 96
Query: 452 LTVFKSTKG 460
L + S +G
Sbjct: 97 LEIDPSNEG 105
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,440,621
Number of Sequences: 26719
Number of extensions: 660460
Number of successful extensions: 2241
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 2090
Number of HSP's gapped (non-prelim): 119
length of query: 721
length of database: 11,318,596
effective HSP length: 106
effective length of query: 615
effective length of database: 8,486,382
effective search space: 5219124930
effective search space used: 5219124930
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0400a.8


