

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.7
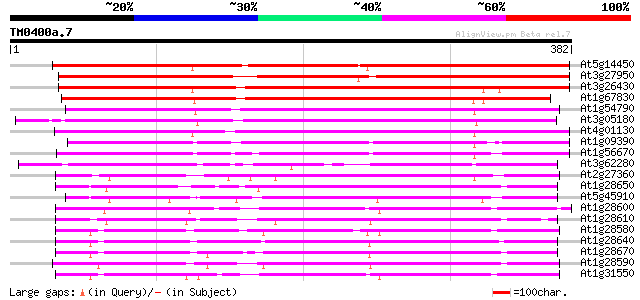
(382 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g14450 early nodule-specific protein - like 370 e-103
At3g27950 early nodule-specific protein, putative 357 8e-99
At3g26430 nodulin, putative 310 6e-85
At1g67830 ENOD8-like protein 298 4e-81
At1g54790 273 1e-73
At3g05180 putative nodulin 267 8e-72
At4g01130 putative acetyltransferase 239 1e-63
At1g09390 putative lipase 234 5e-62
At1g56670 lipase homolog (Lip-4) 231 4e-61
At3g62280 putative protein 198 5e-51
At2g27360 putative lipase 156 2e-38
At1g28650 lipase, putative 152 2e-37
At5g45910 GDSL-motif lipase/hydrolase-like protein 147 7e-36
At1g28600 lipase like protein 147 7e-36
At1g28610 lipase, putative 146 2e-35
At1g28580 lipase like protein 144 6e-35
At1g28640 lipase, putative 141 7e-34
At1g28670 lipase 140 9e-34
At1g28590 putative lipase (At1g28590) 140 1e-33
At1g31550 unknown protein 139 3e-33
>At5g14450 early nodule-specific protein - like
Length = 389
Score = 370 bits (949), Expect = e-103
Identities = 192/362 (53%), Positives = 240/362 (66%), Gaps = 15/362 (4%)
Query: 30 SYSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFIT 89
S S C++PA+YNFGDSNSDTG + AAF ++ P G FF +GR SDGRL IDFI
Sbjct: 30 SVSVQPTCTFPAIYNFGDSNSDTGGISAAFEPIRDPYGQGFFHRPTGRDSDGRLTIDFIA 89
Query: 90 EELEIPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPFYLGMQVAQFIQLQ 143
E L +PYLSAYLNS+GSN+RHGANFA GG++IR YG SPF L MQ+AQF Q +
Sbjct: 90 ERLGLPYLSAYLNSLGSNFRHGANFATGGSTIRRQNETIFQYGISPFSLDMQIAQFDQFK 149
Query: 144 SHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIP 203
+ L Q S + LPR E+F+KALYT DIGQNDL G S +++ +IP
Sbjct: 150 ARSALLFTQIKSRYD----REKLPRQEEFAKALYTFDIGQNDLSVGFRTMSVDQLKATIP 205
Query: 204 EMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFY--EPKKGNLDANGCVIPHNKI 261
+++ + V+ +Y G R FW+HNTGP GCLP ++FY P G LD +GCV N++
Sbjct: 206 DIVNHLASAVRNIYQQGGRTFWVHNTGPFGCLPV-NMFYMGTPAPGYLDKSGCVKAQNEM 264
Query: 262 AQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGY- 320
A EFNR+LK+ V LR+ L +A TYVDVYTAKYE++SN K GF NPL+VCCG + Y
Sbjct: 265 AMEFNRKLKETVINLRKELTQAAITYVDVYTAKYEMMSNPKKLGFANPLKVCCGYHEKYD 324
Query: 321 RIDCGKKAVVNGT-VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQ 379
I CG K VN T +YG C NP +SWDGVHYT+AANK VA +G L+DPPVPI +
Sbjct: 325 HIWCGNKGKVNNTEIYGGSCPNPVMAVSWDGVHYTEAANKHVADRTLNGLLTDPPVPITR 384
Query: 380 AC 381
AC
Sbjct: 385 AC 386
>At3g27950 early nodule-specific protein, putative
Length = 361
Score = 357 bits (915), Expect = 8e-99
Identities = 180/352 (51%), Positives = 236/352 (66%), Gaps = 24/352 (6%)
Query: 34 SSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEELE 93
SS C++PAV+NFGDSNSDTG + AA + P G++FFG +GR SDGRLIIDFITE L
Sbjct: 25 SSSCNFPAVFNFGDSNSDTGAISAAIGEVPPPNGVAFFGRSAGRHSDGRLIIDFITENLT 84
Query: 94 IPYLSAYLNSIGSNYRHGANFAAGGASIRPVYG-FSPFYLGMQVAQFIQLQSHIENLLNQ 152
+PYL+ YL+S+G+NYRHGANFA GG+ IRP FSPF+LG QV+QFI ++ +L NQ
Sbjct: 85 LPYLTPYLDSVGANYRHGANFATGGSCIRPTLACFSPFHLGTQVSQFIHFKTRTLSLYNQ 144
Query: 153 FSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYD 212
DFSKALYT+DIGQNDL G + +EE++ +IP ++ NFT
Sbjct: 145 ----------------TNDFSKALYTLDIGQNDLAIGFQNMTEEQLKATIPLIIENFTIA 188
Query: 213 VQVLYDVGARVFWIHNTGPIGCLP--TSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLK 270
+++LY GAR F IHNTGP GCLP + P+ D GC+ P N +A EFN+QLK
Sbjct: 189 LKLLYKEGARFFSIHNTGPTGCLPYLLKAFPAIPR----DPYGCLKPLNNVAIEFNKQLK 244
Query: 271 DQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRIDCGKKAVV 330
+++ QL++ LP + FTYVDVY+AKY LI+ A GF++P + CC G + CGK +
Sbjct: 245 NKITQLKKELPSSFFTYVDVYSAKYNLITKAKALGFIDPFDYCCVGAIGRGMGCGKTIFL 304
Query: 331 NGT-VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
NGT +Y + C+N ISWDG+HYT+ AN VA I DGS+SDPP+P +AC
Sbjct: 305 NGTELYSSSCQNRKNFISWDGIHYTETANMLVANRILDGSISDPPLPTQKAC 356
>At3g26430 nodulin, putative
Length = 380
Score = 310 bits (795), Expect = 6e-85
Identities = 160/361 (44%), Positives = 223/361 (61%), Gaps = 19/361 (5%)
Query: 34 SSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEELE 93
S C++PA++NFGDSNSDTG + A+F P G +FF + SGR SDGRLIIDFI EEL
Sbjct: 24 SPSCNFPAIFNFGDSNSDTGGLSASFGQAPYPNGQTFFHSPSGRFSDGRLIIDFIAEELG 83
Query: 94 IPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPFYLGMQVAQFIQLQSHIE 147
+PYL+A+L+SIGSN+ HGANFA G+++RP G SP L +Q+ QF + +
Sbjct: 84 LPYLNAFLDSIGSNFSHGANFATAGSTVRPPNATIAQSGVSPISLDVQLVQFSDFITRSQ 143
Query: 148 NLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGL-MHTSEEEVLRSIPEMM 206
+ N+ FK LP+ E FS+ALYT DIGQNDL GL ++ + +++ IP++
Sbjct: 144 LIRNRGG------VFKKLLPKKEYFSQALYTFDIGQNDLTAGLKLNMTSDQIKAYIPDVH 197
Query: 207 RNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFN 266
+ ++ +Y G R FWIHNT P+GCLP + +D +GC IP N+IA+ +N
Sbjct: 198 DQLSNVIRKVYSKGGRRFWIHNTAPLGCLPYVLDRFPVPASQIDNHGCAIPRNEIARYYN 257
Query: 267 RQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYR----I 322
+LK +V +LR+ L +A FTYVD+Y+ K LI+ A K GF PL CCG Y I
Sbjct: 258 SELKRRVIELRKELSEAAFTYVDIYSIKLTLITQAKKLGFRYPLVACCGHGGKYNFNKLI 317
Query: 323 DCGKKAVVNG--TVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQA 380
CG K ++ G V C + S +SWDG+H+T+ N W+ + I DG+ SDPP+P+ A
Sbjct: 318 KCGAKVMIKGKEIVLAKSCNDVSFRVSWDGIHFTETTNSWIFQQINDGAFSDPPLPVKSA 377
Query: 381 C 381
C
Sbjct: 378 C 378
>At1g67830 ENOD8-like protein
Length = 372
Score = 298 bits (762), Expect = 4e-81
Identities = 161/346 (46%), Positives = 212/346 (60%), Gaps = 19/346 (5%)
Query: 36 KCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEELEIP 95
+C +PA++NFGDSNSDTG + AAF P G SFFG+ +GR DGRL+IDFI E L +P
Sbjct: 25 QCHFPAIFNFGDSNSDTGGLSAAFGQAGPPHGSSFFGSPAGRYCDGRLVIDFIAESLGLP 84
Query: 96 YLSAYLNSIGSNYRHGANFAAGGASIRPV------YGFSPFYLGMQVAQFIQLQSHIENL 149
YLSA+L+S+GSN+ HGANFA G+ IR + GFSPF L +Q QF + + +
Sbjct: 85 YLSAFLDSVGSNFSHGANFATAGSPIRALNSTLRQSGFSPFSLDVQFVQFYNFHNRSQTV 144
Query: 150 LNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFG-LMHTSEEEVLRSIPEMMRN 208
++ +K+ LP + FSKALYT DIGQNDL G + + E+V +PE++
Sbjct: 145 RSRGG------VYKTMLPESDSFSKALYTFDIGQNDLTAGYFANKTVEQVETEVPEIISQ 198
Query: 209 FTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQ 268
F ++ +Y G R FWIHNTGPIGCL + K + D++GCV P N +AQ+FN
Sbjct: 199 FMNAIKNIYGQGGRYFWIHNTGPIGCLAYVIERFPNKASDFDSHGCVSPLNHLAQQFNHA 258
Query: 269 LKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCC--GSYYGYR--IDC 324
LK V +LR +L +A TYVDVY+ K+EL +A GF L CC G Y Y I C
Sbjct: 259 LKQAVIELRSSLSEAAITYVDVYSLKHELFVHAQGHGFKGSLVSCCGHGGKYNYNKGIGC 318
Query: 325 GKKAVVNG-TVY-GNPCKNPSQHISWDGVHYTQAANKWVAKHIRDG 368
G K +V G VY G PC P + + WDGVH+TQAANK++ I G
Sbjct: 319 GMKKIVKGKEVYIGKPCDEPDKAVVWDGVHFTQAANKFIFDKIAPG 364
>At1g54790
Length = 383
Score = 273 bits (697), Expect = 1e-73
Identities = 145/346 (41%), Positives = 210/346 (59%), Gaps = 15/346 (4%)
Query: 39 YPAVYNFGDSNSDTG-VVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPYL 97
YPA+ NFGDSNSDTG ++ A + P G ++F SGR DGRLI+DF+ +E+++P+L
Sbjct: 30 YPAIINFGDSNSDTGNLISAGIENVNPPYGQTYFNLPSGRYCDGRLIVDFLLDEMDLPFL 89
Query: 98 SAYLNSIG-SNYRHGANFAAGGASIRPVY--GFSPFYLGMQVAQFIQLQSHIENLLNQFS 154
+ YL+S+G N++ G NFAA G++I P SPF +Q++QFI+ +S LL+
Sbjct: 90 NPYLDSLGLPNFKKGCNFAAAGSTILPANPTSVSPFSFDLQISQFIRFKSRAIELLS--- 146
Query: 155 SNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQ 214
+T ++ YLP + +SK LY IDIGQND+ + ++VL SIP ++ F ++
Sbjct: 147 --KTGRKYEKYLPPIDYYSKGLYMIDIGQNDIAGAFYSKTLDQVLASIPSILETFEAGLK 204
Query: 215 VLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVF 274
LY+ G R WIHNTGP+GCL + + LD GCV HN+ A+ FN QL
Sbjct: 205 RLYEEGGRNIWIHNTGPLGCLAQNIAKFGTDSTKLDEFGCVSSHNQAAKLFNLQLHAMSN 264
Query: 275 QLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCG-----SYYGYRIDCGKKAV 329
+ + P A TYVD+++ K LI+N S+ GF PL CCG Y RI CG+ V
Sbjct: 265 KFQAQYPDANVTYVDIFSIKSNLIANYSRFGFEKPLMACCGVGGAPLNYDSRITCGQTKV 324
Query: 330 VNG-TVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPP 374
++G +V C + S++I+WDG+HYT+AAN++V+ I G SDPP
Sbjct: 325 LDGISVTAKACNDSSEYINWDGIHYTEAANEFVSSQILTGKYSDPP 370
>At3g05180 putative nodulin
Length = 379
Score = 267 bits (682), Expect = 8e-72
Identities = 156/379 (41%), Positives = 220/379 (57%), Gaps = 20/379 (5%)
Query: 5 TLIHALWCFALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQS 64
TL H L L VA + P GS+ S+ +PAV+NFGDSNSDTG + + L
Sbjct: 3 TLFHTLLRLLLFVAISHTLSPLA-GSFRISN--DFPAVFNFGDSNSDTGELSSGLGFLPQ 59
Query: 65 PG-GISFFGN-LSGRASDGRLIIDFITEELEIPYLSAYLNSIG-SNYRHGANFAAGGASI 121
P I+FF + SGR +GRLI+DF+ E ++ PYL YL+SI YR G NFAA ++I
Sbjct: 60 PSYEITFFRSPTSGRFCNGRLIVDFLMEAIDRPYLRPYLDSISRQTYRRGCNFAAAASTI 119
Query: 122 RPVYG--FSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTI 179
+ +SPF G+QV+QFI +S + L+ Q + + YLP FS LY
Sbjct: 120 QKANAASYSPFGFGVQVSQFITFKSKVLQLIQQ------DEELQRYLPSEYFFSNGLYMF 173
Query: 180 DIGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSS 239
DIGQND+ + ++VL +P ++ F ++ LY GAR +WIHNTGP+GCL
Sbjct: 174 DIGQNDIAGAFYTKTVDQVLALVPIILDIFQDGIKRLYAEGARNYWIHNTGPLGCLAQVV 233
Query: 240 IFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELIS 299
+ K LD GCV HN+ A+ FN QL +L + P ++FTYVD+++ K +LI
Sbjct: 234 SIFGEDKSKLDEFGCVSDHNQAAKLFNLQLHGLFKKLPQQYPNSRFTYVDIFSIKSDLIL 293
Query: 300 NASKQGFVNPLEVCCGS-----YYGYRIDCGKKAVVNGTVY-GNPCKNPSQHISWDGVHY 353
N SK GF + + VCCG+ Y ++ CGK A NGT+ PC + S++++WDG+HY
Sbjct: 294 NHSKYGFDHSIMVCCGTGGPPLNYDDQVGCGKTARSNGTIITAKPCYDSSKYVNWDGIHY 353
Query: 354 TQAANKWVAKHIRDGSLSD 372
T+AAN++VA HI G S+
Sbjct: 354 TEAANRFVALHILTGKYSE 372
>At4g01130 putative acetyltransferase
Length = 382
Score = 239 bits (611), Expect = 1e-63
Identities = 130/363 (35%), Positives = 193/363 (52%), Gaps = 18/363 (4%)
Query: 31 YSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITE 90
+ SKC + A++NFGDSNSDTG +AAF P G+++F +GRASDGRLIIDF+ +
Sbjct: 24 HKGDSKCDFEAIFNFGDSNSDTGGFWAAFPAQSGPWGMTYFKKPAGRASDGRLIIDFLAK 83
Query: 91 ELEIPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPFYLGMQVAQFIQLQS 144
L +P+LS YL SIGS++RHGANFA +++ V G SPF L +Q+ Q Q +
Sbjct: 84 SLGMPFLSPYLQSIGSDFRHGANFATLASTVLLPNTSLFVSGISPFSLAIQLNQMKQFKV 143
Query: 145 HIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPE 204
+++ S+ + P LP F K+LYT IGQND L E V +P+
Sbjct: 144 NVD------ESHSLDRPGLKILPSKIVFGKSLYTFYIGQNDFTSNLASIGVERVKLYLPQ 197
Query: 205 MMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQE 264
++ ++ +Y +G R F + N P+GC P Y +LD GC+IP NK +
Sbjct: 198 VIGQIAGTIKEIYGIGGRTFLVLNLAPVGCYPAILTGYTHTDADLDKYGCLIPVNKAVKY 257
Query: 265 FNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCG-----SYYG 319
+N L + Q R L A Y+D + +L + G + ++ CCG +
Sbjct: 258 YNTLLNKTLSQTRTELKNATVIYLDTHKILLDLFQHPKSYGMKHGIKACCGYGGRPYNFN 317
Query: 320 YRIDCGK-KAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIG 378
++ CG K + N + C +P ++SWDG+H T+AAN ++ I DGS+S PP +
Sbjct: 318 QKLFCGNTKVIGNFSTTAKACHDPHNYVSWDGIHATEAANHHISMAILDGSISYPPFILN 377
Query: 379 QAC 381
C
Sbjct: 378 NLC 380
>At1g09390 putative lipase
Length = 370
Score = 234 bits (598), Expect = 5e-62
Identities = 138/350 (39%), Positives = 183/350 (51%), Gaps = 25/350 (7%)
Query: 40 PAVYNFGDSNSDTGVVYAAFA-GLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPYLS 98
P ++NFGDSNSDTG + A + P G SFF +GR SDGRL+IDF+ + L L+
Sbjct: 36 PVIFNFGDSNSDTGGLVAGLGYSIGLPNGRSFFQRSTGRLSDGRLVIDFLCQSLNTSLLN 95
Query: 99 AYLNS-IGSNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFIQLQSHIENLLNQFSSNR 157
YL+S +GS +++GANFA G+S P Y PF L +Q+ QF+ +S L +
Sbjct: 96 PYLDSLVGSKFQNGANFAIVGSSTLPRY--VPFALNIQLMQFLHFKSRALELAS------ 147
Query: 158 TEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMH-TSEEEVLRSIPEMMRNFTYDVQVL 216
P K + F ALY IDIGQND+ S V++ IP ++ +++L
Sbjct: 148 ISDPLKEMMIGESGFRNALYMIDIGQNDIADSFSKGLSYSRVVKLIPNVISEIKSAIKIL 207
Query: 217 YDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQL 276
YD G R FW+HNTGP+GCLP K D +GC+ +N A+ FN L L
Sbjct: 208 YDEGGRKFWVHNTGPLGCLPQKLSMVHSK--GFDKHGCLATYNAAAKLFNEGLDHMCRDL 265
Query: 277 RRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCG-----SYYGYRIDCGKKAVVN 331
R L +A YVD+Y KY+LI+N++ GF PL CCG Y I CG N
Sbjct: 266 RTELKEANIVYVDIYAIKYDLIANSNNYGFEKPLMACCGYGGPPYNYNVNITCG-----N 320
Query: 332 GTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
G C S+ ISWDG+HYT+ AN VA + S PP P C
Sbjct: 321 GG--SKSCDEGSRFISWDGIHYTETANAIVAMKVLSMQHSTPPTPFHFFC 368
>At1g56670 lipase homolog (Lip-4)
Length = 373
Score = 231 bits (590), Expect = 4e-61
Identities = 139/357 (38%), Positives = 192/357 (52%), Gaps = 25/357 (7%)
Query: 33 SSSKCSYPAVYNFGDSNSDTGVVYAAFA-GLQSPGGISFFGNLSGRASDGRLIIDFITEE 91
++S + P ++NFGDSNSDTG + A + P G FF +GR SDGRL+IDF+ +
Sbjct: 32 AASCTARPVIFNFGDSNSDTGGLVAGLGYPIGFPNGRLFFRRSTGRLSDGRLLIDFLCQS 91
Query: 92 LEIPYLSAYLNSIG-SNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFIQLQSHIENLL 150
L L YL+S+G + +++GANFA G+ P PF L +QV QF +S L
Sbjct: 92 LNTSLLRPYLDSLGRTRFQNGANFAIAGSPTLPKN--VPFSLNIQVKQFSHFKSRSLELA 149
Query: 151 NQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMH-TSEEEVLRSIPEMMRNF 209
+ SSN + F S F ALY IDIGQND+ S + ++ IP+++
Sbjct: 150 S--SSNSLKGMFISN----NGFKNALYMIDIGQNDIARSFARGNSYSQTVKLIPQIITEI 203
Query: 210 TYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQL 269
++ LYD G R FWIHNTGP+GCLP + K +LD +GC++ +N A FN+ L
Sbjct: 204 KSSIKRLYDEGGRRFWIHNTGPLGCLPQKLSMVKSK--DLDQHGCLVSYNSAATLFNQGL 261
Query: 270 KDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCG-----SYYGYRIDC 324
+LR L A Y+D+Y KY LI+N+++ GF +PL CCG Y +I C
Sbjct: 262 DHMCEELRTELRDATIIYIDIYAIKYSLIANSNQYGFKSPLMACCGYGGTPYNYNVKITC 321
Query: 325 GKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
G K N C+ S+ ISWDG+HYT+ AN VA + S PP P C
Sbjct: 322 GHKG-------SNVCEEGSRFISWDGIHYTETANAIVAMKVLSMHYSKPPTPFHFFC 371
>At3g62280 putative protein
Length = 343
Score = 198 bits (503), Expect = 5e-51
Identities = 129/370 (34%), Positives = 186/370 (49%), Gaps = 42/370 (11%)
Query: 7 IHALWCFALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFA-GLQSP 65
+ +L CF L + + + + SY S+ K P + NFGDSNSDTG V A + P
Sbjct: 5 VSSLQCFFLVLCLSLLVCSNSETSYKSNKK---PILINFGDSNSDTGGVLAGVGLPIGLP 61
Query: 66 GGISFFGNLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVY 125
GI+FF +GR DGRLI+DF E L++ YLS YL+S+ N++ G NFA GA+ P++
Sbjct: 62 HGITFFHRGTGRLGDGRLIVDFYCEHLKMTYLSPYLDSLSPNFKRGVNFAVSGATALPIF 121
Query: 126 GFSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQND 185
F L +Q+ QF+ ++ + L+ SS R + L F ALY IDIGQND
Sbjct: 122 SFP---LAIQIRQFVHFKNRSQELI---SSGRRD------LIDDNGFRNALYMIDIGQND 169
Query: 186 LGFGL--MHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYE 243
L L + + V+ IP M+ +Q G +HN
Sbjct: 170 LLLALYDSNLTYAPVVEKIPSMLLEIKKAIQ-----GELAIHLHN--------------- 209
Query: 244 PKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASK 303
+LD GC HN++A+ FN+ L +LR A YVD+Y+ KY+L ++
Sbjct: 210 --DSDLDPIGCFRVHNEVAKAFNKGLLSLCNELRSQFKDATLVYVDIYSIKYKLSADFKL 267
Query: 304 QGFVNPLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAK 363
GFV+PL CCG YG R + + G C++ ++ I WDGVHYT+AAN++V
Sbjct: 268 YGFVDPLMACCG--YGGRPNNYDRKATCGQPGSTICRDVTKAIVWDGVHYTEAANRFVVD 325
Query: 364 HIRDGSLSDP 373
+ S P
Sbjct: 326 AVLTNRYSYP 335
>At2g27360 putative lipase
Length = 394
Score = 156 bits (395), Expect = 2e-38
Identities = 115/370 (31%), Positives = 176/370 (47%), Gaps = 50/370 (13%)
Query: 32 SSSSKC-SYPAVYNFGDSNSDTGVVYAAFAGLQSPG----------GISFFGNLSGRASD 80
+S ++C ++ ++ +FGDS +DTG + GL SP G +FF + SGR SD
Sbjct: 23 TSQTRCRNFKSIISFGDSITDTGNLL----GLSSPNDLPESAFPPYGETFFHHPSGRFSD 78
Query: 81 GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFI 140
GRLIIDFI E L IP++ + S N+ G NFA GGA+ ++ +
Sbjct: 79 GRLIIDFIAEFLGIPHVPPFYGSKNGNFEKGVNFAVGGAT------------ALECSVLE 126
Query: 141 QLQSHIE----NLLNQFSSNRTEPPF--KSYLPRPEDFSKALYTI--DIGQNDLGFGLMH 192
+ +H +L NQ S + P+ S P D + + + +IG ND F L
Sbjct: 127 EKGTHCSQSNISLGNQLKSFKESLPYLCGSSSPDCRDMIENAFILIGEIGGNDYNFPLFD 186
Query: 193 TSE-EEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYE-PKKGNLD 250
EEV +P ++ + + L D+GAR F + P+GC YE P K +
Sbjct: 187 RKNIEEVKELVPLVITTISSAISELVDMGARTFLVPGNFPLGCSVAYLTLYETPNKEEYN 246
Query: 251 -ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN- 308
GC+ N + N QL+ ++ +LR P Y D Y L+ SK G ++
Sbjct: 247 PLTGCLTWLNDFSVYHNEQLQAELKRLRNLYPHVNIIYGDYYNTLLRLMQEPSKFGLMDR 306
Query: 309 PLEVCCG----SYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKH 364
PL CCG + + I CG K V C +PS++++WDG+H T+AA KW+++
Sbjct: 307 PLPACCGLGGPYNFTFSIKCGSKGV-------EYCSDPSKYVNWDGIHMTEAAYKWISEG 359
Query: 365 IRDGSLSDPP 374
+ G + PP
Sbjct: 360 VLTGPYAIPP 369
>At1g28650 lipase, putative
Length = 385
Score = 152 bits (385), Expect = 2e-37
Identities = 115/363 (31%), Positives = 168/363 (45%), Gaps = 39/363 (10%)
Query: 32 SSSSKCS-YPAVYNFGDSNSDTGVVYAAFAGLQS-------PGGISFFGNLSGRASDGRL 83
SS S+C Y ++ +FGDS +DTG Y + + + P G SFF SGR SDGRL
Sbjct: 27 SSESRCRRYKSIISFGDSIADTGN-YVHLSNVNNLPQAAFLPYGESFFHPPSGRYSDGRL 85
Query: 84 IIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFIQLQ 143
+IDFI E L +PY+ Y S ++ G NFA VYG + V Q I +
Sbjct: 86 VIDFIAEFLGLPYVPPYFGSQNVSFNQGINFA--------VYGATALDRAFLVKQGI--K 135
Query: 144 SHIENLLNQFSSNRTEPPFKSYLPR---------PEDFSKALYTI-DIGQNDLGFGLMH- 192
S N+ N FK LP E +L + +IG ND +
Sbjct: 136 SDFTNISLSVQLN----TFKQILPNLCASSTRDCREMLGDSLILMGEIGGNDYNYPFFEG 191
Query: 193 TSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDA- 251
S E+ +P +++ + + L D+G + F + PIGC ++ D
Sbjct: 192 KSINEIKELVPLIIKAISSAIVDLIDLGGKTFLVPGNFPIGCSTAYLTLFQTATVEHDPF 251
Query: 252 NGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN-PL 310
GC+ NK + N QLK ++ QL++ P Y D Y + Y L +K GF N PL
Sbjct: 252 TGCIPWLNKFGEHHNEQLKIELKQLQKLYPHVNIIYADYYNSLYGLFQEPAKYGFKNRPL 311
Query: 311 EVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSL 370
CCG Y GK+ NG Y C+NPS++++WDG H T+A + +A+ + +G
Sbjct: 312 AACCGVGGQYNFTIGKECGENGVSY---CQNPSEYVNWDGYHLTEATYQKMAQGLLNGRY 368
Query: 371 SDP 373
+ P
Sbjct: 369 TTP 371
>At5g45910 GDSL-motif lipase/hydrolase-like protein
Length = 372
Score = 147 bits (372), Expect = 7e-36
Identities = 110/360 (30%), Positives = 173/360 (47%), Gaps = 43/360 (11%)
Query: 39 YPAVYNFGDSNSDTGVVYAAFAGLQSPG------GISFFGNLSGRASDGRLIIDFITEEL 92
Y +++NFGDS SDTG + + SP G +FF +GR SDGRLIIDFI E
Sbjct: 28 YESIFNFGDSLSDTGN-FLLSGDVDSPNIGRLPYGQTFFNRSTGRCSDGRLIIDFIAEAS 86
Query: 93 EIPYLSAYLNSIGSN----YRHGANFAAGGASIRPVYGFSPFYLGMQVAQFIQLQSHIEN 148
+PY+ YL S+ +N ++ GANFA GA+ FS F+ ++ + ++
Sbjct: 87 GLPYIPPYLQSLRTNDSVDFKRGANFAVAGATANE---FS-FFKNRGLSVTLLTNKTLDI 142
Query: 149 LLNQF-----SSNRTEPPFKSYLPRPEDFSKALYTI-DIGQNDLGFGLM-HTSEEEVLRS 201
L+ F S +T+P + Y F K+L+ + +IG ND + L+ S + +
Sbjct: 143 QLDWFKKLKPSLCKTKPECEQY------FRKSLFLVGEIGGNDYNYPLLAFRSFKHAMDL 196
Query: 202 IPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNL--DANGCVIPHN 259
+P ++ L + GA + PIGC + G L N C +P N
Sbjct: 197 VPFVINKIMDVTSALIEEGAMTLIVPGNLPIGCSAALLERFNDNSGWLYDSRNQCYMPLN 256
Query: 260 KIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFV-NPLEVCCGSYY 318
+A+ N +LK + LR+ P AK Y D Y++ + ++ SK GF + L+ CCG
Sbjct: 257 NLAKLHNDKLKKGLAALRKKYPYAKIIYADYYSSAMQFFNSPSKYGFTGSVLKACCGGGD 316
Query: 319 GY-----RIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDP 373
G + CG+K C++PS + +WDG+H T+AA + +A + G + P
Sbjct: 317 GRYNVQPNVRCGEKG-------STTCEDPSTYANWDGIHLTEAAYRHIATGLISGRFTMP 369
>At1g28600 lipase like protein
Length = 393
Score = 147 bits (372), Expect = 7e-36
Identities = 110/373 (29%), Positives = 176/373 (46%), Gaps = 44/373 (11%)
Query: 32 SSSSKC-SYPAVYNFGDSNSDTGVVYAAFAGLQ------SPGGISFFGNLSGRASDGRLI 84
SS + C ++ ++ +FGDS +DTG + Q P G +FF + +GR+ DGR+I
Sbjct: 21 SSETPCPNFKSIISFGDSIADTGNLVGLSDRNQLPVTAFPPYGETFFHHPTGRSCDGRII 80
Query: 85 IDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASI--------RPVYGFSPFYLGMQV 136
+DFI E + +PY+ Y S N+ G NFA GA+ R + + LG+Q+
Sbjct: 81 MDFIAEFVGLPYVPPYFGSKNRNFDKGVNFAVAGATALKSSFLKKRGIQPHTNVSLGVQL 140
Query: 137 AQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTI-DIGQNDLGFGLMHTSE 195
F + + NL S R + AL + +IG ND F +
Sbjct: 141 KSF---KKSLPNLCGSPSDCR------------DMIGNALILMGEIGGNDYNFPFFNRKP 185
Query: 196 -EEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLD---- 250
+EV +P ++ + + + L +G + F + PIGC Y K N D
Sbjct: 186 VKEVEELVPFVIASISSTITELIGMGGKTFLVPGEFPIGCSVVYLTLY--KTSNKDEYDP 243
Query: 251 ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFV-NP 309
+ GC+ NK + + +LK ++ +LR+ P Y D Y + + +K GF+ P
Sbjct: 244 STGCLKWLNKFGEYHSEKLKVELNRLRKLYPHVNIIYADYYNSLLRIFKEPAKFGFMERP 303
Query: 310 LEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGS 369
CCG Y + +K G+V CK+PS+++ WDGVH T+AA KW+A I +G
Sbjct: 304 FPACCGIGGPYNFNFTRKC---GSVGVKSCKDPSKYVGWDGVHMTEAAYKWIADGILNGP 360
Query: 370 LSDPPVPIGQACL 382
++P P ++CL
Sbjct: 361 YANP--PFDRSCL 371
>At1g28610 lipase, putative
Length = 383
Score = 146 bits (368), Expect = 2e-35
Identities = 111/366 (30%), Positives = 168/366 (45%), Gaps = 50/366 (13%)
Query: 32 SSSSKC-SYPAVYNFGDSNSDTGVVYAAFAGLQS----------PGGISFFGNLSGRASD 80
SS ++C + ++ +FGDS +DTG + GL P G +FF + +GR+ +
Sbjct: 21 SSQTQCRNLESIISFGDSITDTGNL----VGLSDRNHLPVTAFLPYGETFFHHPTGRSCN 76
Query: 81 GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGA-----SIRPVYGFSPFYLGMQ 135
GR+IIDFI E L +P++ + S N+ G NFA GA SI G +Y
Sbjct: 77 GRIIIDFIAEFLGLPHVPPFYGSKNGNFEKGVNFAVAGATALETSILEKRGI--YYPHSN 134
Query: 136 VAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTI-----DIGQNDLGFGL 190
++ IQL++ E+L N S P D + +IG ND F
Sbjct: 135 ISLGIQLKTFKESLPNLCGS-------------PTDCRDMIGNAFIIMGEIGGNDFNFAF 181
Query: 191 MHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEP--KKGN 248
EV +P ++ + + L D+G R F + P+GC T Y+ K+
Sbjct: 182 FVNKTSEVKELVPLVITKISSAIVELVDMGGRTFLVPGNFPLGCSATYLTLYQTSNKEEY 241
Query: 249 LDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN 308
GC+ N ++ +N +L+ ++ +L + P Y D + A L SK GF++
Sbjct: 242 DPLTGCLTWLNDFSEYYNEKLQAELNRLSKLYPHVNIIYGDYFNALLRLYQEPSKFGFMD 301
Query: 309 -PLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRD 367
PL CCG Y KK G Y C +PS++++WDGVH T+AA KW+A D
Sbjct: 302 RPLPACCGLGGPYNFTLSKKCGSVGVKY---CSDPSKYVNWDGVHMTEAAYKWIA----D 354
Query: 368 GSLSDP 373
G L P
Sbjct: 355 GLLKGP 360
>At1g28580 lipase like protein
Length = 390
Score = 144 bits (364), Expect = 6e-35
Identities = 110/367 (29%), Positives = 173/367 (46%), Gaps = 45/367 (12%)
Query: 32 SSSSKC-SYPAVYNFGDSNSDTG----------VVYAAFAGLQSPGGISFFGNLSGRASD 80
SS +KC + ++ +FGDS +DTG + + AF P G +FF + +GR S+
Sbjct: 27 SSETKCREFKSIISFGDSIADTGNLLGLSDPKDLPHMAFP----PYGENFFHHPTGRFSN 82
Query: 81 GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLGMQVAQFI 140
GRLIIDFI E L +P + + S +N+ G NFA GGA+ +L + F
Sbjct: 83 GRLIIDFIAEFLGLPLVPPFYGSHNANFEKGVNFAVGGAT-----ALERSFLEDRGIHFP 137
Query: 141 QLQSHIENLLNQFSSNRTEPPFKSYLPRPED----FSKALYTI-DIGQNDLGFG-LMHTS 194
+ LN F + S P D AL + +IG ND + +
Sbjct: 138 YTNVSLGVQLNSFKES-----LPSICGSPSDCRDMIENALILMGEIGGNDYNYAFFVDKG 192
Query: 195 EEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFY--EPKKGNLD-- 250
EE+ +P ++ + + L +G R F + P+GC S+ Y + N++
Sbjct: 193 IEEIKELMPLVITTISSAITELIGMGGRTFLVPGEFPVGC----SVLYLTSHQTSNMEEY 248
Query: 251 --ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN 308
GC+ NK + QL+ ++ +L++ P Y D Y A + L +K GF+N
Sbjct: 249 DPLTGCLKWLNKFGENHGEQLRAELNRLQKLYPHVNIIYADYYNALFHLYQEPAKFGFMN 308
Query: 309 -PLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRD 367
PL CCG+ Y G+K GT C +PS++++WDGVH T+AA + +A+ I +
Sbjct: 309 RPLSACCGAGGPYNYTVGRKC---GTDIVESCDDPSKYVAWDGVHMTEAAYRLMAEGILN 365
Query: 368 GSLSDPP 374
G + PP
Sbjct: 366 GPYAIPP 372
>At1g28640 lipase, putative
Length = 390
Score = 141 bits (355), Expect = 7e-34
Identities = 104/359 (28%), Positives = 170/359 (46%), Gaps = 30/359 (8%)
Query: 32 SSSSKCS-YPAVYNFGDSNSDTG----------VVYAAFAGLQSPGGISFFGNLSGRASD 80
SS S+C + ++ +FGDS +DTG + +AF P G SFF SGR SD
Sbjct: 25 SSESRCRRFKSIISFGDSIADTGNYLHLSDVNHLPQSAFL----PYGESFFHPPSGRYSD 80
Query: 81 GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLGMQV-AQF 139
GRLIIDFI E L +PY+ +Y S ++ G NFA GA+ F +G + + F
Sbjct: 81 GRLIIDFIAEFLGLPYVPSYFGSQNVSFDQGINFAVYGATALD----RVFLVGKGIESDF 136
Query: 140 IQLQSHIE-NLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMH-TSEEE 197
+ ++ N+ Q N + D + +IG ND + S E
Sbjct: 137 TNVSLSVQLNIFKQILPNLCTSSSRDCREMLGD--SLILMGEIGVNDYNYPFFEGKSINE 194
Query: 198 VLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYE--PKKGNLDANGCV 255
+ + +P +++ + + L D+G + F + P+GC P ++ ++ + GC+
Sbjct: 195 IKQLVPLVIKAISSAIVDLIDLGGKTFLVPGNFPLGCYPAYLTLFQTAAEEDHDPFTGCI 254
Query: 256 IPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN-PLEVCC 314
N+ + N QLK ++ +L+ Y D Y + + L K GF N PL CC
Sbjct: 255 PRLNEFGEYHNEQLKTELKRLQELYDHVNIIYADYYNSLFRLYQEPVKYGFKNRPLAACC 314
Query: 315 GSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDP 373
G Y GK+ G + C+NPS++++WDG H T+A ++ +A+ I +G+ + P
Sbjct: 315 GVGGQYNFTIGKECGHRGV---SCCQNPSEYVNWDGYHLTEATHQKMAQVILNGTYASP 370
>At1g28670 lipase
Length = 384
Score = 140 bits (354), Expect = 9e-34
Identities = 109/364 (29%), Positives = 168/364 (45%), Gaps = 40/364 (10%)
Query: 32 SSSSKCS-YPAVYNFGDSNSDTG----------VVYAAFAGLQSPGGISFFGNLSGRASD 80
SS S+C + ++ +FGDS +DTG + +AF P G SFF SGRAS+
Sbjct: 25 SSESRCRRFKSIISFGDSIADTGNYLHLSDVNHLPQSAFL----PYGESFFHPPSGRASN 80
Query: 81 GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLG------- 133
GRLIIDFI E L +PY+ Y S ++ G NFA GA+ F LG
Sbjct: 81 GRLIIDFIAEFLGLPYVPPYFGSQNVSFEQGINFAVYGATALD----RAFLLGKGIESDF 136
Query: 134 MQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMH- 192
V+ +QL + + L N +S+ + K L + +IG ND +
Sbjct: 137 TNVSLSVQLDTFKQILPNLCASSTRD--CKEMLG-----DSLILMGEIGGNDYNYPFFEG 189
Query: 193 TSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYE--PKKGNLD 250
S E+ +P +++ + + L D+G + F + P GC ++ +K
Sbjct: 190 KSINEIKELVPLIVKAISSAIVDLIDLGGKTFLVPGGFPTGCSAAYLTLFQTVAEKDQDP 249
Query: 251 ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN-P 309
GC N+ + N QLK ++ +L++ P Y D + + Y +K GF N P
Sbjct: 250 LTGCYPLLNEFGEHHNEQLKTELKRLQKFYPHVNIIYADYHNSLYRFYQEPAKYGFKNKP 309
Query: 310 LEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGS 369
L CCG Y GK+ G Y C+NPS++++WDG H T+AA + + + I +G
Sbjct: 310 LAACCGVGGKYNFTIGKECGYEGVNY---CQNPSEYVNWDGYHLTEAAYQKMTEGILNGP 366
Query: 370 LSDP 373
+ P
Sbjct: 367 YATP 370
>At1g28590 putative lipase (At1g28590)
Length = 403
Score = 140 bits (353), Expect = 1e-33
Identities = 108/369 (29%), Positives = 173/369 (46%), Gaps = 45/369 (12%)
Query: 30 SYSSSSKC-SYPAVYNFGDSNSDTGVVYAAF------AGLQSPGGISFFGNLSGRASDGR 82
S +S ++C ++ ++ +FGDS +DTG + A P G +FF + +GR SDGR
Sbjct: 24 SVNSQTQCRNFKSIISFGDSIADTGNLLGLSDPNDLPASAFPPYGETFFHHPTGRYSDGR 83
Query: 83 LIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLG--------M 134
LIIDFI E L P + + +N++ G NFA GA+ P +L
Sbjct: 84 LIIDFIAEFLGFPLVPPFYGCQNANFKKGVNFAVAGAT-----ALEPSFLEERGIHSTIT 138
Query: 135 QVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPED----FSKALYTI-DIGQNDLGFG 189
V+ +QL+S E+L N S P D AL + +IG ND F
Sbjct: 139 NVSLSVQLRSFTESLPNLCGS-------------PSDCRDMIENALILMGEIGGNDYNFA 185
Query: 190 LMHTSE-EEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEP--KK 246
L +EV +P ++ + + L +G R F + PIG + Y+ K+
Sbjct: 186 LFQRKPVKEVEELVPFVIATISSAITELVCMGGRTFLVPGNFPIGYSASYLTLYKTSNKE 245
Query: 247 GNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGF 306
GC+ N ++ +N+QL++++ LR+ P Y D Y A L +K GF
Sbjct: 246 EYDPLTGCLKWLNDFSEYYNKQLQEELNGLRKLYPHVNIIYADYYNALLRLFQEPAKFGF 305
Query: 307 VN-PLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHI 365
+N PL CCG Y + ++ G Y C +PSQ++++DG+H T+AA + +++ +
Sbjct: 306 MNRPLPACCGVGGSYNFNFSRRCGSVGVEY---CDDPSQYVNYDGIHMTEAAYRLISEGL 362
Query: 366 RDGSLSDPP 374
G + PP
Sbjct: 363 LKGPYAIPP 371
>At1g31550 unknown protein
Length = 394
Score = 139 bits (349), Expect = 3e-33
Identities = 112/369 (30%), Positives = 171/369 (45%), Gaps = 50/369 (13%)
Query: 32 SSSSKC-SYPAVYNFGDSNSDTG----------VVYAAFAGLQSPGGISFFGNLSGRASD 80
SS S+C ++ ++ +FGDS +DTG + +AF P G +FF + +GR SD
Sbjct: 26 SSESQCRNFESIISFGDSIADTGNLLGLSDHNNLPMSAFP----PYGETFFHHPTGRFSD 81
Query: 81 GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGA-----SIRPVYGF---SPFYL 132
GRLIIDFI E L +PY+ Y S N+ G NFA A S G+ F L
Sbjct: 82 GRLIIDFIAEFLGLPYVPPYFGSTNGNFEKGVNFAVASATALESSFLEEKGYHCPHNFSL 141
Query: 133 GMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTI-DIGQNDLGFGLM 191
G+Q+ F Q + NL S R + AL + +IG ND F
Sbjct: 142 GVQLKIFKQ---SLPNLCGLPSDCR------------DMIGNALILMGEIGANDYNFPFF 186
Query: 192 HTSE-EEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLD 250
+EV +P ++ + + L +G R F + P+GC ++ N++
Sbjct: 187 QLRPLDEVKELVPLVISTISSAITELIGMGGRTFLVPGGFPLGCSVAFLTLHQ--TSNME 244
Query: 251 ----ANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGF 306
GC+ NK + + QL++++ +LR+ P Y D Y A L SK GF
Sbjct: 245 EYDPLTGCLKWLNKFGEYHSEQLQEELNRLRKLNPHVNIIYADYYNASLRLGREPSKYGF 304
Query: 307 VN-PLEVCCGSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHI 365
+N L CCG Y + + G+V C +PS++++WDG+H T+AA+K +A +
Sbjct: 305 INRHLSACCGVGGPYNFNLSRSC---GSVGVEACSDPSKYVAWDGLHMTEAAHKSMADGL 361
Query: 366 RDGSLSDPP 374
G + PP
Sbjct: 362 VKGPYAIPP 370
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,456,200
Number of Sequences: 26719
Number of extensions: 427715
Number of successful extensions: 1456
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 979
Number of HSP's gapped (non-prelim): 136
length of query: 382
length of database: 11,318,596
effective HSP length: 101
effective length of query: 281
effective length of database: 8,619,977
effective search space: 2422213537
effective search space used: 2422213537
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0400a.7


