

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0400a.6
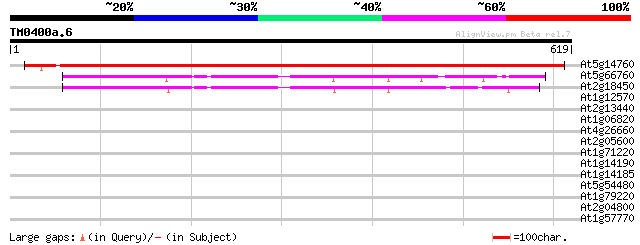
(619 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g14760 L-aspartate oxidase -like protein 907 0.0
At5g66760 succinate dehydrogenase flavoprotein alpha subunit (em... 253 2e-67
At2g18450 putative succinate dehydrogenase flavoprotein subunit 246 2e-65
At1g12570 unknown protein 33 0.36
At2g13440 similar to glucose inhibited division protein A from p... 32 1.0
At1g06820 unknown protein 32 1.4
At4g26660 putative protein 30 5.2
At2g05600 hypothetical protein 30 5.2
At1g71220 putative UDP-glucose:glycoprotein glucosyltransferase 30 5.2
At1g14190 putative mandelonitrile lyase 30 5.2
At1g14185 putative mandelonitrile lyase 30 5.2
At5g54480 unknown protein 29 6.8
At1g79220 unknown protein 29 6.8
At2g04800 unknown protein 29 8.9
At1g57770 unknown protein 29 8.9
>At5g14760 L-aspartate oxidase -like protein
Length = 651
Score = 907 bits (2345), Expect = 0.0
Identities = 456/601 (75%), Positives = 519/601 (85%), Gaps = 9/601 (1%)
Query: 17 SEGVSKVLQIHKCQFSG---TPLYKNRKLVKVISSSCKKDDPTKYFDFVVIGSGIAGLRY 73
S GV K L+ +C +P+ + K ++ +S S TKY+DF VIGSG+AGLRY
Sbjct: 44 SSGVFKALKAERCGCYSRGISPISETSKPIRAVSVSSS----TKYYDFTVIGSGVAGLRY 99
Query: 74 ALEVAKYGSVAVITKAESHECNTNYAQGGVSAVLCPSDSVENHMKDTIVAGAYLCDEESV 133
ALEVAK G+VAVITK E HE NTNYAQGGVSAVLCP DSVE+HM+DT+VAGA+LCDEE+V
Sbjct: 100 ALEVAKQGTVAVITKDEPHESNTNYAQGGVSAVLCPLDSVESHMRDTMVAGAHLCDEETV 159
Query: 134 RVVCTEGPERVRELIAMGASFDHGEDGNLHLMREGGHSHRRIVHAADMTGKEIERALLKA 193
RVVCTEGPER+RELIAMGASFDHGEDGNLHL REGGHSH RIVHAADMTG+EIERALL+A
Sbjct: 160 RVVCTEGPERIRELIAMGASFDHGEDGNLHLAREGGHSHCRIVHAADMTGREIERALLEA 219
Query: 194 AVNNPNIFVFEHHFAIDLLTSQDGSDIICLGADILNTETLEVVRFLSKVTLLASGGAGHI 253
+N+PNI VF+HHFAIDLLTSQDG + +C G D LN +T EVVRF+SKVTLLASGGAGHI
Sbjct: 220 VLNDPNISVFKHHFAIDLLTSQDGLNTVCHGVDTLNIKTNEVVRFISKVTLLASGGAGHI 279
Query: 254 YPKTTNPLVATGDGIAMAHRAQAVISNMEFVQFHPTALADEGLPIKPTKLRDNAFLITEA 313
YP TTNPLVATGDG+AMAHRAQAVISNMEFVQFHPTALADEGLPIK R+NAFLITEA
Sbjct: 280 YPSTTNPLVATGDGMAMAHRAQAVISNMEFVQFHPTALADEGLPIKLQTARENAFLITEA 339
Query: 314 VRGDGGILYNLGMERFMPLYDERAELAPRDVVARSIDDQLKKRDDKYVLLDISHKPKEEI 373
VRGDGGILYNLGMERFMP+YDERAELAPRDVVARSIDDQLKKR++KYVLLDISHKP+E+I
Sbjct: 340 VRGDGGILYNLGMERFMPVYDERAELAPRDVVARSIDDQLKKRNEKYVLLDISHKPREKI 399
Query: 374 LSHFPNIASFCLQYGLDITRHPIPVVPAAHYMCGGVRAGLEGETNVQGLYVAGEVACTGL 433
L+HFPNIAS CL++GLDITR PIPVVPAAHYMCGGVRAGL+GETNV GL+VAGEVACTGL
Sbjct: 400 LAHFPNIASECLKHGLDITRQPIPVVPAAHYMCGGVRAGLQGETNVLGLFVAGEVACTGL 459
Query: 434 HGANRLASNSLLEALVFARRAVQPSVDQMKSSSLDLTASNLWPRPIVPLS--LESDVMNK 491
HGANRLASNSLLEALVFARRAVQPS + MK + LD+ AS W RP+V + L +V+ K
Sbjct: 460 HGANRLASNSLLEALVFARRAVQPSTELMKRTRLDVCASEKWTRPVVATARLLGDEVIAK 519
Query: 492 ILSLTKELRKELQSIMWYYVGIVRSTMRLETAELKIGNLEAKWEEYLFQHGWKPTMVAPE 551
I++LTKE+R+ELQ +MW YVGIVRST+RL TAE KI LEAKWE +LF+HGW+ T+VA E
Sbjct: 520 IIALTKEVRRELQEVMWKYVGIVRSTIRLTTAERKIAELEAKWETFLFEHGWEQTVVALE 579
Query: 552 ICEMRNLFCCAKLVVSSALSRHESRGLHYTIDFPYLEESERLPTIIFPSSRVKSTRSFRQ 611
CEMRNLFCCAKLVVSSAL+RHESRGLHY DFP++EES+R+PTII PSS ++ S R+
Sbjct: 580 ACEMRNLFCCAKLVVSSALARHESRGLHYMTDFPFVEESKRIPTIILPSSPTTASWSSRR 639
Query: 612 L 612
L
Sbjct: 640 L 640
>At5g66760 succinate dehydrogenase flavoprotein alpha subunit
(emb|CAA05025.1)
Length = 634
Score = 253 bits (647), Expect = 2e-67
Identities = 195/573 (34%), Positives = 278/573 (48%), Gaps = 68/573 (11%)
Query: 59 FDFVVIGSGIAGLRYALEVAKYG-SVAVITKAESHECNTNYAQGGVSAVL--CPSDSVEN 115
+D VV+G+G AGLR A+ ++++G + A ITK +T AQGG++A L D
Sbjct: 50 YDAVVVGAGGAGLRAAIGLSEHGFNTACITKLFPTRSHTVAAQGGINAALGNMSEDDWRW 109
Query: 116 HMKDTIVAGAYLCDEESVRVVCTEGPERVRELIAMGASFDHGEDGNLHLMREGGHS---- 171
HM DT+ +L D+++++ +C E P+ V EL G F E+G ++ GG S
Sbjct: 110 HMYDTVKGSDWLGDQDAIQYMCREAPKAVIELENYGLPFSRTEEGKIYQRAFGGQSLDFG 169
Query: 172 ----HRRIVHAADMTGKEIERALLKAAVNNPNIFVFEHHFAIDLLTSQDGSDIICLGADI 227
R AAD TG + L A+ + F F +FA+DLL + DGS C G
Sbjct: 170 KGGQAYRCACAADRTGHALLHTLYGQAMKHNTQF-FVEYFALDLLMASDGS---CQGVIA 225
Query: 228 LNTETLEVVRFLSKVTLLASGGAGHIYPKTTNPLVATGDGIAMAHRAQAVISNMEFVQFH 287
LN E + RF S T+LA+GG G Y T+ TGDG AM RA + ++EFVQFH
Sbjct: 226 LNMEDGTLHRFRSSQTILATGGYGRAYFSATSAHTCTGDGNAMVARAGLPLQDLEFVQFH 285
Query: 288 PTALADEGLPIKPTKLRDNAFLITEAVRGDGGILYNLGMERFMPLYDERA-ELAPRDVVA 346
PT + G LITE RG+GGIL N ERFM Y A +LA RDVV+
Sbjct: 286 PTGIYGAGC------------LITEGSRGEGGILRNSEGERFMERYAPTAKDLASRDVVS 333
Query: 347 RSIDDQLKK-----RDDKYVLLDISHKPKEEILSHFPNIASFCLQY-GLDITRHPIPVVP 400
RS+ ++++ ++ L ++H P E + P I+ + G+D+T+ PIPV+P
Sbjct: 334 RSMTMEIREGRGVGPHKDHIYLHLNHLPPEVLKERLPGISETAAIFAGVDVTKEPIPVLP 393
Query: 401 AAHYMCGGVRAGLEGE----------TNVQGLYVAGEVACTGLHGANRLASNSLLEALVF 450
HY GG+ GE + GL AGE AC +HGANRL +NSLL+ +VF
Sbjct: 394 TVHYNMGGIPTNYHGEVVTIKGDDPDAVIPGLMAAGEAACASVHGANRLGANSLLDIVVF 453
Query: 451 AR----RAVQPSVDQMKSSSLDLTASNLWPRPIVPLSLESDVMNKILSL-TKELRKELQS 505
R R + S K L+ A ++ + N SL T +R +Q
Sbjct: 454 GRACANRVAEISKPGEKQKPLEKDAGE------KTIAWLDRLRNSNGSLPTSTIRLNMQR 507
Query: 506 IMWYYVGIVRSTMRLE-------TAELKIGNLEAKWEEYLFQHGWKPTMVAPEICEMRNL 558
IM + R+ LE A G+++ K + W ++ E E+ NL
Sbjct: 508 IMQNNAAVFRTQETLEEGCQLIDKAWESFGDVQVKDRSMI----WNSDLI--ETLELENL 561
Query: 559 FCCAKLVVSSALSRHESRGLHYTIDFPYLEESE 591
A + + SA +R ESRG H DF E+ E
Sbjct: 562 LINASITMHSAEARKESRGAHAREDFTKREDGE 594
>At2g18450 putative succinate dehydrogenase flavoprotein subunit
Length = 632
Score = 246 bits (629), Expect = 2e-65
Identities = 190/560 (33%), Positives = 273/560 (47%), Gaps = 56/560 (10%)
Query: 59 FDFVVIGSGIAGLRYALEVAKYG-SVAVITKAESHECNTNYAQGGVSAVL--CPSDSVEN 115
+D VV+G+G AGLR A+ ++++G + A ITK +T AQGG++A L D
Sbjct: 48 YDAVVVGAGGAGLRAAIGLSEHGFNTACITKLFPTRSHTVAAQGGINAALGNMSVDDWRW 107
Query: 116 HMKDTIVAGAYLCDEESVRVVCTEGPERVRELIAMGASFDHGEDGNLHLMREGGHSHR-- 173
HM DT+ +L D+++++ +C E P+ V EL G F EDG ++ GG S
Sbjct: 108 HMYDTVKGSDWLGDQDAIQYMCREAPKAVIELENYGLPFSRTEDGKIYQRAFGGQSLEFG 167
Query: 174 ------RIVHAADMTGKEIERALLKAAVNNPNIFVFEHHFAIDLLTSQDGSDIICLGADI 227
R AAD TG + L A+ + F F +FA+DL+ + DG+ C G
Sbjct: 168 IGGQAYRCACAADRTGHALLHTLYGQAMKHNTQF-FVEYFALDLIMNSDGT---CQGVIA 223
Query: 228 LNTETLEVVRFLSKVTLLASGGAGHIYPKTTNPLVATGDGIAMAHRAQAVISNMEFVQFH 287
LN E + RF + T+LA+GG G Y T+ TGDG AM RA + ++EFVQFH
Sbjct: 224 LNMEDGTLHRFHAGSTILATGGYGRAYFSATSAHTCTGDGNAMVARAGLPLQDLEFVQFH 283
Query: 288 PTALADEGLPIKPTKLRDNAFLITEAVRGDGGILYNLGMERFMPLYDERA-ELAPRDVVA 346
PT + G LITE RG+GGIL N E+FM Y A +LA RDVV+
Sbjct: 284 PTGIYGAGC------------LITEGARGEGGILRNSEGEKFMDRYAPTARDLASRDVVS 331
Query: 347 RSIDDQLKKRD-----DKYVLLDISHKPKEEILSHFPNIASFCLQY-GLDITRHPIPVVP 400
RS+ ++++ Y+ L ++H P E + P I+ + G+D+TR PIPV+P
Sbjct: 332 RSMTMEIRQGRGAGPMKDYLYLYLNHLPPEVLKERLPGISETAAIFAGVDVTREPIPVLP 391
Query: 401 AAHYMCGGVRAGLEGE----------TNVQGLYVAGEVACTGLHGANRLASNSLLEALVF 450
HY GG+ GE V GL AGE AC +HGANRL +NSLL+ +VF
Sbjct: 392 TVHYNMGGIPTNYHGEVITLRGDDPDAVVPGLMAAGEAACASVHGANRLGANSLLDIVVF 451
Query: 451 ARRAVQPSVD-QMKSSSLDLTASNLWPRPIVPLSLESDVMNKILSL-TKELRKELQSIMW 508
R + Q L + + I L + N SL T ++R +Q +M
Sbjct: 452 GRACANRVAEIQKPGEKLKPLEKDAGEKSIEWL---DRIRNSNGSLPTSKIRLNMQRVMQ 508
Query: 509 YYVGIVRSTMRLETAELKIGNLEAKWEEYLFQHGWKPTMV----APEICEMRNLFCCAKL 564
+ R+ ET E ++ W+ + +M+ E E+ NL A +
Sbjct: 509 NNAAVFRTQ---ETLEEGCDLIDKTWDSFGDVKVTDRSMIWNSDLIETMELENLLVNACI 565
Query: 565 VVSSALSRHESRGLHYTIDF 584
+ SA +R ESRG H DF
Sbjct: 566 TMHSAEARKESRGAHAREDF 585
>At1g12570 unknown protein
Length = 572
Score = 33.5 bits (75), Expect = 0.36
Identities = 14/42 (33%), Positives = 25/42 (59%)
Query: 56 TKYFDFVVIGSGIAGLRYALEVAKYGSVAVITKAESHECNTN 97
T Y+D+++IG G AG A +++ SV ++ + +S N N
Sbjct: 43 TSYYDYIIIGGGTAGCPLAATLSQNASVLLLERGDSPYNNPN 84
>At2g13440 similar to glucose inhibited division protein A from
prokaryotes
Length = 723
Score = 32.0 bits (71), Expect = 1.0
Identities = 17/51 (33%), Positives = 26/51 (50%)
Query: 36 LYKNRKLVKVISSSCKKDDPTKYFDFVVIGSGIAGLRYALEVAKYGSVAVI 86
L+ NR L SSS +D +V+G+G AG AL A+ G+ ++
Sbjct: 50 LFLNRPLAASFSSSSSGATSDSTYDVIVVGAGHAGCEAALASARLGASTLL 100
>At1g06820 unknown protein
Length = 595
Score = 31.6 bits (70), Expect = 1.4
Identities = 28/76 (36%), Positives = 38/76 (49%), Gaps = 19/76 (25%)
Query: 31 FSGTPLYKNRK---------LVKVISSSC--------KKDDPTKYFDFVVIGSGIAGLRY 73
F+G P+ +NRK VK +SSS K+D +D +VIGSGI GL
Sbjct: 34 FNG-PVLENRKKKKKLPRMVTVKSVSSSVVASTVQGTKRDGGESLYDAIVIGSGIGGLVA 92
Query: 74 ALEVA-KYGSVAVITK 88
A ++A K V V+ K
Sbjct: 93 ATQLAVKEARVLVLEK 108
>At4g26660 putative protein
Length = 806
Score = 29.6 bits (65), Expect = 5.2
Identities = 25/92 (27%), Positives = 45/92 (48%), Gaps = 14/92 (15%)
Query: 80 YGSVAVITKAESHECNTNY--------AQGGVSAVL-CPS--DSVENHMKDTIVAGAYLC 128
+G +A+ AE N + ++GG+S++L C S V+ + T AG+
Sbjct: 380 FGHMALKPCAEGENLNASVQSFRKDKASEGGLSSILLCLSCRKKVDQEAEVTEEAGS--- 436
Query: 129 DEESVRVVCTEGPERVRELIAMGASFDHGEDG 160
+E+ ++ +C E ++ EL + D GEDG
Sbjct: 437 NEKHLKNMCMEQAAKIEELTLLLRKSDDGEDG 468
>At2g05600 hypothetical protein
Length = 239
Score = 29.6 bits (65), Expect = 5.2
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 5/43 (11%)
Query: 463 KSSSLDLTASNLW-----PRPIVPLSLESDVMNKILSLTKELR 500
K S+DL +++ W RP+ PL++ D ILSLT + R
Sbjct: 148 KIFSVDLHSTSTWFGGITSRPLTPLTISKDTNKVILSLTSQER 190
>At1g71220 putative UDP-glucose:glycoprotein glucosyltransferase
Length = 1674
Score = 29.6 bits (65), Expect = 5.2
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query: 34 TPLYK-NRKLVKVISSSCKKDDPTKYFD--FVVIGSGIAGLRYALEVAKY 80
T LY +R +V +I SS +DDP KYF F+ + S +A + E A++
Sbjct: 987 TRLYSLSRLMVLLIFSSSMRDDPIKYFSDVFMFVSSAMATRDRSSESARF 1036
>At1g14190 putative mandelonitrile lyase
Length = 501
Score = 29.6 bits (65), Expect = 5.2
Identities = 15/40 (37%), Positives = 23/40 (57%)
Query: 52 KDDPTKYFDFVVIGSGIAGLRYALEVAKYGSVAVITKAES 91
K+ K FD++V+G G AG A +++ SV VI + S
Sbjct: 9 KEVSGKSFDYIVVGGGTAGCSLAATLSEKYSVLVIERGGS 48
>At1g14185 putative mandelonitrile lyase
Length = 503
Score = 29.6 bits (65), Expect = 5.2
Identities = 15/40 (37%), Positives = 23/40 (57%)
Query: 52 KDDPTKYFDFVVIGSGIAGLRYALEVAKYGSVAVITKAES 91
K+ K FD++V+G G AG A +++ SV VI + S
Sbjct: 30 KEVSGKSFDYIVVGGGTAGCSLAATLSEKYSVLVIERGGS 69
>At5g54480 unknown protein
Length = 720
Score = 29.3 bits (64), Expect = 6.8
Identities = 20/68 (29%), Positives = 32/68 (46%), Gaps = 2/68 (2%)
Query: 491 KILSLTKELRKELQSIMWYYVGIVRSTMRLETAELKIGNLEAKWEEYLFQHGWKPT--MV 548
K+LS+ +E+R Q + + V + MR E ++ + EE G PT MV
Sbjct: 598 KVLSVVEEMRLRFQGLGFKQVEEEKQRMRTERLSKELEKKTKELEEIRGTRGSSPTSNMV 657
Query: 549 APEICEMR 556
PE+ +R
Sbjct: 658 EPELLFLR 665
>At1g79220 unknown protein
Length = 399
Score = 29.3 bits (64), Expect = 6.8
Identities = 20/80 (25%), Positives = 39/80 (48%), Gaps = 6/80 (7%)
Query: 455 VQPSVDQMKS---SSLDLTASNLWPRPIVPLSLESDVMNKILSLTKELRKELQSIMWYYV 511
++P+++ K S DL A + ++P ++ N+ ++ +S M+ YV
Sbjct: 185 IKPAIEYYKGLGFSQQDLVAMLISRPTLIP---RTNFNNEKFEYIEKTGVTRESKMFKYV 241
Query: 512 GIVRSTMRLETAELKIGNLE 531
++ R+ET E K+ NLE
Sbjct: 242 AVIIGVSRMETIEEKVRNLE 261
>At2g04800 unknown protein
Length = 78
Score = 28.9 bits (63), Expect = 8.9
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query: 299 KPTKLRDNAFLITEAVRGDGGILYNLGMERFMPLYDERAELAPRDVVARSI 349
KP+ L D +I E V G+++ + PLY E EL +D V+R+I
Sbjct: 4 KPSSLADEVAIIVEQVDKVMGMVF------WCPLYWESIELLAKDEVSRAI 48
>At1g57770 unknown protein
Length = 574
Score = 28.9 bits (63), Expect = 8.9
Identities = 13/33 (39%), Positives = 19/33 (57%)
Query: 60 DFVVIGSGIAGLRYALEVAKYGSVAVITKAESH 92
D VVIGSGI GL +A+Y ++ ++ H
Sbjct: 54 DVVVIGSGIGGLCCGALLARYDQDVIVLESHDH 86
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,468,786
Number of Sequences: 26719
Number of extensions: 570333
Number of successful extensions: 1381
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1355
Number of HSP's gapped (non-prelim): 16
length of query: 619
length of database: 11,318,596
effective HSP length: 105
effective length of query: 514
effective length of database: 8,513,101
effective search space: 4375733914
effective search space used: 4375733914
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0400a.6


