

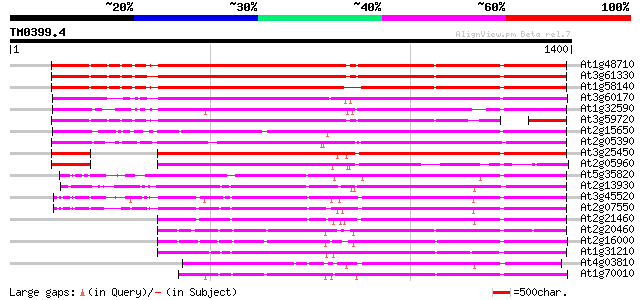
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0399.4
(1400 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g48710 hypothetical protein 1106 0.0
At3g61330 copia-type polyprotein 1098 0.0
At1g58140 hypothetical protein 1091 0.0
At3g60170 putative protein 1000 0.0
At1g32590 hypothetical protein, 5' partial 916 0.0
At3g59720 copia-type reverse transcriptase-like protein 910 0.0
At2g15650 putative retroelement pol polyprotein 906 0.0
At2g05390 putative retroelement pol polyprotein 862 0.0
At3g25450 hypothetical protein 841 0.0
At2g05960 putative retroelement pol polyprotein 670 0.0
At5g35820 copia-like retrotransposable element 643 0.0
At2g13930 putative retroelement pol polyprotein 625 e-179
At3g45520 copia-like polyprotein 616 e-176
At2g07550 putative retroelement pol polyprotein 616 e-176
At2g21460 putative retroelement pol polyprotein 590 e-168
At2g20460 putative retroelement pol polyprotein 568 e-162
At2g16000 putative retroelement pol polyprotein 568 e-162
At1g31210 putative reverse transcriptase 566 e-161
At4g03810 putative retrotransposon protein 555 e-158
At1g70010 hypothetical protein 528 e-150
>At1g48710 hypothetical protein
Length = 1352
Score = 1106 bits (2860), Expect = 0.0
Identities = 584/1292 (45%), Positives = 808/1292 (62%), Gaps = 61/1292 (4%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
A+++KKV+LQT+R +FE LQM+ E ++D+F+R++++TN +K GEK+ D I+EKVLR+
Sbjct: 105 ADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRS 164
Query: 165 LTPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQ-TSKK 223
L KF+H+V IEE+K LE + IE+L GSL+A+E E+ K +Q L Q T ++
Sbjct: 165 LDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYE----EKKKKKEDIIEQVLNMQITKEE 220
Query: 224 GGFS----GKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRK 279
G S G G RG+G+ Y N + ++++ +Q +S RG G + K +
Sbjct: 221 NGQSYQRRGGGQVRGRGRG-GYGNG---RGWRPHEDNTNQRGENSSRGRGKGHPKS---R 273
Query: 280 FDRKKIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEG 339
+D+ ++C+NC K GH++SECKAPS + ++AN +E EE + LM ++
Sbjct: 274 YDKSSVKCYNCGKFGHYASECKAPSNK----KFEEKANYVEEK--IQEEDMLLMASYKKD 327
Query: 340 ESNPATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVR 399
E +E H WYLDSG SNHM G K LDE+ + V
Sbjct: 328 EQ--------EENH---------------KWYLDSGASNHMCGRKSMFAELDESVRGNVA 364
Query: 400 FADDRFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHL 459
D+ + +G G++L++ ++G IS V Y+P MKTN++S+GQLLEK + + +K +L
Sbjct: 365 LGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNL 424
Query: 460 EVFDPNERKIMKVPLTPNRTFQVKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLK 519
+ D I KVP++ NR F + + +QCL ++ SWLWH RFGHLNF L L
Sbjct: 425 SIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLS 484
Query: 520 SKDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIG 579
K+MV GL I P++VCE CL+ KQ + F + +R+ L++I++DVCGP + S+G
Sbjct: 485 RKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSSRAQKSLELIHTDVCGPIKPKSLG 544
Query: 580 GNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSN 639
+ YF FID++SRK WVY LK KSEVF +FK FK +K+SG IK +R+D GGE+ S
Sbjct: 545 KSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSK 604
Query: 640 EMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAV 699
E +CE+NGI ++T P +PQ NGVAER+NRT+L M RSMLK K LP EAV AV
Sbjct: 605 EFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAV 664
Query: 700 YVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIG 759
Y+LN PTKSV + P+ WSGRK V HLR+FG + H H+PD++R KLDDKSE IFIG
Sbjct: 665 YLLNRSPTKSVSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIG 724
Query: 760 Y-SSTGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRVNLDLDHDDSEGIE 818
Y +++ YKLYNP T + SR++VF+E W + D E
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDEPEPTREEPP 784
Query: 819 SAVVDVPGTSQNQNQIQVHNPRPIRTKTLPARFSDYQLIAETEFNSDGDMIHMALLADAN 878
S P TS +QI+ + RT RF Q + E N + ++ L A+
Sbjct: 785 SEEPTTPPTSPTSSQIEESSSE--RT----PRFRSIQELYEVTENQE-NLTLFCLFAECE 837
Query: 879 PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISK 938
P+ F+EAI+ KTWR AM EE+ SI++N TW+L LP I VKWV+K K N G + +
Sbjct: 838 PMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKTIGVKWVYKAKKNSKGEVER 897
Query: 939 HKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLE 998
+KARLV +G++QR G+DY EVFAPVARLETVR+I++LA+ W + Q+DVKSAFLNG LE
Sbjct: 898 YKARLVAKGYIQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLE 957
Query: 999 EEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVY 1058
EEVYI QP G+ +KG E KVL+L+KALYGLKQAPRAWN RID + + F KC EH +Y
Sbjct: 958 EEVYIEQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALY 1017
Query: 1059 VKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTG 1118
+K +L+ CLYVDDL+ TG++ S + K+ + EFEMTD+G +SY+LGIE Q
Sbjct: 1018 IK-IQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQED 1076
Query: 1119 EGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFI 1178
GI + Q Y EVLK+F + NP TP+E +KL EE VD T F+ LVG LR++
Sbjct: 1077 NGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYL 1136
Query: 1179 CHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLH 1238
+R +I Y VG+VSR+M P +H AAKRILRY+KGT N G+ + +
Sbjct: 1137 TCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHY-------STTSDYK 1189
Query: 1239 LVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQG 1298
LV Y+DSDW GD DR+ST G+VF+ G +W SKKQ V LSTCEAEY+AA S C
Sbjct: 1190 LVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVVLSTCEAEYVAATSCVCHA 1249
Query: 1299 LWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIK 1358
+W++ LL+EL L +E ++ VDNKSAI LAKNPV H RSKHI+T+YH++R+ VSK+ ++
Sbjct: 1250 IWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQ 1309
Query: 1359 LQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
L++ T Q+AD+FTKPLK + F ++ ++ V
Sbjct: 1310 LEYVKTHDQVADIFTKPLKREDFIKMRSLLGV 1341
>At3g61330 copia-type polyprotein
Length = 1352
Score = 1098 bits (2841), Expect = 0.0
Identities = 581/1292 (44%), Positives = 804/1292 (61%), Gaps = 61/1292 (4%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
A+++KKV+LQT+R +FE LQM+ E ++D+F+R++++TN +K GEK+ D I+EKVLR+
Sbjct: 105 ADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRS 164
Query: 165 LTPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQ-TSKK 223
L KF+H+V IEE+K LE + IE+L GSL+A+E E+ K +Q L Q T ++
Sbjct: 165 LDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYE----EKKKKKEDIAEQVLNMQITKEE 220
Query: 224 GGFS----GKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRK 279
G S G G RG+G+ Y N + ++++ +Q +S RG G + K +
Sbjct: 221 NGQSYQRRGGGQVRGRGRG-GYGNG---RGWRPHEDNTNQRGENSSRGRGKGHPKS---R 273
Query: 280 FDRKKIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEG 339
+D+ ++C+NC K GH++SECKAPS + ++A+ +E EE + LM ++
Sbjct: 274 YDKSSVKCYNCGKFGHYASECKAPSNK----KFEEKAHYVEEK--IQEEDMLLMASYKKD 327
Query: 340 ESNPATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVR 399
E E H WYLDSG SNHM G K LDE+ + V
Sbjct: 328 EQK--------ENH---------------KWYLDSGASNHMCGRKSMFAELDESVRGNVA 364
Query: 400 FADDRFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHL 459
D+ + +G G++L++ ++G IS V Y+P MKTN++S+GQLLEK + + +K +L
Sbjct: 365 LGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNL 424
Query: 460 EVFDPNERKIMKVPLTPNRTFQVKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLK 519
+ D I KVP++ NR F + + +QCL ++ SWLWH RFGHLNF L L
Sbjct: 425 SIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLS 484
Query: 520 SKDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIG 579
K+MV GL I P++VCE CL+ KQ + F + +R+ L++I++DVCGP + S+G
Sbjct: 485 RKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLG 544
Query: 580 GNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSN 639
+ YF FID++SRK WVY LK KSEVF +FK FK +K+SG IK +R+D GGE+ S
Sbjct: 545 KSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSK 604
Query: 640 EMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAV 699
E +CE+NGI ++T P +PQ NGV ER+NRT+L M RSMLK K LP EAV AV
Sbjct: 605 EFLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWAEAVACAV 664
Query: 700 YVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIG 759
Y+LN PTKSV + P+ WSGRKP V HLR+FG + H H+PD++R KLDDKSE IFIG
Sbjct: 665 YLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIG 724
Query: 760 Y-SSTGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRVNLDLDHDDSEGIE 818
Y +++ YKLYNP T + SR++VF+E W + D E
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDEPEPTREEPP 784
Query: 819 SAVVDVPGTSQNQNQIQVHNPRPIRTKTLPARFSDYQLIAETEFNSDGDMIHMALLADAN 878
S P TS +QI+ + RT RF Q + E N + ++ L A+
Sbjct: 785 SEEPTTPPTSPTSSQIEESSSE--RT----PRFRSIQELYEVTENQE-NLTLFCLFAECE 837
Query: 879 PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISK 938
P+ F++AI+ KTWR AM EE+ SI++N TW+L LP I VKWV+K K N G + +
Sbjct: 838 PMDFQKAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVER 897
Query: 939 HKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLE 998
+KARLV +G+ QR G+DY EVFAPVARLETVR+I++LA+ W + Q+DVKSAFLNG LE
Sbjct: 898 YKARLVAKGYSQRVGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLE 957
Query: 999 EEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVY 1058
EEVYI QP G+ +KG E KVL+L+K LYGLKQAPRAWN RID + + F KC EH +Y
Sbjct: 958 EEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALY 1017
Query: 1059 VKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTG 1118
+K +L+ CLYVDDL+ TG++ S + K+ + EFEMTD+G +SY+LGIE Q
Sbjct: 1018 IK-IQKEDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQED 1076
Query: 1119 EGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFI 1178
GI + Q Y EVLK+F + NP TP+E +KL EE VD T F+ LVG LR++
Sbjct: 1077 NGIFITQEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYL 1136
Query: 1179 CHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLH 1238
+R +I Y VG+VSR+M P +H AAKRILRY+KGT N G+ + +
Sbjct: 1137 TCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHY-------STTSDYK 1189
Query: 1239 LVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQG 1298
LV Y+DSDW GD DR+ST G+VF+ G +W SKKQ V LSTCEAEY+AA S C
Sbjct: 1190 LVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHA 1249
Query: 1299 LWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIK 1358
+W++ LL+EL L +E ++ VDNKSAI LAKNPV H RSKHI+T+YH++R+ VSK+ ++
Sbjct: 1250 IWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQ 1309
Query: 1359 LQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
L++ T Q+AD FTKPLK + F ++ ++ V
Sbjct: 1310 LEYVKTHDQVADFFTKPLKRENFIKMRSLLGV 1341
>At1g58140 hypothetical protein
Length = 1320
Score = 1091 bits (2821), Expect = 0.0
Identities = 576/1292 (44%), Positives = 793/1292 (60%), Gaps = 93/1292 (7%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
A+++KKV+LQT+R +FE LQM+ E ++D+F+R++++TN +K GEK+ D I+EKVLR+
Sbjct: 105 ADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRS 164
Query: 165 LTPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQ-TSKK 223
L KF+H+V IEE+K LE + IE+L GSL+A+E E+ K +Q L Q T ++
Sbjct: 165 LDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYE----EKKKKKEDIVEQVLNMQITKEE 220
Query: 224 GGFS----GKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRK 279
G S G G RG+G+ Y N + ++++ +Q +S RG G + K +
Sbjct: 221 NGQSYQRRGGGQVRGRGRG-GYGNG---RGWRPHEDNTNQRGENSSRGRGKGHPKS---R 273
Query: 280 FDRKKIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEG 339
+D+ ++C+NC K GH++SECKAPS + ++AN +E EE + LM ++
Sbjct: 274 YDKSSVKCYNCGKFGHYASECKAPSNK----KFEEKANYVEEK--IQEEDMLLMASYKKD 327
Query: 340 ESNPATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVR 399
E +E H WYLDSG SNHM G K LDE+ + V
Sbjct: 328 EQ--------EENH---------------KWYLDSGASNHMCGRKSMFAELDESVRGNVA 364
Query: 400 FADDRFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHL 459
D+ + +G G++L++ ++G IS V Y+P MKTN++S+GQLLEK + + +K +L
Sbjct: 365 LGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNL 424
Query: 460 EVFDPNERKIMKVPLTPNRTFQVKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLK 519
+ D I KVP++ NR F + + +QCL ++ SWLWH RFGHLNF L L
Sbjct: 425 SIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLS 484
Query: 520 SKDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIG 579
K+MV GL I P++VCE CL+ KQ + F + +R+ L++I++DVCGP + S+G
Sbjct: 485 RKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLG 544
Query: 580 GNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSN 639
+ YF FID++SRK WVY LK KSEVF +FK FK +K+SG IK +R+D GGE+ S
Sbjct: 545 KSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSK 604
Query: 640 EMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAV 699
E +CE+NGI ++T P +PQ NGVAER+NRT+L M RSMLK K LP EAV AV
Sbjct: 605 EFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAV 664
Query: 700 YVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIG 759
Y+LN PTKSV + P+ WSGRKP V HLR+FG + H H+PD++R KLDDKSE IFIG
Sbjct: 665 YLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIG 724
Query: 760 Y-SSTGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRVNLDLDHDDSEGIE 818
Y +++ YKLYNP T + SR++VF+E W + D E
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTREEPP 784
Query: 819 SAVVDVPGTSQNQNQIQVHNPRPIRTKTLPARFSDYQLIAETEFNSDGDMIHMALLADAN 878
S P TS +QI+
Sbjct: 785 SEEPTTPPTSPTSSQIE---------------------------------------EKCE 805
Query: 879 PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISK 938
P+ F+EAI+ KTWR AM EE+ SI++N TW+L LP I VKWV+K K N G + +
Sbjct: 806 PMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVER 865
Query: 939 HKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLE 998
+KARLV +G+ QR G+DY EVFAPVARLETVR+I++LA+ W + Q+DVKSAFLNG LE
Sbjct: 866 YKARLVAKGYSQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLE 925
Query: 999 EEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVY 1058
EEVYI QP G+ +KG E KVL+L+KALYGLKQAPRAWN RID + + F KC EH +Y
Sbjct: 926 EEVYIEQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALY 985
Query: 1059 VKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTG 1118
+K +L+ CLYVDDL+ TG++ S + K+ + EFEMTD+G +SY+LGIE Q
Sbjct: 986 IK-IQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQED 1044
Query: 1119 EGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFI 1178
GI + Q Y EVLK+F + NP TP+E +KL EE VD T F+ LVG LR++
Sbjct: 1045 NGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYL 1104
Query: 1179 CHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLH 1238
+R +I Y VG+VSR+M P +H AAKRILRY+KGT N G+ + +
Sbjct: 1105 TCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHY-------STTSDYK 1157
Query: 1239 LVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQG 1298
LV Y+DSDW GD DR+ST G+VF+ G +W SKKQ V LSTCEAEY+AA S C
Sbjct: 1158 LVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHA 1217
Query: 1299 LWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIK 1358
+W++ LL+EL L +E ++ VDNKSAI LAKNPV H RSKHI+T+YH++R+ VSK+ ++
Sbjct: 1218 IWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQ 1277
Query: 1359 LQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
L++ T Q+AD+FTKPLK + F ++ ++ V
Sbjct: 1278 LEYVKTHDQVADIFTKPLKREDFIKMRSLLGV 1309
>At3g60170 putative protein
Length = 1339
Score = 1000 bits (2586), Expect = 0.0
Identities = 550/1309 (42%), Positives = 782/1309 (59%), Gaps = 108/1309 (8%)
Query: 107 KLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLT 166
K+K+ +LQ +R++FELL M+ E I F R +++ N MK GE M IV K+LR+LT
Sbjct: 109 KVKRAQLQALRKEFELLAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLT 168
Query: 167 PKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KS--KSTDQALQAQTSKKG 224
PKF++VV +IEES L L I+EL GSL HEQR+ + K T + +Q +G
Sbjct: 169 PKFNYVVCSIEESNDLSTLSIDELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGRGRG 228
Query: 225 GFSGKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRKFDRKK 284
F G G RG+G+ + N R
Sbjct: 229 VFRGSRG-RGRGRGRSGTN--------------------------------------RAI 249
Query: 285 IRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNPA 344
+ C+ C+ +GHF EC AN A+ + EE + LM E+ ++N
Sbjct: 250 VECYKCHNLGHFQYECP---------EWEKNANYAE---LEEEEELLLMAYVEQNQAN-- 295
Query: 345 TCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDR 404
DE W+LDSGCSNHMTG+KEW L+E V+ +D
Sbjct: 296 ----RDE-----------------VWFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDT 334
Query: 405 FISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDP 464
+S G G V VK +G VI EV YVP ++ NL+S+GQL E+ ++ ++ +V+ P
Sbjct: 335 RMSVVGKGSVKVK-VNGVTQVIPEVYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHP 393
Query: 465 NERKIMKVPLTPNRTFQVKLT--AIDSQCLTAE--LEDNSWLWHQRFGHLNFKDLSSLKS 520
++ IM+ ++ NR F + + +S CL E ++ + LWH RFGHLN + L L
Sbjct: 394 SKGAIMETNMSGNRMFFLLASKPQKNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAH 453
Query: 521 KDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGG 580
K MV GL +K ++C CL KQ R S T +S+ L +++SD+CGP S G
Sbjct: 454 KKMVIGLPILKATKEICAICLTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSG 513
Query: 581 NKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNE 640
+Y SFID+++RK WVY L KSE F+ FK+FK +K+ G + LRTD GGE+ SNE
Sbjct: 514 KRYILSFIDDFTRKTWVYFLHEKSEAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNE 573
Query: 641 MSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVY 700
FC +GI ++TA +TPQ NGVAER+NRT++N VRSML + +P F EA +V+
Sbjct: 574 FGEFCRSHGISRQLTAAFTPQQNGVAERKNRTIMNAVRSMLSERQVPKMFWSEATKWSVH 633
Query: 701 VLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGY 760
+ N PT +V+ PE WSGRKP V++ R+FGC+ + HIPDQ+R KLDDKS+ +F+G
Sbjct: 634 IQNRSPTAAVEGMTPEEAWSGRKPVVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGV 693
Query: 761 S-STGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRVNLDL-DHDD---SE 815
S + A++LY+P ++ S+DVVF+E +W + V + V L+ D DD SE
Sbjct: 694 SEESKAWRLYDPVMKKIVISKDVVFDEDKSWDWDQADV---EAKEVTLECGDEDDEKNSE 750
Query: 816 GIESAVVDVPGTSQNQNQIQVH--------NPRPIRTKTL-----PARFSDYQLIAETEF 862
+E V P + N + P P+ K P +DY+ E
Sbjct: 751 VVEPIAVASPNHVGSDNNVSSSPILAPSSPAPSPVAAKVTRERRPPGWMADYETGEGEEI 810
Query: 863 NSDGDMIHMALLADANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISV 922
+ ++ + ++ +A+P++F++A+K+K WR AM+ E+ SI +N TW+L LP TPI V
Sbjct: 811 EENLSVMLLMMMTEADPIQFDDAVKDKIWREAMEHEIESIVKNNTWELTTLPKGFTPIGV 870
Query: 923 KWVFKVKLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWD 982
KWV+K KLN DG + K+KARLV +G+ Q G+DY+EVFAPVARL+TVR I+A++S NW+
Sbjct: 871 KWVYKTKLNEDGEVDKYKARLVAKGYAQCYGIDYTEVFAPVARLDTVRTILAISSQFNWE 930
Query: 983 LWQLDVKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTF 1042
++QLDVKSAFL+G L+EEVY+ QP GF +G E KV KLRKALYGLKQAPRAW RI+ +
Sbjct: 931 IFQLDVKSAFLHGELKEEVYVRQPEGFIREGEEEKVYKLRKALYGLKQAPRAWYSRIEAY 990
Query: 1043 LSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTD 1102
+ F +C EH ++ K+ G +L++ LYVDDL+ TGS + K+S+ EFEM+D
Sbjct: 991 FLKEEFERCPSEHTLFTKT-RVGNILIVSLYVDDLIFTGSDKAMCDEFKKSMMLEFEMSD 1049
Query: 1103 LGKLSYFLGIEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAE 1162
LGK+ +FLGIE Q+ GI + QR+Y EVL RF + N + P+ KL E +
Sbjct: 1050 LGKMKHFLGIEVKQSDGGIFICQRRYAREVLARFGMDESNAVKNPIVPGTKLTKDENGEK 1109
Query: 1163 VDSTMFRQLVGCLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGV 1222
VD TMF+QLVG L ++ +R ++ YGV L+SRFMS PR SH AAKRILRYLKGT G+
Sbjct: 1110 VDETMFKQLVGSLMYLTVTRPDLMYGVCLISRFMSNPRMSHWLAAKRILRYLKGTVELGI 1169
Query: 1223 FFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALS 1282
F+ ++++ +L L+A+TDSD+ GD DRRST G+VF I W+SKKQ VALS
Sbjct: 1170 FY-----RRRKNRSLKLMAFTDSDYAGDLNDRRSTSGFVFLMASGAICWASKKQPVVALS 1224
Query: 1283 TCEAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIE 1342
T EAEYIAA ACQ +W++ +LE+LG + A + DN S I L+K+PV HG+SKHIE
Sbjct: 1225 TTEAEYIAAAFCACQCVWLRKVLEKLGAEEKSATVINCDNSSTIQLSKHPVLHGKSKHIE 1284
Query: 1343 TKYHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNVL 1391
++H+LRD V+ + +KL++C T+ Q+AD+FTKPLK ++F+ L+ ++ ++
Sbjct: 1285 VRFHYLRDLVNGDVVKLEYCPTEDQVADIFTKPLKLEQFEKLRALLGMV 1333
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 916 bits (2367), Expect = 0.0
Identities = 524/1315 (39%), Positives = 746/1315 (55%), Gaps = 151/1315 (11%)
Query: 106 EKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTL 165
++++ +LQ +RR FE+L+M+ E+I +F+R++ +TN M+ GE M D +VEK+LRTL
Sbjct: 65 DRVQSAQLQRLRRSFEVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTL 124
Query: 166 TPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQTS-KKG 224
KF +VV AIEES ++ L ++ LQ SL HEQ + ++ L+A+T +
Sbjct: 125 VEKFTYVVCAIEESNNIKELTVDGLQSSLMVHEQNLSRHDV-----EERVLKAETQWRPD 179
Query: 225 GFSGKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKG-GKRKFDRK 283
G G+GG S RG G Y+G G+ +R
Sbjct: 180 GGRGRGG-------------------------------SPSRGRGRGGYQGRGRGYVNRD 208
Query: 284 KIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNP 343
+ CF C+K+GH+ +EC + EAN + EE + LM E+
Sbjct: 209 TVECFKCHKMGHYKAECPS---------WEKEANY-----VEMEEDLLLMAHVEQ----- 249
Query: 344 ATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADD 403
I DEE W+LDSGCSNHM G +EW + LD K VR DD
Sbjct: 250 ----IGDEEKQI--------------WFLDSGCSNHMCGTREWFLELDSGFKQNVRLGDD 291
Query: 404 RFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFD 463
R ++ EG G + ++ DG+ VIS+V +VPG+K NL S+GQL +K ++ EV+
Sbjct: 292 RRMAVEGKGKLRLE-VDGRIQVISDVYFVPGLKNNLFSVGQLQQKGLRFIIEGDVCEVWH 350
Query: 464 PNERK-IMKVPLTPNRTFQVKLTAI-------DSQCLTAELEDNSWLWHQRFGHLNFKDL 515
E++ +M +T NR F V A+ +++CL + N+ +WH+RFGHLN + L
Sbjct: 351 KTEKRMVMHSTMTKNRMFVV-FAAVKKSKETEETRCLQVIGKANN-MWHKRFGHLNHQGL 408
Query: 516 SSLKSKDMVHGLSQIKLPSK--VCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPF 573
SL K+MV GL + L + VC+ CL KQ R + +ST VL ++++D+CGP
Sbjct: 409 RSLAEKEMVKGLPKFDLGEEEAVCDICLKGKQIRESIPKESAWKSTQVLQLVHTDICGPI 468
Query: 574 ETASIGGNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEG 633
AS G +Y +FID++SRK W YLL KSE F FK FK +++SG+ + LR+D G
Sbjct: 469 NPASTSGKRYILNFIDDFSRKCWTYLLSEKSETFQFFKEFKAEVERESGKKLVCLRSDRG 528
Query: 634 GEYCSNEMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGE 693
GEY S E +C+E GI ++TA YTPQ NGVAER+NR+V+NM R ML S+P +F E
Sbjct: 529 GEYNSREFDEYCKEFGIKRQLTAAYTPQQNGVAERKNRSVMNMTRCMLMEMSVPRKFWPE 588
Query: 694 AVMTAVYVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSE 753
AV AVY+LN P+K+++ PE WS KPSV+HLRIFG L + +P Q+R KLD+KS
Sbjct: 589 AVQYAVYILNRSPSKALNDITPEEKWSSWKPSVEHLRIFGSLAYALVPYQKRIKLDEKSI 648
Query: 754 TMIFIGYS-STGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRVNLDLDHD 812
+ G S + AY+LY+P T ++ SRDV F+E W+ E + + + N D +
Sbjct: 649 KCVMFGVSKESKAYRLYDPATGKILISRDVQFDEERGWEW-EDKSLEEELVWDNSDHEPA 707
Query: 813 DSEGIE---SAVVDVPGTSQNQNQI--QVHNPRP------IRTKTLPARFSDY------Q 855
EG E + D T + + + VH P +R + P DY
Sbjct: 708 GEEGPEINHNGQQDQEETEEEEETVAETVHQNLPAVGTGGVRQRQQPVWMKDYVVGNARV 767
Query: 856 LIAETEFNSDGDMIHMALLADANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPA 915
LI + E D + + +PV FEEA + + WR AM+ E+ SIE N TW+LV+LP
Sbjct: 768 LITQDE----EDEVLALFIGPDDPVCFEEAAQLEVWRKAMEAEITSIEENNTWELVELPE 823
Query: 916 NKTPISVKWVFKVKLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVAL 975
I +KW+FK K N G + K KARLV +G+ QR G+D+ EVFAPVA+ +T+R+I+ L
Sbjct: 824 EAKVIGLKWIFKTKFNEKGEVDKFKARLVAKGYHQRYGVDFYEVFAPVAKWDTIRLILGL 883
Query: 976 ASWKNWDLWQLDVKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAW 1035
A+ K W ++QLDVKSAFL+G L+E+V++ QP GFE++ KV KL+KALYGLKQAPRAW
Sbjct: 884 AAEKGWSVFQLDVKSAFLHGDLKEDVFVEQPKGFEVEEESSKVYKLKKALYGLKQAPRAW 943
Query: 1036 NKRIDTFLSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLN 1095
RI+ F + GF KC EH ++VK + L++ +YVDDL+ TGSS I+ K S+
Sbjct: 944 YSRIEEFFGKEGFEKCYCEHTLFVKK-ERSDFLVVSVYVDDLIYTGSSMEMIEGFKNSMM 1002
Query: 1096 NEFEMTDLGKLSYFLGIEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLG 1155
EF MTDLGK+ YFLG+E +Q GI ++QRKY E++K++ + CN + P+ KL
Sbjct: 1003 EEFAMTDLGKMKYFLGVEVIQDERGIFINQRKYAAEIIKKYGMEGCNSVKNPIVPGQKL- 1061
Query: 1156 LCEEEAEVDSTMFRQLVGCLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLK 1215
G VSR+M P + HL A KRILRY++
Sbjct: 1062 -------------------------------TKAGAVSRYMESPNEQHLLAVKRILRYVQ 1090
Query: 1216 GTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKK 1275
GT + G+ + + G LV + DSD+ GD DR+ST GYVF G I+W+SKK
Sbjct: 1091 GTLDLGIQY-------ERGGATELVGFVDSDYAGDVDDRKSTSGYVFMLGGGAIAWASKK 1143
Query: 1276 QAAVALSTCEAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSH 1335
Q V LST EAE+++A ACQ +W++ +LEE+G + + + DN S I L+KNPV H
Sbjct: 1144 QPIVTLSTTEAEFVSASYGACQAVWLRNVLEEIGCRQEGGTLVFCDNSSTIKLSKNPVLH 1203
Query: 1336 GRSKHIETKYHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
GRSKHI +YHFLR+ V + I+L +C T Q+AD+ TK +K + F+ L+ M V
Sbjct: 1204 GRSKHIHVRYHFLRELVKEGTIRLDYCTTTDQVADIMTKAVKREVFEELRGRMGV 1258
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 910 bits (2353), Expect = 0.0
Identities = 490/1126 (43%), Positives = 681/1126 (59%), Gaps = 80/1126 (7%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
A+++KKV+LQT+R +FE LQM+ E ++D+F+R++++TN +K GEK+ D I+EKVLR+
Sbjct: 105 ADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRS 164
Query: 165 LTPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQ-TSKK 223
L KF+H+V IEE+K LE + IE+L GSL+A+E E+ K +Q L Q T ++
Sbjct: 165 LDLKFEHIVTVIEETKDLEAMTIEQLLGSLQAYE----EKKKKKEDIVEQVLNMQITKEE 220
Query: 224 GGFS----GKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRK 279
G S G G RG+G+ Y N + ++++ +Q +S RG G + K +
Sbjct: 221 NGQSYQRRGGGQVRGRGRG-GYGNG---RGWRPHEDNTNQRGENSSRGRGKGHPKS---R 273
Query: 280 FDRKKIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEG 339
+D+ ++C+NC K GH++SECKAPS + ++AN +E EE + LM ++
Sbjct: 274 YDKSSVKCYNCGKFGHYASECKAPSNK----KFKEKANYVEEK--IQEEDMLLMASYKKD 327
Query: 340 ESNPATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVR 399
E +E H WYLDSG SNHM G K LDE+ + V
Sbjct: 328 EQ--------EENH---------------KWYLDSGASNHMCGRKSMFAELDESVRGNVA 364
Query: 400 FADDRFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHL 459
D+ + +G G++L++ ++G IS V Y+P MKTN++S+GQLLEK + + +K +L
Sbjct: 365 LGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNL 424
Query: 460 EVFDPNERKIMKVPLTPNRTFQVKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLK 519
+ D I KVP++ NR F + + +QCL ++ SWLWH RFGHLNF L L
Sbjct: 425 SIRDKESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLS 484
Query: 520 SKDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIG 579
K+MV GL I P++VCE CL+ Q + F + +R+ L++I++DVCGP + S+G
Sbjct: 485 RKEMVRGLPCINHPNQVCEGCLLGNQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLG 544
Query: 580 GNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSN 639
+ YF FID++SRK WVY LK KSEVF +FK FK +K+SG IK +R+D GGE+ S
Sbjct: 545 KSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDSGGEFTSK 604
Query: 640 EMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAV 699
E +CE+NGI ++T P +PQ NGVAER+NRT+L M RSMLK K LP EAV AV
Sbjct: 605 EFLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAV 664
Query: 700 YVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIG 759
Y+LN PTKSV + P+ WSGRKP V HLR+FG + H H+PD++R KLDDKSE IFIG
Sbjct: 665 YLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRNKLDDKSEKYIFIG 724
Query: 760 Y-SSTGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRVNLDLDHDDSEGIE 818
Y +++ YKLYNP T + SR++VF+E W + D E
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHFEEDKPEPTREEPP 784
Query: 819 SAVVDVPGTSQNQNQIQVHNPRPIRTKTLPARFSDYQLIAETEFNSDGDMIHMALLADAN 878
S P TS +QI+ + RT RF Q + E N + ++ L A+
Sbjct: 785 SEEPTTPPTSPTSSQIEESSSE--RT----PRFRSIQELYEVTENQE-NLTLFCLFAECE 837
Query: 879 PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISK 938
P+ F+EAI+ KTWR AM EE+ SI++N TW+L LP I VKWV+K K N G + +
Sbjct: 838 PMDFQEAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVER 897
Query: 939 HKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLE 998
+KARLV +G+ QR G+DY E+FAPVARLETVR+I++LA+ W + Q+DVKSAFLNG LE
Sbjct: 898 YKARLVAKGYSQRAGIDYDEIFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLE 957
Query: 999 EEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVY 1058
EEVYI QP G+ +KG E KVL+L+K LYGLKQAPRAWN RID + + F KC EH +Y
Sbjct: 958 EEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALY 1017
Query: 1059 VKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTG 1118
+K +L+ CLYVDDL+ TG++ S + K+ + EFEMTD+G +SY+LGIE Q
Sbjct: 1018 IK-IQKEDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQED 1076
Query: 1119 EGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFI 1178
GI + Q Y EVLK+F + NP+ LVG LR++
Sbjct: 1077 NGIFITQEGYAKEVLKKFKMDDSNPS--------------------------LVGSLRYL 1110
Query: 1179 CHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFF 1224
+R +I Y VG+VSR+M P +H AAKRILRY+KGT N G+ +
Sbjct: 1111 TCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHY 1156
Score = 106 bits (265), Expect = 8e-23
Identities = 47/95 (49%), Positives = 71/95 (74%)
Query: 1296 CQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKE 1355
C +W++ LL+EL L +E ++ VDNKSAI LAKNPV H RSKHI+T+YH++R+ VSK+
Sbjct: 1167 CHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKK 1226
Query: 1356 KIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
++L++ T Q+AD+FTKPLK + F ++ ++ V
Sbjct: 1227 DVQLEYVKTHDQVADIFTKPLKREDFIKMRSLLGV 1261
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 906 bits (2341), Expect = 0.0
Identities = 503/1305 (38%), Positives = 757/1305 (57%), Gaps = 92/1305 (7%)
Query: 107 KLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLT 166
+++ VKLQ++RR++E L+M N++I F +++I L + GEK T+ +++K+L +L
Sbjct: 109 QVRLVKLQSLRREYENLKMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLP 168
Query: 167 PKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQTSKKGGF 226
KFD +V +E+++ L+ L + EL G L+A E R+ R +++K+G F
Sbjct: 169 AKFDSIVSVLEQTRDLDALTMSELLGILKAQEARVTARE-------------ESTKEGAF 215
Query: 227 SGKGGYRGKGKSKDYKNSS*KQSQ----FQNQEDHDQPESSSRRGGGSSNYKGGKRKFDR 282
+ R G +D N+ Q + F H + E + N GK K R
Sbjct: 216 YVRSKGRESGFKQDNTNNRVNQDKKWCGFHKSSKHTEEECREK----PKNDDHGKNK--R 269
Query: 283 KKIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESN 342
I+C+ C KIGH+++EC R + + A++ E NE+ +
Sbjct: 270 SNIKCYKCGKIGHYANEC--------RSKNKERAHVTLEEEDVNEDHMLF---------- 311
Query: 343 PATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFAD 402
+ ++EE TL W +DSGC+NHMT + + N++++ K +R +
Sbjct: 312 ----SASEEESTTLR---------EDVWLVDSGCTNHMTKEERYFSNINKSIKVPIRVRN 358
Query: 403 DRFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVF 462
+ G GD+ V GK + I V VPG++ NL+S+ Q++ + + + + +
Sbjct: 359 GDIVMTAGKGDITVMTRHGKRI-IKNVFLVPGLEKNLLSVPQIISSGYWVRFQDKRCIIQ 417
Query: 463 DPNERKIMKVPLTPNRTFQVKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKD 522
D N ++IM + +T +++F++KL++++ + +TA ++ WH+R GH++ K L ++ K+
Sbjct: 418 DANGKEIMNIEMT-DKSFKIKLSSVEEEAMTANVQTEE-TWHKRLGHVSNKRLQQMQDKE 475
Query: 523 MVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNK 582
+V+GL + K+ + C+ C + KQ R F + T++ L+++++DVCGP + SI G++
Sbjct: 476 LVNGLPRFKVTKETCKACNLGKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSR 535
Query: 583 YFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMS 642
Y+ F+D+Y+ WVY LK KSE F+ FK FK + +KQS SIK LR M
Sbjct: 536 YYVLFLDDYTHMCWVYFLKQKSETFATFKKFKALVEKQSNCSIKTLRP----------ME 585
Query: 643 NFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVL 702
FCE+ GI +VT PY+PQ NG AER+NR+++ M RSML + LP + EAV T+ Y+
Sbjct: 586 VFCEDEGINRQVTLPYSPQQNGAAERKNRSLVEMARSMLVEQDLPLKLWAEAVYTSAYLQ 645
Query: 703 NLCPTKSVDSQV-PEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS 761
N P+K+++ V P W G KP+V HLRIFG +C+ HIPDQ+RRKLD K++ I IGYS
Sbjct: 646 NRLPSKAIEDDVTPMEKWCGHKPNVSHLRIFGSICYVHIPDQKRRKLDAKAKCGILIGYS 705
Query: 762 S-TGAYKLYNPRTSQVEFSRDVVFEEHSAW--------KGKETMVVNDSMQ-RVNLDLDH 811
+ T Y+++ +VE SRDVVF+E W K M +ND + R +
Sbjct: 706 NQTKGYRVFLLEDEKVEVSRDVVFQEDKKWDWDKQEEVKKTFVMSINDIQESRDQQETSS 765
Query: 812 DDSEGIESAVVDVPGTSQNQNQIQVHNPRPIRTKTLPARFSDYQLIAETEFNSDGDM--- 868
D I+ + G + + QV++ T P ++ + I E + D
Sbjct: 766 HDLSQIDDHANNGEGETSSHVLSQVNDQEERETSESPKKYKSMKEILEKAPRMENDEAAQ 825
Query: 869 -IHMALLADANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFK 927
I L+A+ P ++EA +K W AM EE+ IE+N+TW LVD P K ISVKW++K
Sbjct: 826 GIEACLVANEEPQTYDEARGDKEWEEAMNEEIKVIEKNRTWKLVDKPEKKNVISVKWIYK 885
Query: 928 VKLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLD 987
+K + G+ KHKARLV RGF Q G+DY E FAPV+R +T+R ++A A+ W L+Q+D
Sbjct: 886 IKTDASGNHVKHKARLVARGFSQEYGIDYLETFAPVSRYDTIRALLAYAAQMKWRLYQMD 945
Query: 988 VKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTG 1047
VKSAFLNG LEEEVY+TQPPGF I+G E KVL+L KALYGLKQAPRAW +RID++ Q G
Sbjct: 946 VKSAFLNGELEEEVYVTQPPGFVIEGKEEKVLRLYKALYGLKQAPRAWYERIDSYFIQNG 1005
Query: 1048 FHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLS 1107
F + + +Y K VL++ LYVDDL+ITG++ I K+++ +EFEMTDLG L+
Sbjct: 1006 FARSMNDAALYSKK-KGEDVLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLN 1064
Query: 1108 YFLGIEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPV--EGNLKLGLCEEEAEVDS 1165
YFLG+E Q GI + Q KY +++ +F + TP+ +G K +++ D
Sbjct: 1065 YFLGMEVNQDDSGIFLSQEKYANKLIDKFGMKESKSVSTPLTPQGKRKGVEGDDKEFADP 1124
Query: 1166 TMFRQLVGCLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFP 1225
T +R++VG L ++C SR ++ Y +SR+MS P H AKR+LRY+KGT N GV F
Sbjct: 1125 TKYRRIVGGLLYLCASRPDVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVLF- 1183
Query: 1226 KHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCE 1285
+ KE LV Y+DSDW G D++ST GYVF G A W S KQ VA ST E
Sbjct: 1184 ----TSKE--TPRLVGYSDSDWGGSLEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAE 1237
Query: 1286 AEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKY 1345
AEYIA C+A Q +W+Q L E+ GLK E + ++ DNKSAI + +NPV H R+KHIE KY
Sbjct: 1238 AEYIAVCAATNQAIWLQRLFEDFGLKFKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIKY 1297
Query: 1346 HFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
HF+R+ K I+L++C + Q+ADV TK L RF+ L++ + V
Sbjct: 1298 HFVREAEHKGLIQLEYCKGEDQLADVLTKALSVSRFEGLRRKLGV 1342
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 862 bits (2226), Expect = 0.0
Identities = 476/1304 (36%), Positives = 746/1304 (56%), Gaps = 100/1304 (7%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
AE++K+ KLQT+ +F+ L M+ NE+I +F RI ++ ++ GE++ + IV+K L++
Sbjct: 63 AERVKEAKLQTLMAEFDRLNMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKS 122
Query: 165 LT-PKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQTSKK 223
L K+ H++ A+E+ L E++ G ++ +E R+ + +Q + +
Sbjct: 123 LPRKKYIHIIAALEQILDLNTTGFEDIVGRMKTYEDRVCDEDD---SPEEQGKLMYANSE 179
Query: 224 GGFSGKGGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRKFDRK 283
+ +GG RG+G+ + SS RG G Y+ + D+
Sbjct: 180 SSYDTRGG-RGRGRGR-----------------------SSGRGRGGYGYQ----QRDKS 211
Query: 284 KIRCFNCNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNP 343
K+ C+ C+K GH++SEC + + + N +++ + +M+ E N
Sbjct: 212 KVICYRCDKTGHYASECLDRLLKLIKAQEQQQNN-------EDDDEIESLMMHEVVYLNE 264
Query: 344 ATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADD 403
+ + E + +WYLD+G SNHMTGN +W L+E +VRF DD
Sbjct: 265 RSVKPKEFEACS-----------DNSWYLDNGASNHMTGNLQWFSKLNEMITGKVRFGDD 313
Query: 404 RFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFD 463
I +G G +++ + G +++V ++P +K+N+IS+GQ E + MK L + D
Sbjct: 314 SRIDIKGKGSIVLITKGGIRKTLTDVYFIPDLKSNIISLGQATEAGCDVRMKDDQLTLHD 373
Query: 464 PNERKIMKVPLTPNRTFQVKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDM 523
+++ + NR ++V L + +CL E +++ K++
Sbjct: 374 REGCLLLRATRSRNRLYKVDLNVENVKCLQLEA-------------------ATMVRKEL 414
Query: 524 VHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKY 583
V G+S I + C +CL+ KQ R PF T R++ VL++++ D+CGP ++ +Y
Sbjct: 415 VIGISNIPKEKETCGSCLLGKQARQPFPKATTYRASQVLELVHGDLCGPITQSTTAKKRY 474
Query: 584 FASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSN 643
ID+++R MW LLK KSE F F+ FKT +++SG IK RTD+GGE+ S E +
Sbjct: 475 ILVLIDDHTRYMWSMLLKEKSEAFEKFRDFKTKVEQESGVKIKTFRTDKGGEFVSQEFQD 534
Query: 644 FCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLN 703
FC + GI +TAPYTPQ NGV ERRNRT+L M RS+LK +P+ GEAV + Y++N
Sbjct: 535 FCAKEGINRHLTAPYTPQQNGVVERRNRTLLGMTRSILKHMKMPNYLWGEAVRHSTYIIN 594
Query: 704 LCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS-S 762
T+S+ +Q P V+ RKP+V+HLR+FGC+ + I RKLDD+S+ ++++G
Sbjct: 595 RVGTRSLQNQTPYEVFKQRKPNVEHLRVFGCIGYAKIEGPHLRKLDDRSKMLVYLGTEPG 654
Query: 763 TGAYKLYNPRTSQV-------EFSRDV---------VFEEHSAWKGKETMVVNDSMQRVN 806
+ AY+L +P ++ +RD+ F + + + + + N
Sbjct: 655 SKAYRLLDPTNRKIIKWNNSDSETRDISGTFSLTLGEFGNNGIQESDDIETEKNGEESEN 714
Query: 807 LDLDHDDSEGIESAVVDVPGTSQNQNQIQVHNPRPIRTKTLPARFSDYQLIAETEFNSDG 866
+ ++E E +D T + R R P DY L+AE E G
Sbjct: 715 SHEEEGENEHNEQEQIDAEETQPSHATPLPTLRRSTRQVGKPNYLDDYVLMAEIE----G 770
Query: 867 DMIHMALLADANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVF 926
+ + +A+ + P F+EA K K WR A KEE+ SIE+NKTW L+DLP + I +KWVF
Sbjct: 771 EQVLLAI--NDEPWDFKEANKLKEWRDACKEEILSIEKNKTWSLIDLPVRRKVIGLKWVF 828
Query: 927 KVKLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQL 986
K+K N DGSI+K+KARLV +G++QR G+DY EVFA VAR+ET+R+I+ALA+ W++ L
Sbjct: 829 KIKRNSDGSINKYKARLVAKGYVQRHGIDYDEVFAHVARIETIRVIIALAASNGWEVHHL 888
Query: 987 DVKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQT 1046
DVK+AFL+G L E+VY+TQP GF K +E KV KL KALYGLKQAPRAWN +++ L +
Sbjct: 889 DVKTAFLHGELREDVYVTQPEGFTNKDNEGKVYKLHKALYGLKQAPRAWNTKLNKILQEL 948
Query: 1047 GFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKL 1106
F KCS E VY + + +L++ +YVDDLL+TGSS I K+ + +FEM+DLG+L
Sbjct: 949 NFVKCSKEPSVYRRQ-EEKKLLIVAIYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQL 1007
Query: 1107 SYFLGIEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDST 1166
+Y+LGIE + GI++ Q +Y +++++ + +CNP P+ L+L +EE +
Sbjct: 1008 TYYLGIEVLHRKNGIILRQERYAMKIIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITER 1067
Query: 1167 MFRQLVGCLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPK 1226
+R+++GCLR+I H+R ++SY VG++SR++ PR+SH A K++LRYLKGT +HG++ +
Sbjct: 1068 DYRRMIGCLRYIVHTRPDLSYCVGVLSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKR 1127
Query: 1227 HLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEA 1286
S LV Y+DS D D +ST G++F+ + PI+W S+KQ VALS+CEA
Sbjct: 1128 GFKS-------GLVGYSDSSHSADLDDGKSTAGHIFYLHQCPITWCSQKQQVVALSSCEA 1180
Query: 1287 EYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYH 1346
E++AA AA Q +W+Q L E+ T E V + VDNKSAI L KN V HGRSKHI +YH
Sbjct: 1181 EFMAATEAAKQAIWLQDLFAEVCGTTSEKVMIRVDNKSAIALTKNLVFHGRSKHIHRRYH 1240
Query: 1347 FLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
F+R+ V +++ H Q AD+ TKPL +F+ +++++ V
Sbjct: 1241 FIRECVENNLVEVDHVPGVEQRADILTKPLGRIKFREMRELVGV 1284
>At3g25450 hypothetical protein
Length = 1343
Score = 841 bits (2172), Expect = 0.0
Identities = 433/1046 (41%), Positives = 651/1046 (61%), Gaps = 39/1046 (3%)
Query: 370 WYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDGKDVVISEV 429
WYLD+G SNHMTGN+ W LDE +VRF DD I+ +G G + + G+ ++ +V
Sbjct: 292 WYLDNGASNHMTGNRAWFCKLDEMITGKVRFGDDSCINIKGKGSIPFISKGGERKILFDV 351
Query: 430 LYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQVKLTAIDS 489
Y+P +K+N++S+GQ E + M++ +L + D ++K + NR ++V L +S
Sbjct: 352 YYIPDLKSNILSLGQATESGCDIRMREDYLTLHDREGNLLIKAQRSRNRLYKVSLEVENS 411
Query: 490 QCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQ-IKLPSKVCENCLVSKQPRA 548
+CL + S +WH R GH++F+ + ++ K++V G+S + + C +CL KQ R
Sbjct: 412 KCLQLTTTNESTIWHARLGHISFETIKAMIKKELVIGISSSVPQEKETCGSCLFGKQARH 471
Query: 549 PFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKTKSEVFS 608
F T R+ VL++I+ D+CGP ++ +Y ID++SR MW LLK KSE F
Sbjct: 472 SFPKATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLKEKSEAFG 531
Query: 609 VFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNGVAER 668
FK FK + +++ G IK RTD GGE+ S+E FC + GI +TAPYTPQ NGV ER
Sbjct: 532 KFKEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGINRHLTAPYTPQQNGVVER 591
Query: 669 RNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGRKPSVKH 728
RNRT+L M RS+LK ++P+ GEAV + Y++N T+S+ +Q P V+ +KP+V+H
Sbjct: 592 RNRTLLGMTRSILKHMNMPNYLWGEAVRHSTYLINRVGTRSLSNQTPYEVFKHKKPNVEH 651
Query: 729 LRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS-STGAYKLYNPRTSQVEFSRDVVFEEH 787
LR+FGC+ + + +KLDD+S ++++G + AY+L +P ++ SRDVVF+E+
Sbjct: 652 LRVFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRIFVSRDVVFDEN 711
Query: 788 SAWKGKETMVVNDSMQRV-NLDLDHDDSEGI----------ESAVVDVPGTSQN---QNQ 833
+W +E+ D + L + G+ E+ ++ G +N + +
Sbjct: 712 RSWMWQESSSETDKESGTFTITLSEFGNNGVTENDISTEPEETEEAEINGEDENIIEEAE 771
Query: 834 IQVHN-----PRPIRTKTL----PARFSDYQLIAETEFNSDGDMIHMALLADANPVKFEE 884
+ H+ P+P+R P DY L AE E H+ L + P F+E
Sbjct: 772 TEEHDQSQEEPQPVRRSQRQVIRPNYLKDYVLCAEIEAE------HLLLAVNDEPWDFKE 825
Query: 885 AIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISKHKARLV 944
A K+K WR A KEE+ SIE+N+TW LVDLP I VKWVFK+K N DGSI+K+KARLV
Sbjct: 826 ANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAIGVKWVFKLKHNSDGSINKYKARLV 885
Query: 945 VRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEVYIT 1004
+G++QR G+D+ EVFAPVAR+ETVR+I+ALA+ W++ LDVK+AFL+G L E+VY++
Sbjct: 886 AKGYVQRHGVDFEEVFAPVARIETVRLIIALAASNGWEIHHLDVKTAFLHGELREDVYVS 945
Query: 1005 QPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDS 1064
QP GF K S+ KV KL KALYGL+QAPRAWN +++ L + F KC E +Y K +
Sbjct: 946 QPEGFTNKESKEKVYKLHKALYGLRQAPRAWNTKLNEILKELKFEKCHKEPSLYRKQ-EG 1004
Query: 1065 GGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTGEGILMH 1124
+L++ +YVDDLL+TGS+ I K+ + +FEM+DLGKL+Y+LGIE +Q+ +GI +
Sbjct: 1005 ENILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYYLGIEVLQSKDGITLK 1064
Query: 1125 QRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFICHSRLE 1184
Q +Y ++L+ + CN TP+ +L+L ++E +D T +R+ +GCLR++ H+R +
Sbjct: 1065 QERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYRRNIGCLRYLLHTRPD 1124
Query: 1185 ISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYTD 1244
+SY VG++SR++ PR+SH AA K+ILRYL+GT +HG++F K+ N L+ Y+D
Sbjct: 1125 LSYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHGLYF-------KKGENAGLIGYSD 1177
Query: 1245 SDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWIQAL 1304
S D D +ST G++F+ PI+W S+KQ V LS+CEAE++AA AA Q +W+Q L
Sbjct: 1178 SSHNVDLDDGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAKQAIWLQEL 1237
Query: 1305 LEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIKLQHCGT 1364
L E+ E V + VDNKSAI L KNPV HGRSKHI +YHF+R+ V +I+++H
Sbjct: 1238 LAEVIGTECEKVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFIRECVENGQIEVEHVPG 1297
Query: 1365 DLQIADVFTKPLKADRFKTLKKMMNV 1390
Q AD+ TK L +F +++++ V
Sbjct: 1298 VRQKADILTKALGKIKFLEMRELIGV 1323
Score = 68.2 bits (165), Expect = 3e-11
Identities = 33/98 (33%), Positives = 62/98 (62%), Gaps = 1/98 (1%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
AE++K+ +LQT+ +F+ L+M+ +E+I D+ RI +T A GE + + IV+K L++
Sbjct: 99 AERVKEARLQTLMAEFDKLKMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKS 158
Query: 165 L-TPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRI 201
L K+ H+V A+E+ L+ E++ G ++ +E R+
Sbjct: 159 LPRKKYIHIVAALEQVLDLKTTTFEDIAGRIKTYEDRV 196
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 670 bits (1728), Expect = 0.0
Identities = 379/1052 (36%), Positives = 567/1052 (53%), Gaps = 152/1052 (14%)
Query: 370 WYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDGKDVVISEV 429
WYLD+G SNHMTGN++W LDE +V+F DD I G G +L ++G+ ++ V
Sbjct: 257 WYLDNGASNHMTGNRDWFCKLDEMVTGKVKFGDDSRIDIRGKGSILFLTKNGEPKTLANV 316
Query: 430 LYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQVKLTAIDS 489
Y+P +K+N+IS+GQ E + +K +L + D + ++K + NR ++V+L ++
Sbjct: 317 YYIPDLKSNIISLGQATEAGCDVRLKDNYLTLHDRDGNLLVKATRSRNRLYRVELKVKNT 376
Query: 490 QCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSKQPRAP 549
+CL ++ WH R GH+N + + ++ +K+ V G+ ++C +C++ KQ R
Sbjct: 377 KCLQLAALNDLTKWHARLGHINLETIKAMVTKEFVIGIPSAPKEKEICASCMLGKQARQV 436
Query: 550 FSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKTKSEVFSV 609
F T R++ +L++I+ D+CGP + +Y ID++SR MW +LLK KSE F
Sbjct: 437 FPKATTYRASQILELIHGDLCGPISPPTAAKRRYILVLIDDHSRFMWSFLLKEKSEAFGK 496
Query: 610 FKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNGVAERR 669
FK FK ++++G IK LRTD GGE+ S E FCEE GI+ +TAPYTPQ NGV ERR
Sbjct: 497 FKTFKATVEQETGEKIKTLRTDRGGEFLSQEFQTFCEEEGIIRHLTAPYTPQQNGVVERR 556
Query: 670 NRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGRKPSVKHL 729
NRT+L M RS++K S+P+ GEA+ A Y++N T+++ +Q P +KP+V+HL
Sbjct: 557 NRTLLGMTRSIMKHMSVPNYLWGEAIRHATYLINRVGTRALINQTPYEALKKKKPNVEHL 616
Query: 730 RIFGCLCHKHIPDQRRRKLDDKSETMIFIGYSSTG--AYKLYNPRTSQVEFSRDVVFEEH 787
R+FGC+ + + RKLD++S ++++G + TG AY L +P T ++ SRDVVF+E+
Sbjct: 617 RVFGCVSYAKVEFPHLRKLDERSRILVYLG-TETGSKAYTLLDPTTRKIIVSRDVVFDEN 675
Query: 788 SAWKGKETMVVNDSMQ-----------RVNLDLDHDDSEGIESAVVDVPGTSQNQNQI-- 834
+WK + ++ + N +++++ SE IE + ++ +
Sbjct: 676 KSWKWANSELIEIQKEPGMFTLAQTEFHNNEEVENETSEEIEENEAEDSTNKDAEDGLDE 735
Query: 835 -QVHNPR-----PIRTKT-----LPARFSDYQLIAETEFNSDGDMIHMALLADANPVKFE 883
V P P+ T++ PA +DY L+AE E + LL + P F+
Sbjct: 736 PSVPEPNHSDDVPVFTRSGRQVIKPAHLNDYVLLAEIEGE------RLLLLINDEPWDFK 789
Query: 884 EAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISKHKARL 943
EA K + WR A EE+ SI +N+TW LV LP PI +KWVFK+K N DG+I+KHKARL
Sbjct: 790 EANKYREWRDACDEEIKSIIKNRTWSLVSLPVGIKPIGLKWVFKIKRNSDGNITKHKARL 849
Query: 944 VVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEVYI 1003
V +G++Q+ +D+ EVFAPVAR+ETVR I+ALA+ W++ LDVK+AFL+G L+E VY+
Sbjct: 850 VAKGYVQKHVVDFDEVFAPVARIETVRFIIALAASNGWEVHHLDVKTAFLHGELKENVYV 909
Query: 1004 TQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCD 1063
TQP GF KGSE KV KL KALY
Sbjct: 910 TQPEGFVTKGSEEKVYKLHKALY------------------------------------- 932
Query: 1064 SGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTGEGILM 1123
+YVDDLL+TGSS I K+ ++ FEM+DLG+L+Y+LGIE Q +GI++
Sbjct: 933 --------VYVDDLLVTGSSPKLIDDFKKGMSKNFEMSDLGRLTYYLGIEVTQEEDGIIL 984
Query: 1124 HQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRFICHSRL 1183
Q +Y ++L+ + C P+ L+L +E +D +R+ +GCL
Sbjct: 985 KQERYAKKILEEAGINECKSILVPMNSGLELSKALDEKSIDGQQYRRSIGCL-------- 1036
Query: 1184 EISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYT 1243
R+M PR+SH AA K++LRYL+GT +G+ F K+ LV Y+
Sbjct: 1037 ----------RYMQDPRESHGAALKQVLRYLQGTTGYGLVF-------KKGDKTGLVGYS 1079
Query: 1244 DSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWIQA 1303
D+ D D
Sbjct: 1080 DASHSVDADD------------------------------------------------DL 1091
Query: 1304 LLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIKLQHCG 1363
L E +G E V L VDNKSAI L KNPV HGRSKHI KYHF+R+ V ++ ++H
Sbjct: 1092 LAEVIGTPC-ERVTLRVDNKSAIALTKNPVFHGRSKHIHRKYHFIRECVENGQVSVEHVP 1150
Query: 1364 TDLQIADVFTKPLKADRFKTLKKMMNVLVL*N 1395
Q A++ TK L +FK ++ ++ V L N
Sbjct: 1151 GVKQRANILTKALAKIKFKDMRDLIGVQDLSN 1182
Score = 63.5 bits (153), Expect = 8e-10
Identities = 30/98 (30%), Positives = 63/98 (63%), Gaps = 1/98 (1%)
Query: 105 AEKLKKVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRT 164
AE++K+ KL+T+ +F+ L+M+ NE+I + R+ ++ + GE + + +V+K L++
Sbjct: 57 AERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEISTKSTSLGEDIEETKVVKKFLKS 116
Query: 165 L-TPKFDHVVVAIEESKKLENLKIEELQGSLEAHEQRI 201
L T K+ H+V A+E+ L+N +++ G ++ +E +I
Sbjct: 117 LPTKKYIHIVAALEQVLDLKNTTFKDIVGRIKTYEDKI 154
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 643 bits (1658), Expect = 0.0
Identities = 423/1307 (32%), Positives = 667/1307 (50%), Gaps = 144/1307 (11%)
Query: 125 METNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLTPKFDHVVVAIEESKKLEN 184
M E++ DFF I L N+ ++ DQAIV +L +L +FD + ++ K
Sbjct: 136 MTMEENVNDFFKLISDLENVKVVVPDE--DQAIV--LLMSLPRQFDQLKETLKYCKT--T 189
Query: 185 LKIEELQGSLEAHEQRIVE--RSS*KSKSTDQALQAQTSKKGGFSGKGGYRGKGKSKDYK 242
L +EE+ ++ + +I+E S K+ L Q + GKG + K +SK
Sbjct: 190 LHLEEITSAIRS---KILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKNKSRSKS-- 244
Query: 243 NSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRKFDRKKIRCFNCNKIGHFSSECKA 302
+G G + C+ C K GHF +C
Sbjct: 245 -----------------------KGAGKT---------------CWICGKEGHFKKQCYV 266
Query: 303 PSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNPATCNITDEEHVTLMMVTKE 362
+ +G TS E GE++ T +TD + +
Sbjct: 267 WKERNKQGSTS-----------------------ERGEASTVTARVTDAAALVVSRALLG 303
Query: 363 GGNY-PGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDG 421
P TW LD+GCS HMT K+W+I+ E +VR +D + +GIGDV +K EDG
Sbjct: 304 FAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKNEDG 363
Query: 422 KDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQ 481
+++++V Y+P M NLIS+G L +K KK L +F N+ ++ F
Sbjct: 364 STILLTDVRYIPEMSKNLISLGTLEDKGCWFESKKGILTIFK-NDLTVLTGKKESTLYFL 422
Query: 482 VKLTAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCL 541
T + + +D + LWH R GH+ K L L SK H I + ++
Sbjct: 423 QGTTLAGEANVIDKEKDETSLWHSRLGHIGAKGLQVLVSKG--HLDKNIMISFGAAKHVT 480
Query: 542 VSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETA-SIGGNKYFASFIDEYSRKMWVYLL 600
K LD ++SD+ G SIG +YF +FID+++R+ W+Y +
Sbjct: 481 KDK-----------------LDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWIYFI 523
Query: 601 KTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTP 660
+TK E FS F +KT + Q + +K+L TD G E+C+ E +FC + G++ T YTP
Sbjct: 524 RTKDEAFSKFVEWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCAYTP 583
Query: 661 QHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWS 720
Q NGVAER NRT++N VR ML L +F EA TAV+++N P+ S++ +PE W+
Sbjct: 584 QQNGVAERMNRTIMNKVRCMLSESGLGKQFWAEAASTAVFLINKSPSSSIEFDIPEEKWT 643
Query: 721 GRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGY-SSTGAYKLYNPRTSQVEFS 779
G P K L+ FG + + H + KL+ +++ IF+GY +K++ + S
Sbjct: 644 GHPPDYKILKKFGSVAYIH---SDQGKLNPRAKKGIFLGYPDGVKRFKVWLLEDRKCVVS 700
Query: 780 RDVVFEEHSAWKG--KETMVVNDS----MQRVNLDL------DHDDSEGIESAVVDVPGT 827
RD+VF+E+ +K K M D ++R ++L D + SEG +++ + T
Sbjct: 701 RDIVFQENQMYKELQKNDMSEEDKQLTEVERTLIELKNLSADDENQSEGGDNSNQEQAST 760
Query: 828 SQNQNQI-QVHNPRP----IRTKTLPARFSDYQLIAETEF-NSDGDMIHMALLADAN--- 878
+++ ++ QV + L Q+ A F D ++ AL +
Sbjct: 761 TRSASKDKQVEETDSDDDCLENYLLARDRIRRQIRAPQRFVEEDDSLVGFALTMTEDGEV 820
Query: 879 --PVKFEEAIKN---KTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLN-P 932
P +EEA+++ + W+ A EE+ S+++N TWD++D P K I KW+FK K P
Sbjct: 821 YEPETYEEAMRSPECEKWKQATIEEMDSMKKNDTWDVIDKPEGKRVIGCKWIFKRKAGIP 880
Query: 933 DGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAF 992
++KARLV +GF QR G+DY E+F+PV + ++R ++++ + +L QLDVK+AF
Sbjct: 881 GVEPPRYKARLVAKGFSQREGIDYQEIFSPVVKHVSIRYLLSIVVQFDMELEQLDVKTAF 940
Query: 993 LNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCS 1052
L+G L+E + ++QP G+E + S KV L+K+LYGLKQ+PR WN+R D+F+ +G+ +
Sbjct: 941 LHGNLDEYILMSQPEGYEDEDSTEKVCLLKKSLYGLKQSPRQWNQRFDSFMINSGYQRSK 1000
Query: 1053 VEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGI 1112
VY + + G + L LYVDD+LI + +IQ +K SLN EFEM DLG LG+
Sbjct: 1001 YNPCVYTQQLNDGSYIYLLLYVDDMLIASQNKDQIQKLKESLNREFEMKDLGPARKILGM 1060
Query: 1113 EFVQTGE-GIL-MHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQ 1170
E + E GIL + Q +Y+ VL+ F + ++TP+ + KL E+ + +
Sbjct: 1061 EITRNREQGILDLSQSEYVAGVLRAFGMDQSKVSQTPLGAHFKLRAANEKTLARDAEYMK 1120
Query: 1171 LV------GCLRF-ICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVF 1223
LV G + + + SR +++Y VG+VSRFMS P + H A K ++RY+KGT + +
Sbjct: 1121 LVPYPNAIGSIMYSMIGSRPDLAYPVGVVSRFMSKPSKEHWQAVKWVMRYMKGTQDTCLR 1180
Query: 1224 FPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALST 1283
F K+D + Y DSD+ D RRS G+VF G ISW S Q VALST
Sbjct: 1181 F-------KKDDKFEIRGYCDSDYATDLDRRRSITGFVFTAGGNTISWKSGLQRVVALST 1233
Query: 1284 CEAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIET 1343
EAEY+A A + +W++ L E+G + D AV++M D++SAI L+KN V H R+KHI+
Sbjct: 1234 TEAEYMALAEAVKEAIWLRGLAAEMGFEQD-AVEVMCDSQSAIALSKNSVHHERTKHIDV 1292
Query: 1344 KYHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
+YHF+R++++ +I++ T AD+FTK + + + K++ V
Sbjct: 1293 RYHFIREKIADGEIQVVKISTTWNPADIFTKTVPVSKLQEALKLLRV 1339
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 625 bits (1613), Expect = e-179
Identities = 410/1315 (31%), Positives = 666/1315 (50%), Gaps = 143/1315 (10%)
Query: 128 NESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLTPKFDHVVVAIEESKKLENLKI 187
+E+I DF + L +L +TD+ +L +L ++D +V ++ S E L++
Sbjct: 109 DENIDDFLKIVADLNHLQI----DVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRL 164
Query: 188 EELQGSLEAHEQRIVERSS*KSKSTDQALQAQTSKKGGFSGKGGYRGKGKSKDYKNSS*K 247
+++ + E+ + + + + +G F+ RG+
Sbjct: 165 DDVMVAARDKERELSQNN-------------RPVVEGHFA-----RGR------------ 194
Query: 248 QSQFQNQEDHDQPESSSRRGGGSSNYKGGKRKFDRKKIRCFNCNKIGHFSSECKA---PS 304
P+ + G + + D K++ C+ C K GHF +C +
Sbjct: 195 ------------PDGKNNNQGNKGKNRSRSKSADGKRV-CWICGKEGHFKKQCYKWIERN 241
Query: 305 GGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNPATCNITDEEHVTLMMVTKEGG 364
+G + E++LAK N P +++ T+E T +TD
Sbjct: 242 KSKQQGSDNGESSLAKSTEAFN--PAMVLLATDE------TLVVTD-------------- 279
Query: 365 NYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDGKDV 424
+ W LD+GCS HMT K+W + E V+ +D + +GIG + ++ DG V
Sbjct: 280 SIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGIGSIKIRNSDGSQV 339
Query: 425 VISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQVKL 484
++++V Y+P M NLIS+G L ++ + L++ I+K +
Sbjct: 340 ILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCST-ILKGQKRDTLYILDGV 398
Query: 485 TAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSK 544
T +AE++D + LWH R GH++ K + L K + ++ + CE+C+ K
Sbjct: 399 TEEGESHSSAEVKDETALWHSRLGHMSQKGMEILVKKGCLR--REVIKELEFCEDCVYGK 456
Query: 545 QPRAPFSSFTPTRSTAV--LDVIYSDVCG-PFETASIGGNKYFASFIDEYSRKMWVYLLK 601
Q R SF P + L ++SD+ G P AS+G ++YF SF+D+YSRK+W+Y L+
Sbjct: 457 QHRV---SFAPAQHVTKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRKVWIYFLR 513
Query: 602 TKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQ 661
K E F F +K M + QS R +K LRTD G EYC++ FC+E GI+ T YTPQ
Sbjct: 514 KKDEAFEKFVEWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVRHKTCAYTPQ 573
Query: 662 HNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSG 721
NG+AER NRT+++ VRSML + +F EA TAVY++N P+ +++ +PE W+G
Sbjct: 574 QNGIAERLNRTIMDKVRSMLSRSGMEKKFWAEAASTAVYLINRSPSTAINFDLPEEKWTG 633
Query: 722 RKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGY-SSTGAYKLYNPRTSQVEFSR 780
P + LR FGCL + H + KL+ +S+ IF Y YK++ + SR
Sbjct: 634 ALPDLSSLRKFGCLAYIHAD---QGKLNPRSKKGIFTSYPEGVKGYKVWVLEDKKCVISR 690
Query: 781 DVVFEEHSAWK----GKETMVVNDSMQRVNLDLDHDDSEGIESAVVDVPGTSQNQNQIQV 836
+V+F E +K + + ++ + ++ D +D E + T N N +
Sbjct: 691 NVIFREQVMFKDLKGDSQNTISESDLEDLRVNPDMNDQEFTDQG----GATQDNSNPSEA 746
Query: 837 ---HNP---RPIRTKTLPAR---------FSDYQLI---------AETEFNSDGDMIHMA 872
HNP P ++ + S YQL+ A ++N + +M+ A
Sbjct: 747 TTSHNPVLNSPTHSQDEESEEEDSDAVEDLSTYQLVRDRVRRTIKANPKYN-ESNMVGFA 805
Query: 873 LLAD----ANPVKFEEAIKN---KTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWV 925
++ P ++EA+ + + W AMKEE+ S+ +N TWDLV P I +WV
Sbjct: 806 YYSEDDGKPEPKSYQEALLDPDWEKWNAAMKEEMVSMSKNHTWDLVTKPEKVKLIGCRWV 865
Query: 926 FKVKLN-PDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLW 984
F K P + ARLV +GF Q+ G+DY+E+F+PV + ++R ++++ N +L
Sbjct: 866 FTRKAGIPGVEAPRFIARLVAKGFTQKEGVDYNEIFSPVVKHVSIRYLLSMVVHYNMELQ 925
Query: 985 QLDVKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLS 1044
Q+DVK+AFL+G LEEE+Y+ QP GFEIK +KV L+++LYGLKQ+PR WN R D F+
Sbjct: 926 QMDVKTAFLHGFLEEEIYMAQPEGFEIKRGSNKVCLLKRSLYGLKQSPRQWNLRFDEFMR 985
Query: 1045 QTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLG 1104
+ + + + VY K C+ + L LYVDD+LI ++ S++ +K+ L+ EFEM DLG
Sbjct: 986 GIKYTRSAYDSCVYFKKCNGDTYIYLLLYVDDMLIASANKSEVNELKQLLSREFEMKDLG 1045
Query: 1105 KLSYFLGIEFVQTGEG--ILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLC----- 1157
LG+E + + + + Q Y+ +VL+ F + + P TP+ + KL
Sbjct: 1046 DAKKILGMEISRDRDAGLLTLSQEGYVKKVLRSFQMDNAKPVSTPLGIHFKLKAATDKEY 1105
Query: 1158 EEEAE-VDSTMFRQLVGCLRF-ICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLK 1215
EE+ E + + +G + + + +R +++Y +G++SRFMS P + H A K +LRY++
Sbjct: 1106 EEQFERMKIVPYANTIGSIMYSMIGTRPDLAYSLGVISRFMSKPLKDHWQAVKWVLRYMR 1165
Query: 1216 GTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKK 1275
GT + F K +ED L Y DSD+ + RRS GYVF G ISW SK
Sbjct: 1166 GTEKKKLCFRK-----QED--FLLRGYCDSDYGSNFDTRRSITGYVFTVGGNTISWKSKL 1218
Query: 1276 QAAVALSTCEAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSH 1335
Q VA+S+ EAEY+A A + LW++ ELG + + V++ D++SAI LAKN V H
Sbjct: 1219 QKVVAISSTEAEYMALTEAVKEALWLKGFAAELG-HSQDYVEVHSDSQSAITLAKNSVHH 1277
Query: 1336 GRSKHIETKYHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
R+KHI+ + HF+RD + IK+ T+ A++FTK + +F+ M+ V
Sbjct: 1278 ERTKHIDIRLHFIRDIICAGLIKVVKIATECNPANIFTKTVPLAKFEGALNMLRV 1332
>At3g45520 copia-like polyprotein
Length = 1363
Score = 616 bits (1589), Expect = e-176
Identities = 420/1339 (31%), Positives = 662/1339 (49%), Gaps = 155/1339 (11%)
Query: 110 KVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLTPKF 169
K KL + + E L +E N I +F + + L NL ++ DQAI+ +L +L F
Sbjct: 119 KQKLYSFKMS-ENLSIEGN--IDEFLHIVADLENLNVLVSDE--DQAIL--LLMSLPKPF 171
Query: 170 DHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQTSKKGGFSGK 229
D + ++ S L ++E+ ++ + E +E S K QA K G+
Sbjct: 172 DQLKDTLKYSSGKTVLSLDEVAAAIYSRE---LEFGSVKKSIKGQAEGLYVKDKAENRGR 228
Query: 230 GGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRKFDRKKIRCFN 289
+ KGK K K S S+RG C+
Sbjct: 229 SEQKDKGKGKRSK-------------------SKSKRG-------------------CWI 250
Query: 290 CNKIGHFSSEC------KAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNP 343
C + GH S C + + G +G +S EGS+ E M V+E S
Sbjct: 251 CGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNFVESAG-MFVSEALSSTD 309
Query: 344 ATCNITDEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADD 403
++ DE W +D+GC HMT +EWL + DE VR +
Sbjct: 310 I--HLEDE------------------WIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNK 349
Query: 404 RFISAEGIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFD 463
+G+G V + ++G V + V Y+P M NL+S+G + + L +
Sbjct: 350 SISRVKGVGTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKS 409
Query: 464 PNERKIMKVPLTPNRTFQVKLT-----AIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSL 518
N+ L R + A D A D++ LWH+R H++ K++S L
Sbjct: 410 GNQ------VLLEGRRYDTLYILHGKPATDESLAVARANDDTVLWHRRLCHMSQKNMSLL 463
Query: 519 KSKDMVHGLSQIKLPSKVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCG-PFETAS 577
K + L + CE+C+ + + F+ + L+ ++SD+ G P S
Sbjct: 464 IKKGFLDKKKVSMLDT--CEDCIYGRAKKIGFN-LAQHDTKKKLEYVHSDLWGAPTVPMS 520
Query: 578 IGGNKYFASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYC 637
+G +YF SFID+Y+RK+WVY LKTK E F F + ++ + QSG +K LRTD G E+C
Sbjct: 521 LGNCQYFISFIDDYTRKVWVYFLKTKDEAFEKFVSWISLVENQSGERVKTLRTDNGLEFC 580
Query: 638 SNEMSNFCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMT 697
+ FCEE G T YTPQ NGV ER NRT++ VRSML LP RF EA T
Sbjct: 581 NRMFDGFCEEKGFQRHRTCAYTPQQNGVVERMNRTIMEKVRSMLCDSGLPKRFWAEATHT 640
Query: 698 AVYVLNLCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIF 757
AV ++N P +++ + P+ WSG+ P +LR +GC+ H KL+ +++ +
Sbjct: 641 AVLLINKTPCSAINFEFPDKRWSGKAPIYSYLRRYGCVTFVHTDG---GKLNLRAKKGVL 697
Query: 758 IGY-SSTGAYKLYNPRTSQVEFSRDVVFEEHSAWKG----KETMVVND-----SMQRVNL 807
IGY S YK++ + SR+V F+E++ +K KE + + S ++L
Sbjct: 698 IGYPSGVKGYKVWLIEEKKCVVSRNVSFQENAVYKDLMQRKEQVSCEEDDHAGSYIDLDL 757
Query: 808 DLDHDDSEGIESAVV--------------------DVPGTSQNQNQIQVH--NPRPIRTK 845
+ D D+S G E + D+ T +Q+ + H R R
Sbjct: 758 EADKDNSSGGEQSQAQVTPATRGAVTSTPPRYETDDIEETDVHQSPLSYHLVRDRERREI 817
Query: 846 TLPARFSDYQLIAETEFNS-DGDMIHMALLADANPVKFEEAIKNKT---WRLAMKEELAS 901
P RF D AE + + DGD + P ++EA++++ WRLAM EE+ S
Sbjct: 818 RAPRRFDDEDYYAEALYTTEDGDAV--------EPADYKEAVRDENWDKWRLAMNEEIES 869
Query: 902 IERNKTWDLVDLPANKTPISVKWVFKVKLN-PDGSISKHKARLVVRGFMQRGGLDYSEVF 960
+N TW V P + I +W++K K P + KARLV +G+ QR G+DY E+F
Sbjct: 870 QLKNDTWTTVTRPEKQRIIGSRWIYKYKQGIPGVEEPRFKARLVAKGYAQREGVDYHEIF 929
Query: 961 APVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLK 1020
APV + ++R+++++ + +N +L QLDVK+AFL+G L+E++Y+ P G E E++V
Sbjct: 930 APVVKHVSIRILLSIVAQENLELEQLDVKTAFLHGELKEKIYMMPPEGCESLFKENEVCL 989
Query: 1021 LRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLIT 1080
L K+LYGLKQAPR WN++ + ++++ GF + + Y K + L YVDD+L+
Sbjct: 990 LNKSLYGLKQAPRQWNEKFNHYMTEIGFKRSDYDSCAYTKKLSDDSTMYLLFYVDDMLVA 1049
Query: 1081 GSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTGE-GIL-MHQRKYILEVLKRFNL 1138
++ I A+K+ L+ +FEM DLG LGIE + E G+L + Q Y+ +VLK FN+
Sbjct: 1050 ANNMQAIDALKKELSIKFEMKDLGAAKKILGIEIIIDREAGVLWLSQESYLNKVLKTFNM 1109
Query: 1139 LSCNPAETPVEGNLKL------GLCEEEAEVDSTMFRQLVGCLRF-ICHSRLEISYGVGL 1191
L PA TP+ +LK+ L EE ++S + VG + + + +R +++Y VG+
Sbjct: 1110 LESKPALTPLGAHLKMKSATEEKLSTEEEYMNSVPYSSAVGSIMYAMIGTRPDLAYPVGV 1169
Query: 1192 VSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQ 1251
VSRFMS P + H K +LRY+KGT + + + K + + + Y D+D+ D
Sbjct: 1170 VSRFMSQPAKEHWLGVKWVLRYIKGTVDTRLCY-------KRNSDFSICGYCDADYAADL 1222
Query: 1252 VDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWIQALLEELGLK 1311
RRS G VF G ISW S Q VA S+ E EY++ A + +W++ LL++ G +
Sbjct: 1223 DKRRSITGLVFTLGGNTISWKSGLQRVVAQSSTECEYMSLTEAVKEAIWLKGLLKDFGYE 1282
Query: 1312 TDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIKLQHCGTDLQIADV 1371
+ V++ D++SAI L+KN V H R+KHI+ K+HF+R+ ++ K+++ T+ AD+
Sbjct: 1283 -QKNVEIFCDSQSAIALSKNNVHHERTKHIDVKFHFIREIIADGKVEVSKISTEKNPADI 1341
Query: 1372 FTKPLKADRFKTLKKMMNV 1390
FTK L ++F+T + V
Sbjct: 1342 FTKVLPVNKFQTALDFLRV 1360
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 616 bits (1588), Expect = e-176
Identities = 423/1341 (31%), Positives = 659/1341 (48%), Gaps = 166/1341 (12%)
Query: 110 KVKLQTMRRQFELLQMETNESIADFFNRIISLTNLMKACGEKMTDQAIVEKVLRTLTPKF 169
K KL + + E L +E N I +F I L N+ ++ DQAI+ +L L F
Sbjct: 119 KQKLYSFKMS-ENLSVEGN--IDEFLQIITDLENMNVIISDE--DQAIL--LLTALPKAF 171
Query: 170 DHVVVAIEESKKLENLKIEELQGSLEAHEQRIVERSS*KSKSTDQALQAQTSKKGGFSGK 229
D + ++ S L ++E+ ++ + E +E S K QA K GK
Sbjct: 172 DQLKDTLKYSSGKSILTLDEVAAAIYSKE---LELGSVKKSIKVQAEGLYVKDKNENKGK 228
Query: 230 GGYRGKGKSKDYKNSS*KQSQFQNQEDHDQPESSSRRGGGSSNYKGGKRKFDRKKIRCFN 289
G +GKGK GK+ +KK C+
Sbjct: 229 GEQKGKGK--------------------------------------GKKGKSKKKPGCWT 250
Query: 290 CNKIGHFSSECKAPSGGDTRGRTSDEANLAKEGSITNEEPVTLMMVTEEGESNPATCNIT 349
C + GHF S C P+ + + S G N V+E S ++
Sbjct: 251 CGEEGHFRSSC--PNQNKPQFKQSQVVKGESSGGKGNLAEAAGYYVSEALSSTEV--HLE 306
Query: 350 DEEHVTLMMVTKEGGNYPGTWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAE 409
DE W LD+GCS HMT +EW +E+ VR +
Sbjct: 307 DE------------------WILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVR 348
Query: 410 GIGDVLVKREDGKDVVISEVLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKI 469
G+G + VK DG +V++ V Y+P M NL+S+G + + + L + N+
Sbjct: 349 GVGTIRVKNSDGLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQ--- 405
Query: 470 MKVPLTPNR---TFQVKLTAIDSQCLTA-ELEDNSWLWHQRFGHLNFKDLSSLKSKDMVH 525
V LT R + + + S+ L + D++ LWHQR H++ K++ L K
Sbjct: 406 --VLLTGRRYDTLYLLNWKPVASESLAVVKRADDTVLWHQRLCHMSQKNMEILVRKGF-- 461
Query: 526 GLSQIKLPS-KVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCG-PFETASIGGNKY 583
L + K+ S VCE+C+ K R FS + L+ I+SD+ G PF S+G +Y
Sbjct: 462 -LDKKKVSSLDVCEDCIYGKAKRKSFS-LAHHDTKEKLEYIHSDLWGAPFVPLSLGKCQY 519
Query: 584 FASFIDEYSRKMWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSN 643
F S ID+++RK+WVY +KTK E F F + + + Q+ R +K LRTD G E+C+
Sbjct: 520 FMSIIDDFTRKVWVYFMKTKDEAFEKFVEWVNLVENQTDRRVKTLRTDNGLEFCNKLFDG 579
Query: 644 FCEENGILHEVTAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLN 703
FCE GI T YTPQ NGVAER NRT++ VRSML LP RF EA T V ++N
Sbjct: 580 FCESIGIHRHRTCAYTPQQNGVAERMNRTIMEKVRSMLSDSGLPKRFWAEATHTTVLLIN 639
Query: 704 LCPTKSVDSQVPEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS-S 762
P+ +++ ++P+ WSG P +LR +GC+ H D KL+ +++ + IGY
Sbjct: 640 KTPSSALNFEIPDKKWSGNPPVYSYLRRYGCVAFVHTDDG---KLEPRAKKGVLIGYPVG 696
Query: 763 TGAYKLYNPRTSQVEFSRDVVFEEHSAWKGKETMVVNDSMQRV-NLDLDHDDSEGI---- 817
YK++ + SR+++F+E++ +K D MQR N+ + DD G
Sbjct: 697 VKGYKVWILDERKCVVSRNIIFQENAVYK--------DLMQRQENVSTEEDDQTGSYLEF 748
Query: 818 -----------------------ESAVVDVPGTSQ---------NQNQIQVH--NPRPIR 843
ES VV P T NQ+ + H R R
Sbjct: 749 DLEAERDVISGGDQEMVNTIPAPESPVVSTPTTQDTNDDEDSDVNQSPLSYHLVRDRDKR 808
Query: 844 TKTLPARFSDYQLIAETEFNS-DGDMIHMALLADANPVKFEEAIKNKT---WRLAMKEEL 899
P RF D AE + + DG+ + P + +A + W+LAM EE+
Sbjct: 809 EIRAPRRFDDEDYYAEALYTTEDGEAVE--------PENYRKAKLDANFDKWKLAMDEEI 860
Query: 900 ASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDG-SISKHKARLVVRGFMQRGGLDYSE 958
S E+N TW +V P N+ I +W+FK KL G + KARLV +G+ Q+ G+DY E
Sbjct: 861 DSQEKNNTWTIVTRPENQRIIGCRWIFKYKLGILGVEEPRFKARLVAKGYAQKEGIDYHE 920
Query: 959 VFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEVYITQPPGFEIKGSEHKV 1018
+FAPV + ++R+++++ + ++ +L QLDVK+AFL+G L+E++Y++ P G+E ++V
Sbjct: 921 IFAPVVKHVSIRVLLSIVAQEDLELEQLDVKTAFLHGELKEKIYMSPPEGYESMFKANEV 980
Query: 1019 LKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLL 1078
L KALYGLKQAP+ WN++ D F+ + F K + + Y K G V+ L +YVDD+L
Sbjct: 981 CLLNKALYGLKQAPKQWNEKFDNFMKEICFVKSAYDSCAYTKVLPDGSVMYLLIYVDDIL 1040
Query: 1079 ITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQTGE-GIL-MHQRKYILEVLKRF 1136
+ + I A+K +L FEM DLG LG+E ++ G+L + Q Y+ ++L+ +
Sbjct: 1041 VASKNKEAITALKANLGMRFEMKDLGAAKKILGMEIIRDRTLGVLWLSQEGYLNKILETY 1100
Query: 1137 NLLSCNPAETPVEGNLKL------GLCEEEAEVDSTMFRQLVGCLRF-ICHSRLEISYGV 1189
N+ PA TP+ + K L +E + S + VG + + + +R +++Y V
Sbjct: 1101 NMAEAKPAMTPLGAHFKFQAATEQKLIRDEDFMKSVPYSSAVGSIMYAMLGTRPDLAYPV 1160
Query: 1190 GLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLVAYTDSDWCG 1249
G++SRFMS P + H K +LRY+KGT + + K+ + +V Y D+D+
Sbjct: 1161 GIISRFMSQPIKEHWLGVKWVLRYIKGTLKTRLCY-------KKSSSFSIVGYCDADYAA 1213
Query: 1250 DQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQGLWIQALLEELG 1309
D RRS G VF G ISW S Q VA ST E+EY++ A + +W++ LL++ G
Sbjct: 1214 DLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSTTESEYMSLTEAVKEAIWLKGLLKDFG 1273
Query: 1310 LKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKIKLQHCGTDLQIA 1369
+ ++V++ D++SAI L+KN V H R+KHI+ KYHF+R+ +S +++ T+ A
Sbjct: 1274 YE-QKSVEIFCDSQSAIALSKNNVHHERTKHIDVKYHFIREIISDGTVEVLKISTEKNPA 1332
Query: 1370 DVFTKPLKADRFKTLKKMMNV 1390
D+FTK L +F+ ++ V
Sbjct: 1333 DIFTKVLAVSKFQAALNLLRV 1353
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 590 bits (1521), Expect = e-168
Identities = 348/1072 (32%), Positives = 577/1072 (53%), Gaps = 79/1072 (7%)
Query: 370 WYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDGKDVVISEV 429
W +D+GCS HMT +EW +L+E+ VR + GIG + VK E G V ++ V
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNV 346
Query: 430 LYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQVKLT---- 485
Y+P M NL+S+G + +S ++ L + + V LT R + + L
Sbjct: 347 RYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDS-----VLLTVRRCYTLYLLQWRP 401
Query: 486 AIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSKQ 545
+ + +D++ LWH+R GH++ K++ L K ++ KL + CE+C+ K
Sbjct: 402 VTEESLSVVKRQDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKL--ETCEDCIYGKA 459
Query: 546 PRAPFSSFTPTRSTAVLDVIYSDVCG-PFETASIGGNKYFASFIDEYSRKMWVYLLKTKS 604
R F+ + L+ ++SD+ G P S+G +YF SFID+Y+RK+ +Y LKTK
Sbjct: 460 KRIGFN-LAQHDTREKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKD 518
Query: 605 EVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNG 664
E F F + + + Q+ + IK LRTD G E+C+ FC + GIL T YTPQ NG
Sbjct: 519 EAFDKFVEWANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNG 578
Query: 665 VAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGRKP 724
VAER NRT++ VRSML LP +F EA T ++N P+ +++ +VP+ WSG+ P
Sbjct: 579 VAERMNRTLMEKVRSMLSDSGLPKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSP 638
Query: 725 SVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS-STGAYKLYNPRTSQVEFSRDVV 783
+LR FGC+ H D KL+ +++ I +GY YK++ + SR+V+
Sbjct: 639 IYSYLRRFGCIAFVHTDD---GKLNPRAKKGILVGYPIGVKGYKIWLLEEKKCVVSRNVI 695
Query: 784 FEEHSAWKGKETMVVNDSMQRVN---------LDLDHDD--SEGIESAVVDV-------P 825
F+E++++ K+ M D+ + N LDLDH++ + G + +V+ P
Sbjct: 696 FQENASY--KDMMQSKDAEKDENEAPPSSYLDLDLDHEEVITSGGDDPIVEAQSPFNPSP 753
Query: 826 GTSQNQNQ--------------IQVHNPRPIRTKTLPARFSDYQLIAETEFNSDGDMIHM 871
T+Q ++ Q+ R RT P RF D +AE + ++
Sbjct: 754 ATTQTYSEGVNSETDIIQSPLSYQLVRDRDRRTIRAPVRFDDEDYLAEALYTTEDS---- 809
Query: 872 ALLADANPVKFEEAIKN---KTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKV 928
+ P + EA ++ W+LAM EE+ S +N TW +V P ++ I +W++K
Sbjct: 810 ---GEIEPADYSEAKRSMNWNKWKLAMNEEMESQIKNHTWTVVKRPQHQKVIGSRWIYKF 866
Query: 929 KLN-PDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLD 987
KL P + KARLV +G+ QR G+DY E+FAPV + ++R+++++ + ++ +L QLD
Sbjct: 867 KLGIPGVEEGRFKARLVAKGYAQRKGIDYHEIFAPVVKHVSIRILMSIVAQEDLELEQLD 926
Query: 988 VKSAFLNGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTG 1047
VK+AFL+G L+E++Y+ P G+E E +V L K+LYGLKQAP+ WN++ + ++S+ G
Sbjct: 927 VKTAFLHGELKEKIYMVPPEGYEEMFKEDEVCLLNKSLYGLKQAPKQWNEKFNAYMSEIG 986
Query: 1048 FHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLS 1107
F + + Y+K G + L LYVDD+L+ + I +K L+ F+M DLG
Sbjct: 987 FIRSLYDSCAYIKELSDGSRVYLLLYVDDMLVAAKNKEDISQLKEELSQRFDMKDLGAAK 1046
Query: 1108 YFLGIEFVQTGE--GILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLC------EE 1159
LG+E ++ E + + Q Y+ ++L+ +N+ TP+ +LK+ ++
Sbjct: 1047 RILGMEIIRNREENTLWLSQNGYLNKILETYNMAESKHVVTPLGAHLKMRAATVEKQEQD 1106
Query: 1160 EAEVDSTMFRQLVGCLRF-ICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTP 1218
E + S + VG + + + +R +++Y VG++SR+MS P + H K +LRY+KG+
Sbjct: 1107 EDYMKSIPYSSAVGSIMYAMIGTRPDLAYPVGIISRYMSQPAREHWLGVKWVLRYIKGSL 1166
Query: 1219 NHGVFFPKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAA 1278
+ + K + +V Y D+D + RRS G VF G + ISW S +Q
Sbjct: 1167 GTKLQY-------KRSSDFKVVGYCDADHAACKDRRRSITGLVFTLGGSTISWKSGQQRV 1219
Query: 1279 VALSTCEAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRS 1338
VALST EAEY++ A + +W++ LL+E G + ++V++ D++SAI L+KN V H R+
Sbjct: 1220 VALSTTEAEYMSLTEAVKEAVWMKGLLKEFGYE-QKSVEIFCDSQSAIALSKNNVHHERT 1278
Query: 1339 KHIETKYHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
KHI+ +Y ++RD ++ + T+ AD+FTK + ++F+ ++ V
Sbjct: 1279 KHIDVRYQYIRDIIANGDGDVVKIDTEKNPADIFTKIVPVNKFQAALTLLQV 1330
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 568 bits (1465), Expect = e-162
Identities = 348/1057 (32%), Positives = 569/1057 (52%), Gaps = 79/1057 (7%)
Query: 369 TWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDGKDVVISE 428
TW +DSG ++H++ +++ LD + S V + G+G VL+ KD+++
Sbjct: 441 TWVIDSGATHHVSHDRKLFQTLDTSIVSFVNLPTGPNVRISGVGTVLIN----KDIILQN 496
Query: 429 VLYVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVP--LTPNRTFQV-KLT 485
VL++P + NLIS+ L D + +FDP+ +I + LT ++ L
Sbjct: 497 VLFIPEFRLNLISISSLTT-DLGTRV------IFDPSCCQIQDLTKGLTLGEGKRIGNLY 549
Query: 486 AIDSQ--CLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIK-LPSKVCENCLV 542
+D+Q ++ + +WH+R GH +F L SL V G ++ K S C C +
Sbjct: 550 VLDTQSPAISVNAVVDVSVWHKRLGHPSFSRLDSLSE---VLGTTRHKNKKSAYCHVCHL 606
Query: 543 SKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKT 602
+KQ + F S + ++++ DV GPF ++ G KYF + +D++SR W+YLLK+
Sbjct: 607 AKQKKLSFPSANNI-CNSTFELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLLKS 665
Query: 603 KSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQH 662
KS+V +VF F + + Q +K +R+D E E F + GI+ + P TP+
Sbjct: 666 KSDVLTVFPAFIDLVENQYDTRVKSVRSDNAKELAFTE---FYKAKGIVSFHSCPETPEQ 722
Query: 663 NGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGR 722
N V ER+++ +LN+ R+++ ++ + G+ V+TAV+++N P+ + ++ P V +G+
Sbjct: 723 NSVVERKHQHILNVARALMFQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGK 782
Query: 723 KPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYS-STGAYKLYNPRTSQVEFSRD 781
P L+ FGCLC+ ++R K +S +F+GY YKL + ++ V SR+
Sbjct: 783 LPDYSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRN 842
Query: 782 VVFEEH---------SAWKGKETMVVNDSMQRVNLDLDHDDSEGIESAVVDVPGTSQNQN 832
V F E SA + D + N H S I P T ++
Sbjct: 843 VEFHEELFPLASSQQSATTASDVFTPMDPLSSGNSITSHLPSPQIS------PSTQISKR 896
Query: 833 QIQVHNPRPIRTKTLPARFSDYQL----------IAETEFNSDGDMIHMALLADAN---- 878
+I TK PA DY I+ + S HM + + +
Sbjct: 897 RI---------TK-FPAHLQDYHCYFVNKDDSHPISSSLSYSQISPSHMLYINNISKIPI 946
Query: 879 PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISK 938
P + EA +K W A+ +E+ ++ER TW++ LP K + KWVF VK + DGS+ +
Sbjct: 947 PQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWVFTVKFHADGSLER 1006
Query: 939 HKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLE 998
KAR+V +G+ Q+ GLDY+E F+PVA++ TV++++ +++ K W L QLD+ +AFLNG LE
Sbjct: 1007 FKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQLDISNAFLNGDLE 1066
Query: 999 EEVYITQPPGF-EIKGSE---HKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVE 1054
E +Y+ P G+ +IKG+ + V +L+K++YGLKQA R W + L GF K +
Sbjct: 1067 ETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSNSLLALGFEKQHGD 1126
Query: 1055 HGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEF 1114
H ++V+ C ++L +YVDD++I ++ Q++ +L F++ +LG L YFLG+E
Sbjct: 1127 HTLFVR-CIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRELGPLKYFLGLEV 1185
Query: 1115 VQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGC 1174
+T EGI + QRKY LE+L ++L C P+ P+ N++L + D M+R+LVG
Sbjct: 1186 ARTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLLEDKEMYRRLVGK 1245
Query: 1175 LRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKED 1234
L ++ +R +I++ V + +F S PR +HLAA ++L+Y+KGT G+F+ +
Sbjct: 1246 LMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGLFY-------SAE 1298
Query: 1235 GNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSA 1294
+L L YTD+DW RRST G+ F G + ISW SKKQ V+ S+ EAEY A A
Sbjct: 1299 DDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYRALALA 1358
Query: 1295 ACQGLWIQALLEELGLKTDEAVQLMV-DNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVS 1353
+C+ W+ LL L L+ V ++ D+ +A+ +A NPV H R+KHIE H +R+++
Sbjct: 1359 SCEMAWLSTLL--LALRVHSGVPILYSDSTAAVYIATNPVFHERTKHIEIDCHTVREKLD 1416
Query: 1354 KEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
++KL H T Q+AD+ TKPL +F L M++
Sbjct: 1417 NGQLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSI 1453
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 568 bits (1464), Expect = e-162
Identities = 341/1054 (32%), Positives = 564/1054 (53%), Gaps = 67/1054 (6%)
Query: 369 TWYLDSGCSNHMTGNKEWLINLDENKKSRVRFADDRFISAEGIGDVLVKREDGKDVVISE 428
TW +DSG ++H++ ++ +LD + S V + G+G + + D+++
Sbjct: 430 TWVIDSGATHHVSHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKLN----DDILLKN 485
Query: 429 VLYVPGMKTNLISMGQLLEKDFS-MSMKKRHLEVFDPNERKIMKVPLTPNRTFQVKLTAI 487
VL++P + NLIS+ L + S + K E+ D + +++ R + L +
Sbjct: 486 VLFIPEFRLNLISISSLTDDIGSRVIFDKNSCEIQDLIKGRMLG---QGRRVANLYLLDV 542
Query: 488 DSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSKQPR 547
Q ++ + +WH+R GH + + L ++ D + S C C ++KQ +
Sbjct: 543 GDQSISVNAVVDISMWHRRLGHASLQRLDAIS--DSLGTTRHKNKGSDFCHVCHLAKQRK 600
Query: 548 APFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKTKSEVF 607
F + + D+++ DV GPF ++ G KYF + +D++SR W+YLLKTKSEV
Sbjct: 601 LSFPTSNKV-CKEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKSEVL 659
Query: 608 SVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNGVAE 667
+VF F + Q +K +R+D E + ++F E GI+ + P TP+ N V E
Sbjct: 660 TVFPAFIQQVENQYKVKVKAVRSDNAPEL---KFTSFYAEKGIVSFHSCPETPEQNSVVE 716
Query: 668 RRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGRKPSVK 727
R+++ +LN+ R+++ +P G+ V+TAV+++N P++ + ++ P + +G P +
Sbjct: 717 RKHQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPVYE 776
Query: 728 HLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGY-SSTGAYKLYNPRTSQVEFSRDVVFEE 786
LR FGCLC+ ++R K +S +F+GY S YKL + ++ V SR+V F E
Sbjct: 777 QLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHE 836
Query: 787 H-----------SAWKGKETMVVNDSMQRVNLDLDHDDSEGIESAVVDVPGTSQNQNQIQ 835
S+ K MV S + D H S + S + D+P +Q
Sbjct: 837 EVFPLAKNPGSESSLKLFTPMVPVSS--GIISDTTHSPSS-LPSQISDLPPQISSQ---- 889
Query: 836 VHNPRPIRTKTLPARFSDYQL----------IAETEFNSDGDMIHMALLADAN----PVK 881
R + PA +DY I+ T S HM + + P
Sbjct: 890 -------RVRKPPAHLNDYHCNTMQSDHKYPISSTISYSKISPSHMCYINNITKIPIPTN 942
Query: 882 FEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISKHKA 941
+ EA K W A+ E+ ++E+ TW++ LP K + KWVF +K DG++ ++KA
Sbjct: 943 YAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKA 1002
Query: 942 RLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEV 1001
RLV +G+ Q+ GLDY++ F+PVA++ T+++++ +++ K W L QLDV +AFLNG LEEE+
Sbjct: 1003 RLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEI 1062
Query: 1002 YITQPPGF-EIKG---SEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGV 1057
++ P G+ E KG + VL+L++++YGLKQA R W K+ + L GF K +H +
Sbjct: 1063 FMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTL 1122
Query: 1058 YVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFVQT 1117
++K D G +++ +YVDD++I +S + + L+ F++ DLG L YFLG+E +T
Sbjct: 1123 FLKMYD-GEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVART 1181
Query: 1118 GEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVGCLRF 1177
GI + QRKY LE+L+ +L+C P P+ NLK+ + + D +R++VG L +
Sbjct: 1182 TAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMY 1241
Query: 1178 ICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNL 1237
+ +R +I++ V + +F S PR +HL AA R+L+Y+KGT G+F+ +L
Sbjct: 1242 LTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFY-------SASSDL 1294
Query: 1238 HLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQ 1297
L + DSDW Q RRST + F G + ISW SKKQ V+ S+ EAEY A A C+
Sbjct: 1295 TLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCE 1354
Query: 1298 GLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEKI 1357
+W+ LL L + L D+ +AI +A NPV H R+KHI+ H +R+++ ++
Sbjct: 1355 MVWLFTLLVSLQASPPVPI-LYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGEL 1413
Query: 1358 KLQHCGTDLQIADVFTKPLKADRFKTLKKMMNVL 1391
KL H T+ Q+AD+ TKPL +F+ LK M++L
Sbjct: 1414 KLLHVRTEDQVADILTKPLFPYQFEHLKSKMSIL 1447
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 566 bits (1458), Expect = e-161
Identities = 359/1057 (33%), Positives = 535/1057 (49%), Gaps = 53/1057 (5%)
Query: 370 WYLDSGCSNHMTGNKEWLINLDENK-KSRVRFADDRFISAEGIGDVLVKREDGKDVVISE 428
W+ DS + H+T + L + E + V D ++ G +K +GK + ++E
Sbjct: 322 WHPDSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGK-IPLNE 380
Query: 429 VLYVPGMKTNLISMGQLLEK-DFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQVK---L 484
VL VP ++ +L+S+ +L + + + + D +K++ N + ++
Sbjct: 381 VLVVPNIQKSLLSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQEF 440
Query: 485 TAIDSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSK 544
A+ S A E+ +WH R GH N K L L++ + S VCE C + K
Sbjct: 441 VALYSNRQCAATEE---VWHHRLGHANSKALQHLQNSKAIQ--INKSRTSPVCEPCQMGK 495
Query: 545 QPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKTKS 604
R PF + +R LD I+ D+ GP S G KY+A F+D+YSR W Y L KS
Sbjct: 496 SSRLPFL-ISDSRVLHPLDRIHCDLWGPSPVVSNQGLKYYAIFVDDYSRYSWFYPLHNKS 554
Query: 605 EVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNG 664
E SVF F+ + + Q IKV ++D GGE+ SN++ E+GI H ++ PYTPQ NG
Sbjct: 555 EFLSVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTPQQNG 614
Query: 665 VAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGRKP 724
+AER++R ++ + SML P +F E+ TA Y++N P+ + + P G KP
Sbjct: 615 LAERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALFGEKP 674
Query: 725 SVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGYSST-GAYKLYNPRTSQVEFSRDVV 783
LR+FG C+ + + K D +S +F+GY+S Y+ + P T +V SR+V+
Sbjct: 675 DYSSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYISRNVI 734
Query: 784 FEEHS--------------------AWKGKETMVVNDSMQRVNL-----DLDHDDSEGIE 818
F E AW+ + ++ V L DL+ +
Sbjct: 735 FNESELPFKEKYQSLVPQYSTPLLQAWQHNKISEISVPAAPVQLFSKPIDLNTYAGSQVT 794
Query: 819 SAVVDVPGTSQNQNQIQVHNPRPIRTKTLPAR-FSDYQLIAETEFNSDGDMIHMALLAD- 876
+ D TS N+ + NP + + + + ++ AL+
Sbjct: 795 EQLTDPEPTSNNEGSDEEVNPVAEEIAANQEQVINSHAMTTRSKAGIQKPNTRYALITSR 854
Query: 877 ---ANPVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPD 933
A P A+K+ W A+ EE+ + TW LV + +S KWVFK KL+PD
Sbjct: 855 MNTAEPKTLASAMKHPGWNEAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTKLHPD 914
Query: 934 GSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFL 993
GSI K KARLV +GF Q G+DY E F+PV R T+R+++ +++ K W + QLDV +AFL
Sbjct: 915 GSIDKLKARLVAKGFDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVSNAFL 974
Query: 994 NGPLEEEVYITQPPGFEIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSV 1053
+G L+E V++ QP GF V +L KA+YGLKQAPRAW FL GF
Sbjct: 975 HGELQEPVFMYQPSGFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKS 1034
Query: 1054 EHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIE 1113
+ ++V D G +L L LYVDD+L+TGS S ++ + ++L N F M DLG YFLGI+
Sbjct: 1035 DPSLFVCHQD-GKILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQ 1093
Query: 1114 FVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVDSTMFRQLVG 1173
G+ +HQ Y ++L++ + CNP TP+ +L E + T FR L G
Sbjct: 1094 IEDYANGLFLHQTAYATDILQQAGMSDCNPMPTPLP--QQLDNLNSELFAEPTYFRSLAG 1151
Query: 1174 CLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKE 1233
L+++ +R +I + V + + M P S KRILRY+KGT G LP K
Sbjct: 1152 KLQYLTITRPDIQFAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMG------LPI-KR 1204
Query: 1234 DGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACS 1293
+ L L AY+DSD G + RRST G+ G ISWS+K+Q V+ S+ EAEY A
Sbjct: 1205 NSTLTLSAYSDSDHAGCKNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTY 1264
Query: 1294 AACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVS 1353
AA + WI LL +LG+ Q+ DN SA+ L+ NP H RSKH +T YH++R+QV+
Sbjct: 1265 AAREITWISFLLRDLGIPQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVA 1324
Query: 1354 KEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNV 1390
I+ QH Q+ADVFTK L F L+ + V
Sbjct: 1325 LGLIETQHISATFQLADVFTKSLPRRAFVDLRSKLGV 1361
>At4g03810 putative retrotransposon protein
Length = 964
Score = 555 bits (1429), Expect = e-158
Identities = 347/980 (35%), Positives = 536/980 (54%), Gaps = 78/980 (7%)
Query: 431 YVPGMKTNLISMGQLLEKDFSMSMKKRHLEVFDPNERKIMKVPLTPNRTFQVKLTAIDSQ 490
YVP + N+IS+ L + F S+K + FD ++ PL N + +
Sbjct: 10 YVPAINKNIISVSCLDMEGFHFSIKNKCCS-FDRDDMFYGSAPLD-NGLHVLNQSMPIYN 67
Query: 491 CLTAELEDN----SWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPSKVCENCLVSKQP 546
T + + N ++LWH R GH+N K + L S +++ + CE+CL+ K
Sbjct: 68 IRTKKFKSNDLNPTFLWHCRLGHINEKHIQKLHSDGLLNSFDYESY--ETCESCLLGKMT 125
Query: 547 RAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRKMWVYLLKTKSEV 606
+APF+ + R++ +L +I++DVCGP T++ G +YF +F D++SR +VYL+K KS+
Sbjct: 126 KAPFTGHSE-RASDLLGLIHTDVCGPMSTSARGNYQYFITFTDDFSRYGYVYLMKHKSKS 184
Query: 607 FSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEVTAPYTPQHNGVA 666
F FK F+ + Q G+SIK LR+D GGEY S S+ E GI+ ++T P TPQ NGV+
Sbjct: 185 FENFKEFQNEVQNQFGKSIKALRSDRGGEYLSQVFSDHLRECGIVSQLTPPGTPQWNGVS 244
Query: 667 ERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQVPEAVWSGRKPSV 726
ERRNRT+L+MVRSM+ LP F G A+ T+ ++LN CP+KSV+ + P +W+G+ P++
Sbjct: 245 ERRNRTLLDMVRSMMSHTDLPSPFWGYALETSAFMLNRCPSKSVE-KTPYEIWTGKVPNL 303
Query: 727 KHLRIFGCLCH--KHIPDQRRRKLDDKSETMIFIGY-SSTGAYKLYNPRTSQVEFSRDVV 783
L+I+GC + + I D KL KS+ F+GY T Y Y+P ++V R+
Sbjct: 304 SFLKIWGCESYAKRLITD----KLGPKSDKCYFVGYPKETKGYYFYHPTDNKVFVVRNGA 359
Query: 784 FEEHSAWKGKETMVVNDSMQRVNLDLDHDDSEGIESAVVDVPGTSQNQNQIQVHNP---- 839
F E +E + S +V L E + DVP TSQ ++Q+ +
Sbjct: 360 FLE------REFLSKGTSGSKVLL-------EEVREPQGDVP-TSQEEHQLDLRRVVEPI 405
Query: 840 -------RPIRTKTLPARFSDYQLIAETEFNSDGDMIHMALLADANPVKFEEAIK---NK 889
R R++ P RF D+ + F + D P +EEA+ +
Sbjct: 406 LVEPEVRRSERSRHEPDRFRDWVMDDHALFMIESD----------EPTSYEEALMGPDSD 455
Query: 890 TWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKVKLNPDGSISKHKARLVVRGFM 949
W A K E+ S+ +NK W LVDLP PI KW+FK K++ DG+I +KA LV +G+
Sbjct: 456 KWLEAAKSEMESMSQNKVWTLVDLPDGVKPIECKWIFKKKIDMDGNIQIYKAGLVAKGYK 515
Query: 950 QRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDVKSAFLNGPLEEEVYITQPPGF 1009
Q G+DY E ++PVA L+++R+++A A+ ++++WQ+DVK+AFLNG LEE VY+TQP GF
Sbjct: 516 QVHGIDYDETYSPVAMLKSIRILLATAAHYDYEIWQMDVKTAFLNGNLEEHVYMTQPEGF 575
Query: 1010 EIKGSEHKVLKLRKALYGLKQAPRAWNKRIDTFLSQTGFHKCSVEHGVYVKSCDSGGVLL 1069
+ + KV KL +++YGLKQA R+WN R + + + F + E VY K+ S V
Sbjct: 576 TVPEAARKVCKLHRSIYGLKQASRSWNLRFNEAIKEFDFIRNEEEPCVYKKTSGS-AVAF 634
Query: 1070 LCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLGKLSYFLGIEFV--QTGEGILMHQRK 1127
L LYVDD+L+ G+ +Q+VK L + F M D+G+ +Y LGI + + I + Q
Sbjct: 635 LVLYVDDILLLGNDIPLLQSVKTWLGSCFSMKDMGEAAYILGIRIYRDRLNKIIGLSQDT 694
Query: 1128 YILEVLKRFNLLSCNPAETPVEGNLKLGLC------EEEAEVDSTMFRQLVGCLRF-ICH 1180
YI +VL RFN+ P+ + L +E + + +G + + + +
Sbjct: 695 YIDKVLHRFNMHDSKKGFIPMSHGITLSKTQCPSTHDERERMSKIPYASAIGSIMYAMLY 754
Query: 1181 SRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFFPKHLPSQKEDGNLHLV 1240
+R +++ + + SR+ S P +SH + I +YL+ T + + + G+ LV
Sbjct: 755 TRPDVACALSMTSRYQSDPGESHWIVVRNIFKYLRRTKDKFLVY---------GGSEELV 805
Query: 1241 --AYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTCEAEYIAACSAACQG 1298
YTD+ + D+ D RS G+ F +SW S KQ+ VA ST EAEYIAA AA +
Sbjct: 806 VSGYTDASFQTDKDDFRSQSGFFFCLNGGAVSWKSTKQSTVADSTTEAEYIAASEAAKEV 865
Query: 1299 LWIQALLEELGL--KTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETKYHFLRDQVSKEK 1356
+WI+ + ELG+ + L DN AI AK P SH +SKHI+ +YH +R+ + +
Sbjct: 866 VWIRKFITELGVVPSISGPIDLYCDNNGAIAQAKEPKSHQKSKHIQRRYHLIREIIDRGD 925
Query: 1357 IKLQHCGTDLQIADVFTKPL 1376
+K+ TD +AD FTKPL
Sbjct: 926 VKISRVSTDANVADHFTKPL 945
>At1g70010 hypothetical protein
Length = 1315
Score = 528 bits (1360), Expect = e-150
Identities = 324/1007 (32%), Positives = 531/1007 (52%), Gaps = 53/1007 (5%)
Query: 421 GKDVVISEVLYVPGMKTNLISMGQLLEK-DFSMSMKKRHLEVFDPNERKIMKVPLTPNRT 479
G+ +++++VL++P K NL+S+ L + + + + D ++ +
Sbjct: 320 GRHLILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMGKQVANL 379
Query: 480 FQVKLTAI-----DSQCLTAELEDNSWLWHQRFGHLNFKDLSSLKSKDMVHGLSQIKLPS 534
+ V L ++ DS A + + LWH+R GH + + L + S ++ Q
Sbjct: 380 YIVDLDSLSHPGTDSSITVASVTSHD-LWHKRLGHPSVQKLQPMSS--LLSFPKQKNNTD 436
Query: 535 KVCENCLVSKQPRAPFSSFTPTRSTAVLDVIYSDVCGPFETASIGGNKYFASFIDEYSRK 594
C C +SKQ PF S +S+ D+I+ D GPF + G +YF + +D+YSR
Sbjct: 437 FHCRVCHISKQKHLPFVSHN-NKSSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRA 495
Query: 595 MWVYLLKTKSEVFSVFKVFKTMAKKQSGRSIKVLRTDEGGEYCSNEMSNFCEENGILHEV 654
WVYLL+ KS+V +V F TM + Q +IK +R+D E + F GI+
Sbjct: 496 TWVYLLRNKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPEL---NFTQFYHSKGIVPYH 552
Query: 655 TAPYTPQHNGVAERRNRTVLNMVRSMLKGKSLPHRFCGEAVMTAVYVLNLCPTKSVDSQV 714
+ P TPQ N V ER+++ +LN+ RS+ +P + G+ ++TAVY++N P ++ +
Sbjct: 553 SCPETPQQNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDKC 612
Query: 715 PEAVWSGRKPSVKHLRIFGCLCHKHIPDQRRRKLDDKSETMIFIGY-SSTGAYKLYNPRT 773
P V + P+ H+++FGCLC+ + R K +++ FIGY S YKL + T
Sbjct: 613 PFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLET 672
Query: 774 SQVEFSRDVVFEEH------SAWKGKETMVVND-----SMQRVNLDLDHDDSEGIESAVV 822
+ SR VVF E S +E D MQR + D H + S+V
Sbjct: 673 HSIIVSRHVVFHEELFPFLGSDLSQEEQNFFPDLNPTPPMQRQSSD--HVNPSDSSSSVE 730
Query: 823 DVPGTSQNQNQIQVHNPRPIRTKTLPARFSDY---QLIAETEFN----------SDGDMI 869
+P + N + R PA DY +++ T +D +
Sbjct: 731 ILPSANPTNNVPEPSVQTSHRKAKKPAYLQDYYCHSVVSSTPHEIRKFLSYDRINDPYLT 790
Query: 870 HMALLADAN-PVKFEEAIKNKTWRLAMKEELASIERNKTWDLVDLPANKTPISVKWVFKV 928
+A L P + EA K + WR AM E +E TW++ LPA+K I +W+FK+
Sbjct: 791 FLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKI 850
Query: 929 KLNPDGSISKHKARLVVRGFMQRGGLDYSEVFAPVARLETVRMIVALASWKNWDLWQLDV 988
K N DGS+ ++KARLV +G+ Q+ G+DY+E F+PVA+L +V++++ +A+ L QLD+
Sbjct: 851 KYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDI 910
Query: 989 KSAFLNGPLEEEVYITQPPGFEIKGSE----HKVLKLRKALYGLKQAPRAWNKRIDTFLS 1044
+AFLNG L+EE+Y+ P G+ + + + V +L+K+LYGLKQA R W + + L
Sbjct: 911 SNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLL 970
Query: 1045 QTGFHKCSVEHGVYVKSCDSGGVLLLCLYVDDLLITGSSYSKIQAVKRSLNNEFEMTDLG 1104
GF + +H ++K D G L + +Y+DD++I ++ + + +K + + F++ DLG
Sbjct: 971 GLGFIQSYCDHTCFLKISD-GIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLG 1029
Query: 1105 KLSYFLGIEFVQTGEGILMHQRKYILEVLKRFNLLSCNPAETPVEGNLKLGLCEEEAEVD 1164
+L YFLG+E V++ +GI + QRKY L++L L C P+ P++ ++ V+
Sbjct: 1030 ELKYFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVE 1089
Query: 1165 STMFRQLVGCLRFICHSRLEISYGVGLVSRFMSCPRQSHLAAAKRILRYLKGTPNHGVFF 1224
+R+L+G L ++ +R +I++ V +++F PR++HL A +IL+Y+KGT G+F+
Sbjct: 1090 VGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFY 1149
Query: 1225 PKHLPSQKEDGNLHLVAYTDSDWCGDQVDRRSTMGYVFFFGKAPISWSSKKQAAVALSTC 1284
L L Y ++D+ + RRST GY F G + I W S+KQ V+ S+
Sbjct: 1150 -------SATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSA 1202
Query: 1285 EAEYIAACSAACQGLWIQALLEELGLKTDEAVQLMVDNKSAIDLAKNPVSHGRSKHIETK 1344
EAEY + A + +W+ L+EL + + L DN++AI +A N V H R+KHIE+
Sbjct: 1203 EAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESD 1262
Query: 1345 YHFLRDQVSKEKIKLQHCGTDLQIADVFTKPLKADRFKTLKKMMNVL 1391
H +R+++ K +L H T+LQIAD FTKPL F L M +L
Sbjct: 1263 CHSVRERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMGLL 1309
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,039,577
Number of Sequences: 26719
Number of extensions: 1418521
Number of successful extensions: 4714
Number of sequences better than 10.0: 204
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 45
Number of HSP's that attempted gapping in prelim test: 3775
Number of HSP's gapped (non-prelim): 429
length of query: 1400
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1288
effective length of database: 8,326,068
effective search space: 10723975584
effective search space used: 10723975584
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0399.4


