

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
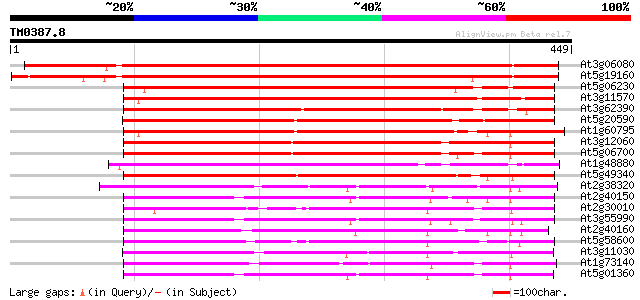
Query= TM0387.8
(449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g06080 unknown protein 607 e-174
At5g19160 putative protein 602 e-172
At5g06230 unknown protein 367 e-102
At3g11570 hypothetical protein 364 e-101
At3g62390 unknown protein 337 1e-92
At5g20590 unknown protein 335 3e-92
At1g60795 unknown protein 327 1e-89
At3g12060 hypothetical protein 312 3e-85
At5g06700 unknown protein 310 1e-84
At1g48880 hypothetical protein 308 4e-84
At5g49340 putative protein 304 7e-83
At2g38320 unknown protein 256 1e-68
At2g40150 unknown protein 256 2e-68
At2g30010 unknown protein 254 5e-68
At3g55990 unknown protein 253 1e-67
At2g40160 unknown protein 244 9e-65
At5g58600 unknown protein 243 2e-64
At3g11030 unknown protein 241 6e-64
At1g73140 hypothetical protein 241 6e-64
At5g01360 unknown protein 237 9e-63
>At3g06080 unknown protein
Length = 469
Score = 607 bits (1566), Expect = e-174
Identities = 281/439 (64%), Positives = 339/439 (77%), Gaps = 17/439 (3%)
Query: 13 VFEGFRRVKRFRL-FEPSLGVLGFVVVTAIVGCCFFYLGNRDVAARFGFLDQPQSFPWLR 71
+ E RR KR RL FEPSLGVLGF +V + C FF+ R VA +G D+ + F WL+
Sbjct: 16 ICEALRRFKRSRLVFEPSLGVLGFFLVGVCLVCSFFFFDYRSVAKSYGLSDKSERFVWLK 75
Query: 72 LKEQS-----------RVEFLGENGVVVGGGCDLFDGEWVWDESYPLYQSKDCMFLDEGF 120
S RV FL E+G GCD+FDG+WVWDESYPLYQSKDC FLDEGF
Sbjct: 76 FDNISSSSSSSSNSSKRVGFLEESG----SGCDVFDGDWVWDESYPLYQSKDCRFLDEGF 131
Query: 121 RCSENGRRDLFYTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCML 180
RCS+ GR DLFYTQWRWQP+ CN+PRF+A MLE+LR+KR+VF GDSIGRNQWESLLC+L
Sbjct: 132 RCSDFGRSDLFYTQWRWQPRHCNLPRFDAKLMLEKLRDKRLVFVGDSIGRNQWESLLCLL 191
Query: 181 SSGVSNKESIYEVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRT 240
SS V N+ IYE+NG+PITKHKGFLVFKF ++NCTVEYYR+PFLV QSRPP G GK++T
Sbjct: 192 SSAVKNESLIYEINGSPITKHKGFLVFKFEEYNCTVEYYRSPFLVPQSRPPIGSPGKVKT 251
Query: 241 TLKVDKMDWNSLKWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSM 300
+LK+D MDW S KW +AD+L+LNTGHWWN KT R GCYFQEG EVKLK++V+DAYKR++
Sbjct: 252 SLKLDTMDWTSSKWRDADVLVLNTGHWWNEGKTTRTGCYFQEGEEVKLKMNVDDAYKRAL 311
Query: 301 ETVLNWMQSSVNPSKTQVFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRF 360
TV+ W+ + ++ +KTQVFFRT APVHFRGGDW+ GG CH ETLP++G+SL ++ W +
Sbjct: 312 NTVVKWIHTELDSNKTQVFFRTFAPVHFRGGDWKTGGTCHMETLPEIGTSLASSETWEQL 371
Query: 361 KAANAVLSSHTNASEIMKIKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCL 420
K VLS ++N SE +K+K+LNIT M AQRKDGH S+YYLG GPAPLHRQDCSHWCL
Sbjct: 372 KILRDVLSHNSNRSETVKVKLLNITAMAAQRKDGHPSLYYLG-PHGPAPLHRQDCSHWCL 430
Query: 421 PGVPDTWNELLYAMLLKHE 439
PGVPDTWNEL YA+ +K E
Sbjct: 431 PGVPDTWNELFYALFMKQE 449
>At5g19160 putative protein
Length = 464
Score = 602 bits (1551), Expect = e-172
Identities = 286/449 (63%), Positives = 345/449 (76%), Gaps = 17/449 (3%)
Query: 2 SEEVEAMMPMTVFEGFRRVKRFRL-FEPSLGVLGFVVVTAIVGCCFFYLGNRDVAAR--F 58
S+E + MP++ +R KR RL FEPSLGVLGF +V + FFYL R VA
Sbjct: 6 SQEEDEAMPISEVV-IKRFKRLRLVFEPSLGVLGFFLVGLCLVFSFFYLDYRTVAKTKSH 64
Query: 59 GFLDQPQSFPWLRLKE-----QSRVEFLGENGVVVGGGCDLFDGEWVWDESYPLYQSKDC 113
F DQ + F WL+ + ++V FL E+G GCDLF+G+WVWDESYPLYQSKDC
Sbjct: 65 DFSDQSERFLWLKELDGFEVNNTKVGFLEESG----NGCDLFNGKWVWDESYPLYQSKDC 120
Query: 114 MFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQW 173
F+DEGFRC+E GR DLFYT+WRWQP C++PRF+A MLE+LRNKR+VF GDSIGRNQW
Sbjct: 121 TFIDEGFRCTEFGRPDLFYTKWRWQPNHCDLPRFDAKLMLEKLRNKRLVFVGDSIGRNQW 180
Query: 174 ESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTG 233
ESLLCML+S +SNK +YEVN PITKH GF VF+F D+NCTVEYYRAPFLVLQSRPP G
Sbjct: 181 ESLLCMLASAISNKNLVYEVNNRPITKHMGFFVFRFHDYNCTVEYYRAPFLVLQSRPPEG 240
Query: 234 VSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVE 293
K++TTLK++ M+W + KW +ADIL+ NTGHWWNYEKTIRGGCYFQEG +V++++ +E
Sbjct: 241 SPEKVKTTLKLETMEWTADKWRDADILVFNTGHWWNYEKTIRGGCYFQEGEKVRMRMKIE 300
Query: 294 DAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVP 353
AY+R+M+TV+ W+Q V+ +KTQVFFRT APVHFRGGDWR GG CH ETLPD G+SLVP
Sbjct: 301 HAYRRAMKTVMKWIQEEVDANKTQVFFRTFAPVHFRGGDWRTGGTCHMETLPDFGASLVP 360
Query: 354 NDNWSRFKAANAVLSS---HTNASEIMKIKVLNITHMTAQRKDGHSSIYYLGRVAGPAPL 410
+ W K VLSS ++N SE +K+KVLNIT M AQR DGH S+YYLG +AGPAP
Sbjct: 361 AETWDHIKLLQDVLSSSLYYSNISETVKLKVLNITAMAAQRNDGHPSLYYLG-LAGPAPF 419
Query: 411 HRQDCSHWCLPGVPDTWNELLYAMLLKHE 439
HRQDCSHWCLPGVPD+WNELLYA+ LKHE
Sbjct: 420 HRQDCSHWCLPGVPDSWNELLYALFLKHE 448
>At5g06230 unknown protein
Length = 413
Score = 367 bits (942), Expect = e-102
Identities = 175/354 (49%), Positives = 233/354 (65%), Gaps = 18/354 (5%)
Query: 92 CDLFDGEWVWDESYP------LYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMP 145
CD G+WV S L+ ++C FLD GFRC ++GR+D Y WRWQP C++P
Sbjct: 62 CDYSKGKWVRRASSSSSSVNGLFYGEECRFLDSGFRCHKHGRKDSGYLDWRWQPHGCDLP 121
Query: 146 RFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFL 205
RFNA+ +LER RN R+VF GDSIGRNQWESL+CMLS + NK IYEVNGNPITKHKGFL
Sbjct: 122 RFNASDLLERSRNGRIVFVGDSIGRNQWESLMCMLSQAIPNKSEIYEVNGNPITKHKGFL 181
Query: 206 VFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTG 265
+F N TVEY+R+PFLV+ RPP +I+TT++VD+ +W S +W +D+L+ N+G
Sbjct: 182 SMRFPRENLTVEYHRSPFLVVIGRPPDKSPKEIKTTVRVDEFNWQSKRWVGSDVLVFNSG 241
Query: 266 HWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAP 325
HWWN +KT+ GCYF+EG +V + V +A+ +S++T +W+ ++P K+ VFFR+ +P
Sbjct: 242 HWWNEDKTVLTGCYFEEGRKVNKTMGVMEAFGKSLKTWKSWVLEKLDPDKSYVFFRSYSP 301
Query: 326 VHFRGGDWRNGGNCHSETLPDLGSSLVPND---NWSRFKAANAVLSSHTNASEIMKIKVL 382
VH+R G W GG C +E P+ + D N +K + H+ K+K L
Sbjct: 302 VHYRNGTWNTGGLCDAEIEPETDKRKLEPDASHNEYIYKVIEEMRYRHS------KVKFL 355
Query: 383 NITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
NIT++T RKDGH S Y R G + QDCSHWCLPGVPDTWNE+LYA LL
Sbjct: 356 NITYLTEFRKDGHISRY---REQGTSVDVPQDCSHWCLPGVPDTWNEILYAQLL 406
>At3g11570 hypothetical protein
Length = 427
Score = 364 bits (934), Expect = e-101
Identities = 173/348 (49%), Positives = 224/348 (63%), Gaps = 9/348 (2%)
Query: 92 CDLFDGEWVW---DESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFN 148
CD G WV D Y ++C FLD GFRC NGR+D + QWRWQP C++PRFN
Sbjct: 79 CDYSYGRWVRRRRDVDETSYYGEECRFLDPGFRCLNNGRKDSGFRQWRWQPHGCDLPRFN 138
Query: 149 ATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFK 208
A+ LER RN R+VF GDSIGRNQWESLLCMLS VSNK IYEVNGNPI+KHKGFL +
Sbjct: 139 ASDFLERSRNGRIVFVGDSIGRNQWESLLCMLSQAVSNKSEIYEVNGNPISKHKGFLSMR 198
Query: 209 FRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWW 268
F + N TVEY+R PFLV+ RPP ++ T++VD+ +W S KW +D+L+ NTGHWW
Sbjct: 199 FPEQNLTVEYHRTPFLVVVGRPPENSPVDVKMTVRVDEFNWQSKKWVGSDVLVFNTGHWW 258
Query: 269 NYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHF 328
N +KT GCYFQEG ++ + V + +++S++T +W+ ++ ++ VFFR+ +PVH+
Sbjct: 259 NEDKTFIAGCYFQEGGKLNKTMGVMEGFEKSLKTWKSWVLERLDSERSHVFFRSFSPVHY 318
Query: 329 RGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMT 388
R G W GG C ++T P+ + D + A+ S K+K LNIT++T
Sbjct: 319 RNGTWNLGGLCDADTEPETDMKKMEPDPIHNNYISQAIQEMRYEHS---KVKFLNITYLT 375
Query: 389 AQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
RKD H S Y AP QDCSHWCLPGVPDTWNE+LYA LL
Sbjct: 376 EFRKDAHPSRYREPGTPEDAP---QDCSHWCLPGVPDTWNEILYAQLL 420
>At3g62390 unknown protein
Length = 475
Score = 337 bits (863), Expect = 1e-92
Identities = 162/350 (46%), Positives = 219/350 (62%), Gaps = 20/350 (5%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
CD+ G+WV+D YPLY + C F+DEGF C NGR DL Y WRW+P+DC+ PRFNATK
Sbjct: 138 CDVTKGKWVYDSDYPLYTNASCPFIDEGFGCQSNGRLDLNYMNWRWEPQDCHAPRFNATK 197
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
MLE +R KR+VF GDSI RNQWES+LC+L V + + +YE + ITK KG F+F D
Sbjct: 198 MLEMIRGKRLVFVGDSINRNQWESMLCLLFQAVKDPKRVYETHNRRITKEKGNYSFRFVD 257
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+ CTVE+Y FLV + R G K R TL++D MD S +W A+IL+ NT HWW++
Sbjct: 258 YKCTVEFYVTHFLVREGRARIG--KKRRETLRIDAMDRTSSRWKGANILVFNTAHWWSHY 315
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT G Y+QEG + KL V A+K++++T +W+ +V+P KT+VFFR+ AP HF GG
Sbjct: 316 KTKSGVNYYQEGDLIHPKLDVSTAFKKALQTWSSWVDKNVDPKKTRVFFRSAAPSHFSGG 375
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTAQR 391
+W +GG+C +P L + P+ + + + + T + +LN++ ++ R
Sbjct: 376 EWNSGGHCREANMP-LNQTFKPSYSSKKSIVEDVLKQMRT------PVTLLNVSGLSQYR 428
Query: 392 KDGHSSIYYLGRVAGPAPLHR-----QDCSHWCLPGVPDTWNELLYAMLL 436
D H SIY G P +R QDCSHWCLPGVPDTWN LY LL
Sbjct: 429 IDAHPSIY------GTKPENRRSRAVQDCSHWCLPGVPDTWNHFLYLHLL 472
>At5g20590 unknown protein
Length = 485
Score = 335 bits (859), Expect = 3e-92
Identities = 159/348 (45%), Positives = 218/348 (61%), Gaps = 9/348 (2%)
Query: 91 GCDLFDGEWV-WDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNA 149
GCDL+ G WV D+ YPLYQ C ++D+ F C NGRRD Y WRW+P C++PRFNA
Sbjct: 140 GCDLYKGSWVKGDDEYPLYQPGSCPYVDDAFDCQRNGRRDSDYLNWRWKPDGCDLPRFNA 199
Query: 150 TKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKF 209
T L +LR K ++ GDS+ RNQ+ES+LC+L G+S+K +YEV+G+ ITK +G+ VFKF
Sbjct: 200 TDFLVKLRGKSLMLVGDSMNRNQFESMLCVLREGLSDKSRMYEVHGHNITKGRGYFVFKF 259
Query: 210 RDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWN 269
D+NCTVE+ R+ FLV + G TL +D++D + KW ADIL+ NTGHWW
Sbjct: 260 EDYNCTVEFVRSHFLVREG-VRANAQGNTNPTLSIDRIDKSHAKWKRADILVFNTGHWWV 318
Query: 270 YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
+ KT RG Y++EG + K +AY+RS++T W+ +VNP K VF+R + HFR
Sbjct: 319 HGKTARGKNYYKEGDYIYPKFDATEAYRRSLKTWAKWIDQNVNPKKQLVFYRGYSSAHFR 378
Query: 330 GGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTA 389
GG+W +GG+C+ E P S++ + + ++ ++ I +LN+T +T
Sbjct: 379 GGEWDSGGSCNGEVEPVKKGSIIDS-----YPLKMKIVQEAIKEMQVPVI-LLNVTKLTN 432
Query: 390 QRKDGHSSIYYLGRVAG-PAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
RKDGH SIY G RQDCSHWCLPGVPD WN L+YA LL
Sbjct: 433 FRKDGHPSIYGKTNTDGKKVSTRRQDCSHWCLPGVPDVWNHLIYASLL 480
>At1g60795 unknown protein
Length = 541
Score = 327 bits (837), Expect = 1e-89
Identities = 162/360 (45%), Positives = 220/360 (61%), Gaps = 15/360 (4%)
Query: 92 CDLFDGEWVW--DESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNA 149
CD++DG WV DE+ P Y C ++D F C NGR D Y +WRWQP C++PR N
Sbjct: 190 CDIYDGSWVRADDETMPYYPPGSCPYIDRDFNCHANGRPDDAYVKWRWQPNGCDIPRLNG 249
Query: 150 TKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKF 209
T LE+LR K++VF GDSI RN WESL+C+L + +K+ +YE++G K KGF F+F
Sbjct: 250 TDFLEKLRGKKLVFVGDSINRNMWESLICILRHSLKDKKRVYEISGRREFKKKGFYAFRF 309
Query: 210 RDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWN 269
D+NCTV++ +PF V +S GV+G TL++D MD + + +ADILI NTGHWW
Sbjct: 310 EDYNCTVDFVGSPFFVRES-SFKGVNGTTLETLRLDMMDKTTSMYRDADILIFNTGHWWT 368
Query: 270 YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
++KT G Y+QEG V +L V +AYKR++ T W+ +++ S+T + FR + HFR
Sbjct: 369 HDKTKLGENYYQEGNVVYPRLKVLEAYKRALITWAKWVDKNIDRSQTHIVFRGYSVTHFR 428
Query: 330 GGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKV--LNITHM 387
GG W +GG CH ET P +S + S+ KA +L + MK V +NI+ +
Sbjct: 429 GGPWNSGGQCHKETEPIFNTSYLAKYP-SKMKALEYIL------RDTMKTPVIYMNISRL 481
Query: 388 TAQRKDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLLKHEVDQRW 444
T RKDGH SIY Y + QDCSHWCLPGVPDTWN+LLY LLK + +W
Sbjct: 482 TDFRKDGHPSIYRMVYRTEKEKREAVSHQDCSHWCLPGVPDTWNQLLYVSLLKAGLASKW 541
>At3g12060 hypothetical protein
Length = 556
Score = 312 bits (799), Expect = 3e-85
Identities = 148/348 (42%), Positives = 213/348 (60%), Gaps = 10/348 (2%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
C+ F+G+WV D+SYPLY+ C +DE F C NGR D+ + + +W+PK C++PR N K
Sbjct: 196 CEFFEGDWVKDDSYPLYKPGSCNLIDEQFNCISNGRPDVDFQKLKWKPKQCSLPRLNGGK 255
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
+LE +R +R+VF GDS+ RN WESL+C+L V ++ ++E +G + + F F+D
Sbjct: 256 LLEMIRGRRLVFVGDSLNRNMWESLVCILKGSVKDESQVFEAHGRHQFRWEAEYSFVFKD 315
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+NCTVE++ +PFLV Q T +G + TL++D + +S ++ ADIL+ NTGHWW +E
Sbjct: 316 YNCTVEFFASPFLV-QEWEVTEKNGTKKETLRLDLVGKSSEQYKGADILVFNTGHWWTHE 374
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT +G Y+QEG+ V KL V++A+++++ T W+ +VNP K+ VFFR +P HF GG
Sbjct: 375 KTSKGEDYYQEGSTVHPKLDVDEAFRKALTTWGRWVDKNVNPKKSLVFFRGYSPSHFSGG 434
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHMTAQR 391
W GG C ET P + N+ + + + LNIT +T R
Sbjct: 435 QWNAGGACDDETEP------IKNETYLTPYMLKMEILERVLRGMKTPVTYLNITRLTDYR 488
Query: 392 KDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
KD H SIY L +PL QDCSHWCLPGVPD+WNE+ YA LL
Sbjct: 489 KDAHPSIYRKQKLSAEESKSPLLYQDCSHWCLPGVPDSWNEIFYAELL 536
>At5g06700 unknown protein
Length = 608
Score = 310 bits (793), Expect = 1e-84
Identities = 151/353 (42%), Positives = 216/353 (60%), Gaps = 20/353 (5%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
C+ FDGEW+ D+SYPLY+ C +DE F C NGR D + + +W+PK C++PR N
Sbjct: 255 CEFFDGEWIKDDSYPLYKPGSCNLIDEQFNCITNGRPDKDFQKLKWKPKKCSLPRLNGAI 314
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFRD 211
+LE LR +R+VF GDS+ RN WESL+C+L V ++ +YE G + + F F+D
Sbjct: 315 LLEMLRGRRLVFVGDSLNRNMWESLVCILKGSVKDETKVYEARGRHHFRGEAEYSFVFQD 374
Query: 212 FNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNYE 271
+NCTVE++ +PFLV Q G + TL++D + +S ++ AD+++ NTGHWW +E
Sbjct: 375 YNCTVEFFVSPFLV-QEWEIVDKKGTKKETLRLDLVGKSSEQYKGADVIVFNTGHWWTHE 433
Query: 272 KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRGG 331
KT +G Y+QEG+ V +L V +A+++++ T W++ +VNP+K+ VFFR + HF GG
Sbjct: 434 KTSKGEDYYQEGSNVYHELAVLEAFRKALTTWGRWVEKNVNPAKSLVFFRGYSASHFSGG 493
Query: 332 DWRNGGNCHSETLPDLGSSLVPNDNW-----SRFKAANAVLSSHTNASEIMKIKVLNITH 386
W +GG C SET P + ND + S+ K VL + LNIT
Sbjct: 494 QWNSGGACDSETEP------IKNDTYLTPYPSKMKVLEKVLRGMKT-----PVTYLNITR 542
Query: 387 MTAQRKDGHSSIY---YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
+T RKDGH S+Y L +PL QDCSHWCLPGVPD+WNE+LYA L+
Sbjct: 543 LTDYRKDGHPSVYRKQSLSEKEKKSPLLYQDCSHWCLPGVPDSWNEILYAELI 595
>At1g48880 hypothetical protein
Length = 485
Score = 308 bits (789), Expect = 4e-84
Identities = 157/368 (42%), Positives = 221/368 (59%), Gaps = 26/368 (7%)
Query: 80 FLGENGV------VVGGG-CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFY 132
FLG NG VVG CD+FDG WV D++YPLY + +C F+++GF C NGR Y
Sbjct: 96 FLGYNGALEFNSSVVGDTECDIFDGNWVVDDNYPLYNASECPFVEKGFNCLGNGRGHDEY 155
Query: 133 TQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYE 192
+WRW+PK C +PRF +L+RLR KR+VF GDS+ R QWESL+CML +G+ +K S+YE
Sbjct: 156 LKWRWKPKHCTVPRFEVRDVLKRLRGKRIVFVGDSMSRTQWESLICMLMTGLEDKRSVYE 215
Query: 193 VNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSL 252
VNGN ITK FL +F +N TVE+YR+ FLV R ++++TLK+D +D +
Sbjct: 216 VNGNNITKRIRFLGVRFSSYNFTVEFYRSVFLVQPGRLRWHAPKRVKSTLKLDVLDVINH 275
Query: 253 KWSNADILILNTGHWWNYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVN 312
+WS+AD LI NTG WW K GCYFQ G ++L + + AY+ ++ET +W++S+V+
Sbjct: 276 EWSSADFLIFNTGQWWVPGKLFETGCYFQVGNSLRLGMSIPAAYRVALETWASWIESTVD 335
Query: 313 PSKTQVFFRTLAPVHFRGGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTN 372
P+KT+V FRT P H W + +C+ P D R K+ + +
Sbjct: 336 PNKTRVLFRTFEPSH-----WSDHRSCNVTKYP-------APDTEGRDKSIFSEMIKEVV 383
Query: 373 ASEIMKIKVLNITHMTAQRKDGHSSIYYLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLY 432
+ + + +L++T M+A R DGH ++ PL DCSHWCLPGVPD WNE+L
Sbjct: 384 KNMTIPVSILDVTSMSAFRSDGHVGLW------SDNPL-VPDCSHWCLPGVPDIWNEILL 436
Query: 433 AMLLKHEV 440
L + V
Sbjct: 437 FFLFRQPV 444
>At5g49340 putative protein
Length = 457
Score = 304 bits (778), Expect = 7e-83
Identities = 156/352 (44%), Positives = 217/352 (61%), Gaps = 16/352 (4%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNATK 151
CD+FDG WV+D+S P+Y C F+++ F C +NGR D + + RWQP C++PRF+ K
Sbjct: 100 CDIFDGTWVFDDSEPVYLPGYCPFVEDKFNCFKNGRPDSGFLRHRWQPHGCSIPRFDGKK 159
Query: 152 MLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPIT-KHKGFLVFKFR 210
ML+ LR KRVVF GDS+ RN WESL+C L S + +K + ++ G ++GF F+F
Sbjct: 160 MLKMLRGKRVVFVGDSLNRNMWESLVCSLRSTLEDKNRVSKIIGKQSNLPNEGFYGFRFN 219
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLK-WSNADILILNTGHWWN 269
DF C++++ ++PFLV +S V GK R TL++D + + K + NADI+I NTGHWW
Sbjct: 220 DFECSIDFIKSPFLVQESE-VVDVYGKRRETLRLDMIQRSMTKIYKNADIVIFNTGHWWT 278
Query: 270 YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFR 329
++KT G Y+QEG V +L V++AY +++ T +W+ S++N +KT+VFF + HFR
Sbjct: 279 HQKTYEGKGYYQEGNRVYERLEVKEAYTKAIHTWADWVDSNINSTKTRVFFVGYSSSHFR 338
Query: 330 GGDWRNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKV--LNITHM 387
G W +GG C ET P + W K +V+S MK V +NIT M
Sbjct: 339 KGAWNSGGQCDGETRPIQNETYTGVYPW-MMKVVESVISE-------MKTPVFYMNITKM 390
Query: 388 TAQRKDGHSSIYYL---GRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
T R DGH S+Y R PA QDCSHWCLPGVPD+WN+LLYA LL
Sbjct: 391 TWYRTDGHPSVYRQPADPRGTSPAAGMYQDCSHWCLPGVPDSWNQLLYATLL 442
>At2g38320 unknown protein
Length = 410
Score = 256 bits (655), Expect = 1e-68
Identities = 140/380 (36%), Positives = 220/380 (57%), Gaps = 26/380 (6%)
Query: 73 KEQSRVEFLGENGVVVGGGCDLFDGEWVWDE-SYPLYQSKDCMFLDEGFRCSENGRRDLF 131
KE + + + G G C+LF+G+WV+D SYPLY+ +DC F+ + C + GR+DL
Sbjct: 40 KENPQSHGVTDRGGDSGRECNLFEGKWVFDNVSYPLYKEEDCKFMSDQLACEKFGRKDLS 99
Query: 132 YTQWRWQPKDCNMPRFNATKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIY 191
Y WRWQP C++PRFN TK+LERLRNKR+V+ GDS+ R QW S++CM+SS ++N +++Y
Sbjct: 100 YKFWRWQPHTCDLPRFNGTKLLERLRNKRMVYVGDSLNRGQWVSMVCMVSSVITNPKAMY 159
Query: 192 EVNGNPITKHKGFLVFKFRDFNCTVEYYRAPFLV-LQSRPPTGVSGKIRTTLKVDKMDWN 250
N + FK ++N T++YY AP LV S PT R +++ ++ +
Sbjct: 160 MHNNG-----SNLITFKALEYNATIDYYWAPLLVESNSDDPTNHRFPDR-IVRIQSIEKH 213
Query: 251 SLKWSNADILILNTGHWWN--YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQ 308
+ W+N+DI++ N+ WW + K++ G F++ + ++ + Y+ +++T+ W++
Sbjct: 214 ARHWTNSDIIVFNSYLWWRMPHIKSLWGS--FEKLDGIYKEVEMVRVYEMALQTLSQWLE 271
Query: 309 SSVNPSKTQVFFRTLAPVHFRGGDWRNGG----NCHSE-TLPDLGSSLVPNDNWSRFKAA 363
VNP+ T++FF +++P H R +W GG NC+ E +L D + +
Sbjct: 272 VHVNPNITKLFFMSMSPTHERAEEW--GGILNQNCYGEASLIDKEGYTGRGSDPKMMRVL 329
Query: 364 NAVLSSHTNASEIMKIKVLNITHMTAQRKDGHSSIY--YLGRVAG---PAPLHRQDCSHW 418
VL N + ++++NIT ++ RK+GH SIY G V P DC HW
Sbjct: 330 ENVLDGLKNRG--LNMQMINITQLSEYRKEGHPSIYRKQWGTVKENEISNPSSNADCIHW 387
Query: 419 CLPGVPDTWNELLYAMLLKH 438
CLPGVPD WNELLYA +L H
Sbjct: 388 CLPGVPDVWNELLYAYILDH 407
>At2g40150 unknown protein
Length = 424
Score = 256 bits (654), Expect = 2e-68
Identities = 133/362 (36%), Positives = 204/362 (55%), Gaps = 28/362 (7%)
Query: 92 CDLFDGEWVWD-ESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNAT 150
CDLF G+WV+D ++YPLY+ ++C FL E C NGR+D + WRWQP+DC++P+FNA
Sbjct: 71 CDLFTGQWVFDNKTYPLYKEEECEFLTEQVTCLRNGRKDSLFQNWRWQPRDCSLPKFNAR 130
Query: 151 KMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFR 210
+LE+LRNKR++F GDS+ RNQWES++C++ +S+ + + VFK +
Sbjct: 131 VLLEKLRNKRLMFVGDSLNRNQWESMVCLV-------QSVIPPGRKSLNQTGSLTVFKIQ 183
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNY 270
D+N TVE+Y APFLV + I + + ++ + + W D L+ N+ WW
Sbjct: 184 DYNATVEFYWAPFLVESNSDDPEKHSIIDRIIMPESIEKHGVNWIGVDFLVFNSYIWWMN 243
Query: 271 E---KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVH 327
K +RG F +G ++ AY+R + T+ +W+ +++P T VFF +++P+H
Sbjct: 244 TVSIKVLRGS--FDDGDTEYDEIKRPIAYERVLRTLGDWVDHNIDPLSTTVFFMSMSPLH 301
Query: 328 FRGGDWRN--GGNCHSETLPDLGSSLVPNDNWSRFKAANA---VLSSHTNASEIMKIKV- 381
+ DW N G C ET P L S N + +F A + N ++ +K+ +
Sbjct: 302 IKSSDWANPEGIRCALETTPILNMSF--NVAYGQFSAVGTDYRLFPVAENVTQSLKVPIH 359
Query: 382 -LNITHMTAQRKDGHSSIY------YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAM 434
LNIT ++ RKD H+S+Y L R P + DC HWCLPG+PDTWNE LY
Sbjct: 360 FLNITALSEYRKDAHTSVYTIKQGKLLTREQQNDPANFADCIHWCLPGLPDTWNEFLYTH 419
Query: 435 LL 436
++
Sbjct: 420 II 421
>At2g30010 unknown protein
Length = 398
Score = 254 bits (650), Expect = 5e-68
Identities = 136/360 (37%), Positives = 211/360 (57%), Gaps = 33/360 (9%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCM--FLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNA 149
CDLF GEWV DE+YPLY+SK+C +D GF C GR D Y ++RW+P +CN+PRFN
Sbjct: 56 CDLFAGEWVRDETYPLYRSKECGRGIIDPGFDCQTYGRPDSDYLKFRWKPFNCNVPRFNG 115
Query: 150 TKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKF 209
K L+ +R+K ++F GDS+GRNQWESL+CM+SS + + + ++ +P++ FK
Sbjct: 116 VKFLQEMRDKTIMFVGDSLGRNQWESLICMISSSAPSINT-HIIHEDPLS------TFKI 168
Query: 210 RDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWN-SLKWSNADILILNTGHWW 268
D+N V +YRAP+LV + ++GK TTLK+D++ + S W AD+L+ NTGHWW
Sbjct: 169 LDYNVKVSFYRAPYLVDIDK----INGK--TTLKLDEISVDASNAWRTADVLLFNTGHWW 222
Query: 269 NYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHF 328
++ ++RG + G + A ++ + T +W+ +N T+VFF +++P H+
Sbjct: 223 SHTGSLRGWEQMETGGRYYGDMDRLVALRKGLGTWSSWVLRYINSPLTRVFFLSVSPTHY 282
Query: 329 RGGDW----------RNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMK 378
+W + G +C+ +T P G++ + ++ K + V+ +
Sbjct: 283 NPNEWTSRSKTSTITQGGKSCYGQTTPFSGTTYPTSSYVNQKKVIDDVVKEMKS-----H 337
Query: 379 IKVLNITHMTAQRKDGHSSIYY--LGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
+ +++IT ++A R DGH SIY L P DCSHWCLPG+PDTWN+L YA LL
Sbjct: 338 VSLMDITMLSALRVDGHPSIYSGDLNPSLKRNPDRSSDCSHWCLPGLPDTWNQLFYAALL 397
>At3g55990 unknown protein
Length = 487
Score = 253 bits (647), Expect = 1e-67
Identities = 133/359 (37%), Positives = 199/359 (55%), Gaps = 28/359 (7%)
Query: 92 CDLFDGEWVWD-ESYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNAT 150
CDLF GEWV+D E++PLY+ C FL C NGRRD Y WRWQP+DC++P+F A
Sbjct: 140 CDLFTGEWVFDNETHPLYKEDQCEFLTAQVTCMRNGRRDSLYQNWRWQPRDCSLPKFKAK 199
Query: 151 KMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFR 210
+LE+LRNKR++F GDS+ RNQWES++C++ +S+ + K VF+
Sbjct: 200 LLLEKLRNKRMMFVGDSLNRNQWESMVCLV-------QSVVPPGRKSLNKTGSLSVFRVE 252
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNY 270
D+N TVE+Y APFLV + + + + + ++ + + W D L+ NT WW
Sbjct: 253 DYNATVEFYWAPFLVESNSDDPNMHSILNRIIMPESIEKHGVNWKGVDFLVFNTYIWWMN 312
Query: 271 E---KTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVH 327
K +RG F +G ++ AY+R M T +W++ +++P +T VFF +++P+H
Sbjct: 313 TFAMKVLRGS--FDKGDTEYEEIERPVAYRRVMRTWGDWVERNIDPLRTTVFFASMSPLH 370
Query: 328 FRGGDWRN--GGNCHSETLPDLGSSL--VPNDNWSRFKAANAVLSSHTNASEIMKIKVLN 383
+ DW N G C ET P L S+ ++ F A V S + + LN
Sbjct: 371 IKSLDWENPDGIKCALETTPILNMSMPFSVGTDYRLFSVAENVTHSLN-----VPVYFLN 425
Query: 384 ITHMTAQRKDGHSSIYYL--GRVAGPA----PLHRQDCSHWCLPGVPDTWNELLYAMLL 436
IT ++ RKD H+S++ + G++ P P DC HWCLPG+PDTWNE LY ++
Sbjct: 426 ITKLSEYRKDAHTSVHTIRQGKMLTPEQQADPNTYADCIHWCLPGLPDTWNEFLYTRII 484
>At2g40160 unknown protein
Length = 427
Score = 244 bits (622), Expect = 9e-65
Identities = 133/356 (37%), Positives = 198/356 (55%), Gaps = 29/356 (8%)
Query: 92 CDLFDGEWVWDE-SYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNAT 150
CD+F G+WV D ++PLY+ +C FL E C+ NGR D Y +WRWQP+DC++PRF++
Sbjct: 77 CDVFTGKWVLDNVTHPLYKEDECEFLSEWVACTRNGRPDSKYQKWRWQPQDCSLPRFDSK 136
Query: 151 KMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFL-VFKF 209
+LE+LR K+++F GDSI NQW+S++CM+ S + +G KH + +F
Sbjct: 137 LLLEKLRGKKLMFIGDSIHYNQWQSMVCMVQSVIP--------SGKKTLKHTAQMSIFNI 188
Query: 210 RDFNCTVEYYRAPFLV-LQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWW 268
++N T+ +Y APFLV + PP GK + + + + W +AD LI NT WW
Sbjct: 189 EEYNATISFYWAPFLVESNADPPDKRDGKTDPVIIPNSISKHGENWKDADYLIFNTYIWW 248
Query: 269 NYEKTIR--GGCYFQEGTEVKL-KLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAP 325
TI+ F +G + ++ + YK+ + T W++ ++NPS+T +FF +++P
Sbjct: 249 TRHSTIKVLKQESFNKGDSKEYNEIGIYIVYKQVLSTWTKWLEQNINPSQTSIFFSSMSP 308
Query: 326 VHFRGGDW--RNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKV-- 381
H R DW G C ET P L S N +R + NA++ K+ +
Sbjct: 309 THIRSSDWGFNEGSKCEKETEPILNMSKPINVGTNR-----RLYEIALNATKSTKVPIHF 363
Query: 382 LNITHMTAQRKDGHSSIY--YLGRVAGPA----PLHRQDCSHWCLPGVPDTWNELL 431
LNIT M+ RKDGH+S Y G++ P P DC HWCLPG+PD+WNELL
Sbjct: 364 LNITTMSEYRKDGHTSFYGSINGKLMTPEQKLDPRTFADCYHWCLPGLPDSWNELL 419
>At5g58600 unknown protein
Length = 402
Score = 243 bits (620), Expect = 2e-64
Identities = 130/355 (36%), Positives = 202/355 (56%), Gaps = 29/355 (8%)
Query: 92 CDLFDGEWVWDESYPLYQSKDCM-FLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNAT 150
C LF G WV D SYPLY+ DC ++ F C GR D Y ++RWQP++CN+P FN
Sbjct: 66 CSLFLGTWVRDNSYPLYKPADCPGVVEPEFDCQMYGRPDSDYLKYRWQPQNCNLPTFNGA 125
Query: 151 KMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFR 210
+ L +++ K ++FAGDS+G+NQWESL+C++ S + + +T+ F+F
Sbjct: 126 QFLLKMKGKTIMFAGDSLGKNQWESLICLIVSSAPSTRT-------EMTRGLPLSTFRFL 178
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNY 270
D+ T+ +Y+APFLV V GK LK+D++ N+ W +AD+LI NTGHWW++
Sbjct: 179 DYGITMSFYKAPFLV----DIDAVQGK--RVLKLDEISGNANAWHDADLLIFNTGHWWSH 232
Query: 271 EKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRG 330
+++G Q G + A ++++ T W+++ V+ S+TQV F +++P H
Sbjct: 233 TGSMQGWDLIQSGNSYYQDMDRFVAMEKALRTWAYWVETHVDRSRTQVLFLSISPTHDNP 292
Query: 331 GDW-----RNGGNCHSETLPDLGSSLVPNDNWSRFKAANA-VLSSHTNASEIMKIKVLNI 384
DW NC+ ET P G++ + + ++ VL N + + L+I
Sbjct: 293 SDWAASSSSGSKNCYGETEPITGTAYPVSSYTDQLRSVIVEVLHGMHNPAFL-----LDI 347
Query: 385 THMTAQRKDGHSSIYYLGRVAG---PAPLHRQDCSHWCLPGVPDTWNELLYAMLL 436
T +++ RKDGH S+ Y G ++G P DCSHWCLPG+PDTWN+LLY +L+
Sbjct: 348 TLLSSLRKDGHPSV-YSGLISGSQRSRPDQSADCSHWCLPGLPDTWNQLLYTLLI 401
>At3g11030 unknown protein
Length = 451
Score = 241 bits (615), Expect = 6e-64
Identities = 125/355 (35%), Positives = 194/355 (54%), Gaps = 21/355 (5%)
Query: 91 GCDLFDGEWVWDESY-PLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNA 149
GCD+F G WV D S PLY+ +C ++ C +GR D Y WRW+P C++P FNA
Sbjct: 105 GCDVFKGNWVKDWSTRPLYRESECPYIQPQLTCRTHGRPDSDYQSWRWRPDSCSLPSFNA 164
Query: 150 TKMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKF 209
T MLE LR K+++F GDS+ R + SL+C+L S + + G+ VF
Sbjct: 165 TVMLESLRGKKMMFVGDSLNRGMYVSLICLLHSQIPENSKSMDTFGS-------LTVFSL 217
Query: 210 RDFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWW- 268
+D+N T+E+Y APFL+ + V ++ ++ + W ADI++ NT WW
Sbjct: 218 KDYNATIEFYWAPFLLESNSDNATVHRVSDRIVRKGSINKHGRHWRGADIVVFNTYLWWR 277
Query: 269 -NYEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVH 327
++ I G + E + +++ EDAY+ +++T++ W++ +++P KT+VFF T++P H
Sbjct: 278 TGFKMKILEGSFKDEKKRI-VEMESEDAYRMALKTMVKWVKKNMDPLKTRVFFATMSPTH 336
Query: 328 FRGGDW--RNGGNCHSETLPDLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNIT 385
++G DW G NC+++T P + P+D K V+ + + VLNIT
Sbjct: 337 YKGEDWGGEQGKNCYNQTTPIQDMNHWPSD---CSKTLMKVIGEELDQRAEFPVTVLNIT 393
Query: 386 HMTAQRKDGHSSIYY-----LGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAML 435
++ RKD H+SIY L + P DC HWCLPG+ DTWNEL +A L
Sbjct: 394 QLSGYRKDAHTSIYKKQWSPLTKEQLANPASYSDCIHWCLPGLQDTWNELFFAKL 448
>At1g73140 hypothetical protein
Length = 403
Score = 241 bits (615), Expect = 6e-64
Identities = 128/356 (35%), Positives = 197/356 (54%), Gaps = 25/356 (7%)
Query: 92 CDLFDGEWVWDE-SYPLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNAT 150
C++F+G+WVWD SYPLY K C +L + C NGR D +Y WRW+P C++PRFNA
Sbjct: 57 CNVFEGQWVWDNVSYPLYTEKSCPYLVKQTTCQRNGRPDSYYQNWRWKPSSCDLPRFNAL 116
Query: 151 KMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFR 210
K+L+ LRNKR++F GDS+ R+ +ES++CM+ S + K+ + + +FK
Sbjct: 117 KLLDVLRNKRLMFIGDSVQRSTFESMVCMVQSVIPEKKKSFH-------RIPPMKIFKAE 169
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWNY 270
++N ++EYY APF+V + +K+D ++ +S W D+L+ + + +
Sbjct: 170 EYNASIEYYWAPFIVESISDHATNHTVHKRLVKLDAIEKHSKSWEGVDVLVFES--YKSL 227
Query: 271 EKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVHFRG 330
E + C + + +EV+ + +V AYK ++ET W ++ +N K +VFF +++P H
Sbjct: 228 EGKLIRICRYGDTSEVR-EYNVTTAYKMALETWAKWFKTKINSEKQKVFFTSMSPTHLWS 286
Query: 331 GDWRNG--GNCHSETLP-DLGSSLVPNDNWSRFKAANAVLSSHTNASEIMKIKVLNITHM 387
+W G G C+ E P D S N K VLS + LNIT +
Sbjct: 287 WEWNPGSDGTCYDELYPIDKRSYWGTGSNQEIMKIVGDVLSRVGE-----NVTFLNITQL 341
Query: 388 TAQRKDGHSSIY------YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAMLLK 437
+ RKDGH+++Y L + P + DC HWCLPGVPDTWNE+LYA LL+
Sbjct: 342 SEYRKDGHTTVYGERRGKLLTKEQRADPKNYGDCIHWCLPGVPDTWNEILYAYLLR 397
>At5g01360 unknown protein
Length = 434
Score = 237 bits (605), Expect = 9e-63
Identities = 126/357 (35%), Positives = 192/357 (53%), Gaps = 27/357 (7%)
Query: 92 CDLFDGEWVWDESY-PLYQSKDCMFLDEGFRCSENGRRDLFYTQWRWQPKDCNMPRFNAT 150
C++ G+WV++ S PLY + C ++D F C +NG+ + Y +W WQP DC +PRF+
Sbjct: 92 CNVAAGKWVYNSSIEPLYTDRSCPYIDRQFSCMKNGQPETDYLRWEWQPDDCTIPRFSPK 151
Query: 151 KMLERLRNKRVVFAGDSIGRNQWESLLCMLSSGVSNKESIYEVNGNPITKHKGFLVFKFR 210
+ +LR KR++F GDS+ R+QWES +C++ ESI + + + + VFK +
Sbjct: 152 LAMNKLRGKRLLFVGDSLQRSQWESFVCLV-------ESIIPEGEKSMKRSQKYFVFKAK 204
Query: 211 DFNCTVEYYRAPFLVLQSRPPTGVSGKIRTTLKVDKMDWNSLKWSNADILILNTGHWWN- 269
++N T+E+Y AP++V + +S + +KVD + + W ADIL+ NT WW
Sbjct: 205 EYNATIEFYWAPYIVESNTDIPVISDPKKRIVKVDSVKDRAKFWEGADILVFNTYVWWMS 264
Query: 270 --YEKTIRGGCYFQEGTEVKLKLHVEDAYKRSMETVLNWMQSSVNPSKTQVFFRTLAPVH 327
K + G F G L + AY+ ++T NW+ S+V+P+KT+VFF T++P H
Sbjct: 265 GLRMKALWGS--FGNGESGAEALDTQVAYRLGLKTWANWVDSTVDPNKTRVFFTTMSPTH 322
Query: 328 FRGGDW--RNGGNCHSETLPDLGSSL-VPNDNWSRFKAANAVLSSHTNASEIMKIKVLNI 384
R DW NG C +ET P N K ++V+ T + V+NI
Sbjct: 323 TRSADWGKPNGTKCFNETKPIKDKKFWGTGSNKQMMKVVSSVIKHMTT-----HVTVINI 377
Query: 385 THMTAQRKDGHSSIY------YLGRVAGPAPLHRQDCSHWCLPGVPDTWNELLYAML 435
T ++ R D H+S+Y L P+H DC HWCLPG+PDTWN +L A L
Sbjct: 378 TQLSEYRIDAHTSVYTETGGKILTAEQRADPMHHADCIHWCLPGLPDTWNRILLAHL 434
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,117,913
Number of Sequences: 26719
Number of extensions: 503829
Number of successful extensions: 1176
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 942
Number of HSP's gapped (non-prelim): 57
length of query: 449
length of database: 11,318,596
effective HSP length: 103
effective length of query: 346
effective length of database: 8,566,539
effective search space: 2964022494
effective search space used: 2964022494
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0387.8


