

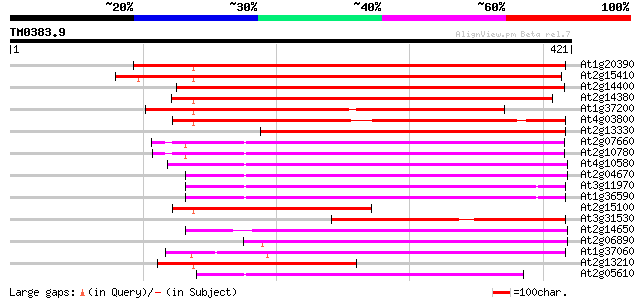
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0383.9
(421 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 358 3e-99
At2g15410 putative retroelement pol polyprotein 350 6e-97
At2g14400 putative retroelement pol polyprotein 325 3e-89
At2g14380 putative retroelement pol polyprotein 324 5e-89
At1g37200 hypothetical protein 281 4e-76
At4g03800 265 5e-71
At2g13330 F14O4.9 253 1e-67
At2g07660 putative retroelement pol polyprotein 193 1e-49
At2g10780 pseudogene 192 3e-49
At4g10580 putative reverse-transcriptase -like protein 179 2e-45
At2g04670 putative retroelement pol polyprotein 174 1e-43
At3g11970 hypothetical protein 172 3e-43
At1g36590 hypothetical protein 172 3e-43
At2g15100 putative retroelement pol polyprotein 168 4e-42
At3g31530 hypothetical protein 168 6e-42
At2g14650 putative retroelement pol polyprotein 166 2e-41
At2g06890 putative retroelement integrase 164 6e-41
At1g37060 Athila retroelment ORF 1, putative 159 3e-39
At2g13210 pseudogene 153 2e-37
At2g05610 putative retroelement pol polyprotein 152 3e-37
>At1g20390 hypothetical protein
Length = 1791
Score = 358 bits (919), Expect = 3e-99
Identities = 174/326 (53%), Positives = 231/326 (70%), Gaps = 2/326 (0%)
Query: 94 GAEIWREDTQDRDMLLGENLDLFAWSHKDMPGIDPNFICLALN--PGIEPVTQTRRQMGD 151
GAEI + LL N FAWS +DM GIDP LN P +PV Q RR++G
Sbjct: 728 GAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNVDPTFKPVKQKRRKLGP 787
Query: 152 VKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKWRMCVDYTDLNKACPKNSYP 211
+ +A+++EV KLL A I E+KYP W+ N V+VKK NGKWR+CVDYTDLNKACPK+SYP
Sbjct: 788 ERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVCVDYTDLNKACPKDSYP 847
Query: 212 LPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNA 271
LP ID+LV+ SGN L+S +DA+SGY+QI MH D++KT+F+T R YCY+ M FGLKNA
Sbjct: 848 LPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNA 907
Query: 272 GATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKC 331
GATYQR ++++ Q+GR +EVY+DDM+VKS+ H E L+K F L + M+LNP KC
Sbjct: 908 GATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKCFDVLNTYGMKLNPTKC 967
Query: 332 SFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGD 391
+FG+ G+FLG+++T RGIE N + +AI E+ SP + +EVQ L GRIAAL+RF+ S D
Sbjct: 968 TFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRLTGRIAALNRFISRSTD 1027
Query: 392 KSAPFFKCLKKNAAFEWNSECEEAFK 417
K PF+ LK+ A F+W+ + EEAF+
Sbjct: 1028 KCLPFYNLLKRRAQFDWDKDSEEAFE 1053
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 350 bits (899), Expect = 6e-97
Identities = 169/343 (49%), Positives = 236/343 (68%), Gaps = 8/343 (2%)
Query: 80 RGTSQQAHADRGDQGA------EIWREDTQDRDMLLGENLDLFAWSHKDMPGIDPNFICL 133
+GT Q + D D ++ E + L +N FAWS +DMPGID C
Sbjct: 713 KGTVVQVNIDESDPSRCVGIRIDLPSELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCH 772
Query: 134 ALN--PGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGK 191
LN P +P+ Q RR++G + K +++EV KLL A I E++YP W+ N V+VKK NGK
Sbjct: 773 ELNVDPTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGK 832
Query: 192 WRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTA 251
WR+C+D+TDLNKACPK+S+PLP ID+LV+ +GNEL+S +DA+SGY+QI MH +D +KT
Sbjct: 833 WRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTV 892
Query: 252 FMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHED 311
F+T + YCY+ MPFGLKNAGATY RL++++F Q+ +MEVY+DDM+VKS+ H
Sbjct: 893 FITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITH 952
Query: 312 LTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKE 371
L + F L ++NM+LNP KC+FG+ G+FLG+++T RGIE N + AI ++ SP + +E
Sbjct: 953 LRQCFQVLNRYNMKLNPSKCTFGVTSGEFLGYLVTRRGIEANPKQISAIIDLPSPRNTRE 1012
Query: 372 VQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEE 414
VQ LIGRIAAL+RF+ S DK PF++ L+ N FEW+ +CEE
Sbjct: 1013 VQRLIGRIAALNRFISRSTDKCLPFYQLLRANKRFEWDEKCEE 1055
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 325 bits (833), Expect = 3e-89
Identities = 152/291 (52%), Positives = 211/291 (72%)
Query: 126 IDPNFICLALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMV 185
+D F+ GI RR++G + KA++ EV+KLL IRE++YP W+ N V+V
Sbjct: 449 VDLIFLSTLERMGISRADVNRRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVV 508
Query: 186 KKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPS 245
KK NGK R+C+D+TDLNKACPK+S+PLP ID+LV+ +GNEL+S +DA+SGY+QI M+P
Sbjct: 509 KKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPE 568
Query: 246 DEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLG 305
D++KT F+T R YCY+ MPFGL+NAGATY RL++++F VG+ MEVY+DDM++KS+
Sbjct: 569 DQEKTLFITDRGIYCYKVMPFGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKK 628
Query: 306 SSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKS 365
H + L + FA L ++ M+LNP KC+FG+ G+FLG+++T RGIE N ++ A M S
Sbjct: 629 EDHVKHLEECFAILNQYQMKLNPAKCTFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPS 688
Query: 366 PGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAF 416
P + KEVQ L GRIAAL+RF+ S DKS PF++ LK N F W+ +CEEAF
Sbjct: 689 PKNFKEVQRLTGRIAALNRFISRSTDKSLPFYQILKGNKEFLWDEKCEEAF 739
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 324 bits (831), Expect = 5e-89
Identities = 154/289 (53%), Positives = 210/289 (72%), Gaps = 3/289 (1%)
Query: 122 DMPGIDPNFICLALN--PGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWM 179
DM GIDP C LN P + V Q RR++G + KA++ EV+KLL A I E+KYP W+
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 180 TNVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQ 239
N V+VKK N KWR+C+D+TDLNKACPK+S+PLP ID++V+ +GNEL+S +DA+SGY+Q
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 240 IKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMI 299
I MH D++KT+F+ R YCY+ MPFGLKN GA YQRL++++F Q+G+ MEVY+DDM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 300 VKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKA 359
VKS + H + L F L K+NM+LNP KC FG+ G+FLG+++T RGIE N + +A
Sbjct: 656 VKSTRSADHIDHLKACFETLNKYNMKLNPAKCLFGVTSGEFLGYIVTKRGIEANPKQIRA 715
Query: 360 IQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKK-NAAFE 407
I +++SP + KEVQ L GRIA L+RF+ S DKS PF++ L+ N FE
Sbjct: 716 ILDLQSPRNKKEVQRLTGRIAGLNRFIARSTDKSLPFYQLLRSANKTFE 764
>At1g37200 hypothetical protein
Length = 1564
Score = 281 bits (720), Expect = 4e-76
Identities = 136/271 (50%), Positives = 184/271 (67%), Gaps = 6/271 (2%)
Query: 103 QDRDMLLGENLDLFAWSHKDMPGIDPNFICLALN--PGIEPVTQTRRQMGDVKEKAIHQE 160
+D L EN D FAWS ++ GI LN P P+ Q RR++G + KA+ E
Sbjct: 645 EDFITFLKENKDSFAWSSANLQGISLEVTSHELNVDPTYRPIKQKRRKLGPERAKAVQDE 704
Query: 161 VNKLLAADFIREIKYPTWMTNVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVD 220
V++LL IRE+KYP W+ N V+VK NGKWR+C+D+ DLNKACPK+S+PLP ID+LV
Sbjct: 705 VDRLLKIGSIREVKYPDWLANPVVVKNKNGKWRVCIDFMDLNKACPKDSFPLPHIDRLVK 764
Query: 221 GASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMD 280
+G+EL+S +DA+SGY+QI M P D++KTAF+T CY+ MPFGLKN GATYQRL++
Sbjct: 765 ATAGHELLSFMDAFSGYNQILMRPDDQEKTAFITD----CYKVMPFGLKNTGATYQRLVN 820
Query: 281 RVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKF 340
R+F Q+G+ ME+Y+DDM+VKS H L + F L K M+LNPEKCSFG+ G+F
Sbjct: 821 RMFADQLGKTMELYIDDMLVKSAHEKDHLPQLRECFKILNKFEMKLNPEKCSFGVPSGEF 880
Query: 341 LGFMITTRGIEINLDKCKAIQEMKSPGSVKE 371
LG+++T RGIE N + A +M SP ++
Sbjct: 881 LGYLVTQRGIEANPKQIAAFIDMPSPKMARD 911
>At4g03800
Length = 637
Score = 265 bits (676), Expect = 5e-71
Identities = 134/297 (45%), Positives = 197/297 (66%), Gaps = 23/297 (7%)
Query: 123 MPGIDPNFICLALN--PGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMT 180
MP IDP+ IC LN P +P+ Q RR++G + KA++ +++KLL IRE++YP W+
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 181 NVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQI 240
V+VKK NGK R+C+D+TDLNKACPK+S+PLP ID+LV+ +GNEL++ +DA+ GY+QI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 241 KMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIV 300
M+P D++KT+F+T R GATYQ L++++F + + MEV +DD +V
Sbjct: 441 MMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 301 KSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAI 360
KS+ H + L + F L ++ M+LN KC+FG+ G+FLG+++T RGIE N ++ A
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNLAKCTFGVPSGEFLGYIVTKRGIEANPNQINAF 545
Query: 361 QEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFK 417
+ S + KEVQ L GRIA+ S DKS PF++ LK N F W+ +CEEAF+
Sbjct: 546 LKTPSLRNFKEVQRLTGRIAS------RSTDKSLPFYQILKGNNGFLWDEKCEEAFR 596
>At2g13330 F14O4.9
Length = 889
Score = 253 bits (646), Expect = 1e-67
Identities = 119/229 (51%), Positives = 161/229 (69%)
Query: 189 NGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDED 248
NGKWR+C+D+ DLNKACPK+S+PLP ID+LV+ +E +S +DA+ GY+QI M ++
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 249 KTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSH 308
KTAF+T R Y Y+ MPFGLKNAG TYQRL++R+F Q+G+ +EVY+DDM+VKS H
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 309 HEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGS 368
L + F L K M+LNPEKCSF +Q +FL +++T RGIE N + A EM SP
Sbjct: 411 VPQLRECFKILIKFEMKLNPEKCSFEVQSREFLEYLVTERGIEANPKQIAAFIEMPSPKM 470
Query: 369 VKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFK 417
+EVQ L GRI AL+ F+ S +K PF++ L+K F+WN +CE+AFK
Sbjct: 471 AREVQRLTGRIPALNGFISRSANKCVPFYQPLRKGKEFDWNKDCEQAFK 519
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 193 bits (491), Expect = 1e-49
Identities = 105/313 (33%), Positives = 172/313 (54%), Gaps = 8/313 (2%)
Query: 107 MLLGENLDLFAWSHKDMPGIDPNF---ICLALNPGIEPVTQTRRQMGDVKEKAIHQEVNK 163
+++ E D+FA + G+ P+ + L PG P+++ +M + + +++ +
Sbjct: 121 LIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAELKKQLEE 176
Query: 164 LLAADFIREIKYPTWMTNVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGAS 223
LLA FIR P W V+ VKK +G +R+C+DY LNK KN YPLP ID+L+D
Sbjct: 177 LLAKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLG 235
Query: 224 GNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVF 283
G + S ID SGYHQI + P+D KTAF T ++ + MPFGL NA A + ++M+ VF
Sbjct: 236 GAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVF 295
Query: 284 EGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGF 343
+ + +++DD++V S +H E L RLR+H + K SF + FLG
Sbjct: 296 RDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQRSVGFLGH 355
Query: 344 MITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKN 403
+I+ +G+ ++ +K ++I+E P + E++ +G RF+ + P + K+
Sbjct: 356 VISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKD 415
Query: 404 AAFEWNSECEEAF 416
AF W+ ECE++F
Sbjct: 416 TAFNWSDECEKSF 428
>At2g10780 pseudogene
Length = 1611
Score = 192 bits (488), Expect = 3e-49
Identities = 104/312 (33%), Positives = 171/312 (54%), Gaps = 8/312 (2%)
Query: 108 LLGENLDLFAWSHKDMPGIDPNF---ICLALNPGIEPVTQTRRQMGDVKEKAIHQEVNKL 164
++ E D+FA + G+ P+ + L PG P+++ +M + + +++ +L
Sbjct: 627 IVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEEL 682
Query: 165 LAADFIREIKYPTWMTNVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASG 224
L FIR P W V+ VKK +G +R+C+DY LNK KN YPLP ID+L+D G
Sbjct: 683 LDKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGG 741
Query: 225 NELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFE 284
+ S ID SGYHQI + P+D KTAF T ++ + MPFGL NA A + ++M+ VF
Sbjct: 742 AQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFR 801
Query: 285 GQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFM 344
+ + ++++D++V S +H E L RLR+H + KCSF + FLG +
Sbjct: 802 DFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHV 861
Query: 345 ITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNA 404
I+ +G+ ++ +K ++I+E P + E++ +G RF+ + P + K+
Sbjct: 862 ISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDT 921
Query: 405 AFEWNSECEEAF 416
AF W+ ECE++F
Sbjct: 922 AFNWSDECEKSF 933
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 179 bits (455), Expect = 2e-45
Identities = 98/300 (32%), Positives = 158/300 (52%), Gaps = 1/300 (0%)
Query: 119 SHKDMPGIDPNFICLALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTW 178
S + +P + + L PG P+++ +M + + +++ LL FIR P W
Sbjct: 469 SLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSP-W 527
Query: 179 MTNVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYH 238
V+ VKK +G +R+C+DY +LN+ KN YPLP ID+L+D G S ID SGYH
Sbjct: 528 GAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYH 587
Query: 239 QIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDM 298
QI + +D KTAF T ++ + MPFGL NA A + RLM+ VF+ + + +++DD+
Sbjct: 588 QIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDI 647
Query: 299 IVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCK 358
+V S L + +LR+ + KCSF + FLG +++ G+ ++ +K +
Sbjct: 648 LVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIE 707
Query: 359 AIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFKS 418
AI++ P + E++ +G RF+ + P K K+ F W+ ECEE F S
Sbjct: 708 AIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVS 767
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 174 bits (440), Expect = 1e-43
Identities = 95/286 (33%), Positives = 150/286 (52%), Gaps = 1/286 (0%)
Query: 133 LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKW 192
+ L PG P+++ +M + + +++ LL FIR P W V+ VKK +G +
Sbjct: 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 567
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAF 252
R+C+DY LN KN YPLP ID+L+D G S ID SGYHQI + +D KTAF
Sbjct: 568 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 627
Query: 253 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDL 312
T ++ + MPF L NA A + RLM+ VF+ + + +++DD++V S H L
Sbjct: 628 RTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHL 687
Query: 313 TKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEV 372
+ +LR+ + KCSF + FLG +++ G+ ++ +K +AI++ P + E+
Sbjct: 688 RRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEI 747
Query: 373 QHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFKS 418
+ + RF+ + P K K+ F W+ ECEE F S
Sbjct: 748 RSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVS 793
>At3g11970 hypothetical protein
Length = 1499
Score = 172 bits (436), Expect = 3e-43
Identities = 98/285 (34%), Positives = 158/285 (55%), Gaps = 2/285 (0%)
Query: 133 LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKW 192
+ L G PV Q + ++ I + V LL ++ P + + VV+VKK +G W
Sbjct: 593 IKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTW 651
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAF 252
R+CVDY +LN K+S+P+P I+ L+D G + S ID +GYHQ++M P D KTAF
Sbjct: 652 RLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAF 711
Query: 253 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDL 312
T ++ Y MPFGL NA AT+Q LM+ +F+ + + + V+ DD++V S H + L
Sbjct: 712 KTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHL 771
Query: 313 TKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEV 372
+ F +R + + KC+F + ++LG I+ +GIE + K KA++E P ++K++
Sbjct: 772 KQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQL 831
Query: 373 QHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFK 417
+ +G RF+ G + P L K AFEW + ++AF+
Sbjct: 832 RGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFE 875
>At1g36590 hypothetical protein
Length = 1499
Score = 172 bits (436), Expect = 3e-43
Identities = 98/285 (34%), Positives = 158/285 (55%), Gaps = 2/285 (0%)
Query: 133 LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKW 192
+ L G PV Q + ++ I + V LL ++ P + + VV+VKK +G W
Sbjct: 593 IKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDGTW 651
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAF 252
R+CVDY +LN K+S+P+P I+ L+D G + S ID +GYHQ++M P D KTAF
Sbjct: 652 RLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAF 711
Query: 253 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDL 312
T ++ Y MPFGL NA AT+Q LM+ +F+ + + + V+ DD++V S H + L
Sbjct: 712 KTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQHL 771
Query: 313 TKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEV 372
+ F +R + + KC+F + ++LG I+ +GIE + K KA++E P ++K++
Sbjct: 772 KQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAVKEWPQPTTLKQL 831
Query: 373 QHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFK 417
+ +G RF+ G + P L K AFEW + ++AF+
Sbjct: 832 RGFLGLAGYYRRFVRSFGVIAGP-LHALTKTDAFEWTAVAQQAFE 875
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 168 bits (426), Expect = 4e-42
Identities = 80/151 (52%), Positives = 110/151 (71%), Gaps = 2/151 (1%)
Query: 123 MPGIDPNFICLALN--PGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMT 180
M GI I LN P +PV Q RR++G + +A++ EV +LL IRE+KYP W+
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 181 NVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQI 240
N V+VKK NGKWR+CVD+TDLNKAC K+ +PLP ID+LV+ +G+E++S +DA+SGY+QI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 241 KMHPSDEDKTAFMTARVNYCYQTMPFGLKNA 271
M+P D++KT+F+T Y Y+ MPFGLKNA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
>At3g31530 hypothetical protein
Length = 831
Score = 168 bits (425), Expect = 6e-42
Identities = 80/176 (45%), Positives = 117/176 (66%), Gaps = 11/176 (6%)
Query: 242 MHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVK 301
M+P D++KTAF T + +CY+ MPFGLKNAGATY+R ++++F Q+G+ MEVY+DDM+VK
Sbjct: 96 MNPEDQEKTAFYTEQGIFCYRVMPFGLKNAGATYERFVNKIFALQIGKTMEVYIDDMLVK 155
Query: 302 SVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQ 361
S+ H L + F +L +N++LNP KC FG++ GIE N + +A+
Sbjct: 156 SMTEKDHISHLRECFKQLNLYNVKLNPAKCRFGVRS-----------GIEANPKQIEALL 204
Query: 362 EMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFK 417
M SP + +EVQ L GR+AAL+RF+ S +K F+ L++N FEW + CEEAF+
Sbjct: 205 GMASPQNKREVQCLTGRVAALNRFISRSTEKCLAFYDVLRRNKKFEWTTRCEEAFQ 260
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 166 bits (420), Expect = 2e-41
Identities = 92/286 (32%), Positives = 148/286 (51%), Gaps = 13/286 (4%)
Query: 133 LALNPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKW 192
+ L PG P+++ +M + + +++ LL A V+ VKK +G +
Sbjct: 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDLLGAP-------------VLFVKKKDGSF 516
Query: 193 RMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAF 252
R+C+DY LN KN YPLP ID+L+D G S ID SGYH I + +D KTAF
Sbjct: 517 RLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAF 576
Query: 253 MTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDL 312
T ++ + MPFGL NA A + RLM+ VF+ + + +++DD++V S H L
Sbjct: 577 RTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHL 636
Query: 313 TKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEV 372
+ +LR+ + KCSF + FLG +++ G+ ++ +K +AI++ +P + E+
Sbjct: 637 RRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEI 696
Query: 373 QHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSECEEAFKS 418
+ +G RF+ + P K K+ F W+ ECEE F S
Sbjct: 697 RSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVS 742
>At2g06890 putative retroelement integrase
Length = 1215
Score = 164 bits (416), Expect = 6e-41
Identities = 92/247 (37%), Positives = 134/247 (54%), Gaps = 4/247 (1%)
Query: 176 PTWMTNVVMVKKA----NGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMSLI 231
P + TN V K+ +G WRMC D +N K +P+P +D ++D G+ + S I
Sbjct: 449 PAYRTNPVETKELQRQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKI 508
Query: 232 DAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQRLMDRVFEGQVGRNM 291
D SGYHQI+M+ DE KTAF T Y + MPFGL +A +T+ RLM+ V +G +
Sbjct: 509 DLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFV 568
Query: 292 EVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQGGKFLGFMITTRGIE 351
VY DD++V S H E L LRK + N +KC+F FLGF+++ G++
Sbjct: 569 IVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVK 628
Query: 352 INLDKCKAIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPFFKCLKKNAAFEWNSE 411
++ +K KAI++ SP +V EV+ G RF AP + +KK+ F+W
Sbjct: 629 VDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKA 688
Query: 412 CEEAFKS 418
EEAF+S
Sbjct: 689 QEEAFQS 695
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 159 bits (402), Expect = 3e-39
Identities = 100/321 (31%), Positives = 156/321 (48%), Gaps = 22/321 (6%)
Query: 118 WSHKDMPGIDPNFICLAL---NPGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIK 174
+S D+ GI P + N + RR ++KE + +E+ KLL A I I
Sbjct: 874 YSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKE-VVKKEILKLLDAGVIYPIS 932
Query: 175 YPTWMTNVVMVKKANGKW------------------RMCVDYTDLNKACPKNSYPLPSID 216
TW++ V V K G RMC++Y LN A K +PLP ID
Sbjct: 933 DSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFID 992
Query: 217 KLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVNYCYQTMPFGLKNAGATYQ 276
+++ + + +D+YSG+ QI +HP+D+ KT F + Y+ MPFGL NA AT+Q
Sbjct: 993 HMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQ 1052
Query: 277 RLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFARLRKHNMRLNPEKCSFGIQ 336
R M +F + +EV++DD V SS +L + R + N+ LN EKC F ++
Sbjct: 1053 RCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKRCEETNLVLNWEKCHFMVR 1112
Query: 337 GGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGRIAALSRFLPHSGDKSAPF 396
G LG I+ GIE++ K + +++ P +VK+++ +G F+ + P
Sbjct: 1113 EGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGHAGFYRIFIKDFSKLARPL 1172
Query: 397 FKCLKKNAAFEWNSECEEAFK 417
+ L K F ++ EC AFK
Sbjct: 1173 TRLLCKETEFAFDDECLTAFK 1193
>At2g13210 pseudogene
Length = 152
Score = 153 bits (386), Expect = 2e-37
Identities = 69/151 (45%), Positives = 103/151 (67%), Gaps = 2/151 (1%)
Query: 112 NLDLFAWSHKDMPGIDPNFICLALN--PGIEPVTQTRRQMGDVKEKAIHQEVNKLLAADF 169
++ F WS +D+PG+ + LN P +PV Q RR++G + +A+ EV KLL
Sbjct: 2 DVSTFVWSPEDLPGVSVEIVAHELNVDPTFKPVKQKRRKLGRERAEAVKAEVEKLLRIGS 61
Query: 170 IREIKYPTWMTNVVMVKKANGKWRMCVDYTDLNKACPKNSYPLPSIDKLVDGASGNELMS 229
I + KYP W+ N V+VKK NGKWR+C+D+TDLNKACPK+S+P+ I +LV+ SG++L+S
Sbjct: 62 ITKAKYPEWLANPVVVKKKNGKWRVCIDFTDLNKACPKDSFPMSHIGRLVESTSGDKLLS 121
Query: 230 LIDAYSGYHQIKMHPSDEDKTAFMTARVNYC 260
+DA++GY+QI M+ D++KT F T + C
Sbjct: 122 FMDAFAGYNQIMMNSEDQEKTTFYTEQGIVC 152
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 152 bits (384), Expect = 3e-37
Identities = 85/245 (34%), Positives = 138/245 (55%), Gaps = 1/245 (0%)
Query: 141 PVTQTRRQMGDVKEKAIHQEVNKLLAADFIREIKYPTWMTNVVMVKKANGKWRMCVDYTD 200
PV Q + ++ I + V+ +LA+ I+ P + + VV+VKK +G WR+CVDY +
Sbjct: 43 PVNQRPYRYVVHQKNEIDKIVDDMLASGTIQASSSP-YASPVVLVKKKDGTWRLCVDYRE 101
Query: 201 LNKACPKNSYPLPSIDKLVDGASGNELMSLIDAYSGYHQIKMHPSDEDKTAFMTARVNYC 260
LN K+ +P+P I+ L+D G+ + S ID +GYHQ++M P D KTAF T +Y
Sbjct: 102 LNGMTVKDRFPIPLIEDLMDELGGSNVYSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYE 161
Query: 261 YQTMPFGLKNAGATYQRLMDRVFEGQVGRNMEVYVDDMIVKSVLGSSHHEDLTKAFARLR 320
Y MPFGL NA A++Q LM+ F+ + + + V+ DD+++ S H + L F +R
Sbjct: 162 YLVMPFGLTNAPASFQSLMNSFFKPFLRKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMR 221
Query: 321 KHNMRLNPEKCSFGIQGGKFLGFMITTRGIEINLDKCKAIQEMKSPGSVKEVQHLIGRIA 380
H++ KC+F + ++LG I+ GI + K KA+Q+ P ++K++ +G
Sbjct: 222 VHHLFAKMSKCAFAVPRVEYLGHFISGEGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTG 281
Query: 381 ALSRF 385
RF
Sbjct: 282 YYRRF 286
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,004,926
Number of Sequences: 26719
Number of extensions: 432073
Number of successful extensions: 895
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 814
Number of HSP's gapped (non-prelim): 65
length of query: 421
length of database: 11,318,596
effective HSP length: 102
effective length of query: 319
effective length of database: 8,593,258
effective search space: 2741249302
effective search space used: 2741249302
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0383.9


