

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0383.7
(316 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
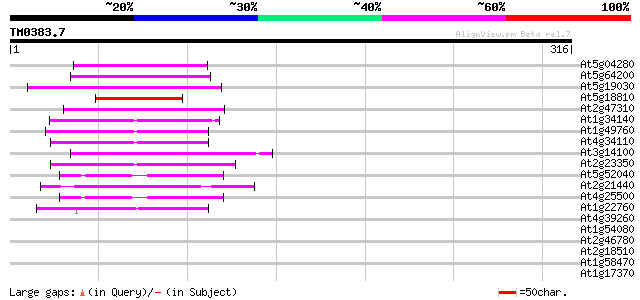
Score E
Sequences producing significant alignments: (bits) Value
At5g04280 RNA-binding protein-like 49 4e-06
At5g64200 unknown protein 47 1e-05
At5g19030 unknown protein 47 2e-05
At5g18810 Serine/arginine rich protein - like 46 2e-05
At2g47310 putative FCA-related protein 46 2e-05
At1g34140 putative protein 45 4e-05
At1g49760 Putative Poly-A Binding Protein (F14J22.3) 45 7e-05
At4g34110 poly(A)-binding protein 43 2e-04
At3g14100 oligouridylate binding protein, putative 43 2e-04
At2g23350 putative poly(A) binding protein 43 3e-04
At5g52040 arginine/serine-rich splicing factor RSP41 homolog 42 6e-04
At2g21440 unknown protein (At2g21440) 42 6e-04
At4g25500 splicing factor At-SRp40 41 7e-04
At1g22760 putative polyA-binding protein, PAB3 41 7e-04
At4g39260 glycine-rich protein (clone AtGRP8) 40 0.001
At1g54080 unknown protein 40 0.001
At2g46780 putative RNA-binding protein 40 0.002
At2g18510 putative spliceosome associated protein 40 0.002
At1g58470 unknown protein 40 0.002
At1g17370 oligouridylate binding protein, putative 40 0.002
>At5g04280 RNA-binding protein-like
Length = 310
Score = 48.9 bits (115), Expect = 4e-06
Identities = 28/75 (37%), Positives = 39/75 (51%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
IFV G+S V+ DL R F R+G + + R GR F FI F + E+IR +
Sbjct: 9 IFVGGLSPEVTDRDLERAFSRFGDILDCQIMLERDTGRSRGFGFITFADRRAMDESIREM 68
Query: 97 NGSWFFESFLMVNRA 111
+G F + + VNRA
Sbjct: 69 HGRDFGDRVISVNRA 83
>At5g64200 unknown protein
Length = 303
Score = 47.4 bits (111), Expect = 1e-05
Identities = 24/79 (30%), Positives = 44/79 (55%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIR 94
+++ V ++ + DL +F +YG + VF+ R R+ G FAF+R+KY+ +A +A+
Sbjct: 16 YSLLVLNITFRTTADDLYPLFAKYGKVVDVFIPRDRRTGDSRGFAFVRYKYKDEAHKAVE 75
Query: 95 SLNGSWFFESFLMVNRARY 113
L+G + V A+Y
Sbjct: 76 RLDGRVVDGREITVQFAKY 94
>At5g19030 unknown protein
Length = 172
Score = 46.6 bits (109), Expect = 2e-05
Identities = 25/109 (22%), Positives = 52/109 (46%)
Query: 11 PVSRPEVVGSHGVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRR 70
P+ V R+ + ++FV G SD+VS L+++F +G + V +
Sbjct: 53 PLRTISCVAGDDETREASSLPSSISSLFVKGFSDSVSEGRLKKVFSEFGQVTNVKIIANE 112
Query: 71 KQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRARYDYTNRR 119
+ + + ++ F ++DA A+ ++NG +F F++V + + RR
Sbjct: 113 RTRQSLGYGYVWFNSKEDAQSAVEAMNGKFFDGRFILVKFGQPGLSRRR 161
>At5g18810 Serine/arginine rich protein - like
Length = 147
Score = 46.2 bits (108), Expect = 2e-05
Identities = 19/49 (38%), Positives = 31/49 (62%)
Query: 49 HDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLN 97
+DLR FER+G L ++L R G F F++++Y +DA EA++ +N
Sbjct: 68 NDLRDSFERFGPLKDIYLPRNYYTGEPRGFGFVKYRYAEDAAEAMKRMN 116
>At2g47310 putative FCA-related protein
Length = 324
Score = 46.2 bits (108), Expect = 2e-05
Identities = 20/91 (21%), Positives = 48/91 (51%)
Query: 31 DKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDAC 90
D + ++V +S + +D+R++FE+YG + + L + + G ++ + FI++K ++
Sbjct: 106 DGSIAKLYVAPISKTATEYDIRQVFEKYGNVTEIILPKDKMTGERAAYCFIKYKKVEEGN 165
Query: 91 EAIRSLNGSWFFESFLMVNRARYDYTNRRNV 121
AI +L + F ++ + R+ R +
Sbjct: 166 AAIAALTEQFTFPGEMLPVKVRFAEAERERI 196
Score = 30.8 bits (68), Expect = 1.0
Identities = 14/62 (22%), Positives = 36/62 (57%), Gaps = 1/62 (1%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
++V ++ + ++ +F RYG++ +++ + + +AF++F ++ A AI++L
Sbjct: 213 LYVRCLNKQTTKMEVNEVFSRYGIIEDIYMALDDMKICRG-YAFVQFSCKEMALAAIKAL 271
Query: 97 NG 98
NG
Sbjct: 272 NG 273
>At1g34140 putative protein
Length = 398
Score = 45.4 bits (106), Expect = 4e-05
Identities = 28/96 (29%), Positives = 47/96 (48%), Gaps = 2/96 (2%)
Query: 23 VDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIR 82
V R QW + ++V + + + DL+R+F +G + + + +G+ RF F+
Sbjct: 107 VSRGQWDKSRVFTNVYVKNLVETATDADLKRLFGEFGEITSAVVMKDG-EGKSRRFGFVN 165
Query: 83 FKYEQDACEAIRSLNGSWFFESFLMVNRARYDYTNR 118
F+ + A AI +NG E L V RA+ TNR
Sbjct: 166 FEKAEAAVTAIEKMNGVVVDEKELHVGRAQRK-TNR 200
>At1g49760 Putative Poly-A Binding Protein (F14J22.3)
Length = 671
Score = 44.7 bits (104), Expect = 7e-05
Identities = 24/92 (26%), Positives = 45/92 (48%), Gaps = 1/92 (1%)
Query: 21 HGVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAF 80
H + R K ++V +S+++S +L ++F +GV + R +G+ F F
Sbjct: 210 HKLQRDPSGEKVKFTNVYVKNLSESLSDEELNKVFGEFGVTTSCVIMRDG-EGKSKGFGF 268
Query: 81 IRFKYEQDACEAIRSLNGSWFFESFLMVNRAR 112
+ F+ DA A+ +LNG F + V +A+
Sbjct: 269 VNFENSDDAARAVDALNGKTFDDKEWFVGKAQ 300
>At4g34110 poly(A)-binding protein
Length = 629
Score = 43.1 bits (100), Expect = 2e-04
Identities = 23/89 (25%), Positives = 42/89 (46%), Gaps = 1/89 (1%)
Query: 24 DRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRF 83
+R K ++V ++++ + DL+ F YG + + + +G+ F F+ F
Sbjct: 204 ERDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDG-EGKSKGFGFVNF 262
Query: 84 KYEQDACEAIRSLNGSWFFESFLMVNRAR 112
+ DA A+ SLNG F + V RA+
Sbjct: 263 ENADDAARAVESLNGHKFDDKEWYVGRAQ 291
>At3g14100 oligouridylate binding protein, putative
Length = 427
Score = 43.1 bits (100), Expect = 2e-04
Identities = 31/114 (27%), Positives = 49/114 (42%), Gaps = 1/114 (0%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIR 94
F IFV +S V+ L + F + + +K GR F F+ F+ +QDA AI
Sbjct: 144 FNIFVGDLSPEVTDATLYQSFSVFSSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQTAIN 203
Query: 95 SLNGSWFFESFLMVNRARYDYTNRRNVTNRHSKVWKQKFVGEKQAFGQETVVSE 148
+NG W + N A T+ + + K + G + G+ET+ E
Sbjct: 204 EMNGKWLSSRQIRCNWATKGATSGDDKLSSDGKSVVELTTGSSED-GKETLNEE 256
>At2g23350 putative poly(A) binding protein
Length = 662
Score = 42.7 bits (99), Expect = 3e-04
Identities = 25/105 (23%), Positives = 51/105 (47%), Gaps = 2/105 (1%)
Query: 24 DRQQWRTDKKVFT-IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIR 82
+ ++ DK FT ++V +S+ + +L+ F +YG + + R G+ F F+
Sbjct: 213 EERESAADKMKFTNVYVKNLSEATTDDELKTTFGQYGSISSAVVMRDG-DGKSRCFGFVN 271
Query: 83 FKYEQDACEAIRSLNGSWFFESFLMVNRARYDYTNRRNVTNRHSK 127
F+ +DA A+ +LNG F + V +A+ ++ R+ +
Sbjct: 272 FENPEDAARAVEALNGKKFDDKEWYVGKAQKKSERELELSRRYEQ 316
>At5g52040 arginine/serine-rich splicing factor RSP41 homolog
Length = 356
Score = 41.6 bits (96), Expect = 6e-04
Identities = 29/92 (31%), Positives = 44/92 (47%), Gaps = 10/92 (10%)
Query: 29 RTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQD 88
R K +F I D + N DL R FE YG + V ++R FAFI+++ ++D
Sbjct: 93 RPSKTLFVINFD--AQNTRTRDLERHFEPYGKIVNVRIRRN--------FAFIQYEAQED 142
Query: 89 ACEAIRSLNGSWFFESFLMVNRARYDYTNRRN 120
A A+ + N S + + V A D +R N
Sbjct: 143 ATRALDATNSSKLMDKVISVEYAVKDDDSRGN 174
Score = 32.3 bits (72), Expect = 0.35
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 11/67 (16%)
Query: 50 DLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVN 109
DL R+F +YG + V + K+ FAF+ + E+DA +AIR+L+ FE
Sbjct: 17 DLERLFRKYGKVERVDM--------KAGFAFVYMEDERDAEDAIRALDR---FEYGRTGR 65
Query: 110 RARYDYT 116
R R ++T
Sbjct: 66 RLRVEWT 72
>At2g21440 unknown protein (At2g21440)
Length = 1003
Score = 41.6 bits (96), Expect = 6e-04
Identities = 27/121 (22%), Positives = 53/121 (43%), Gaps = 12/121 (9%)
Query: 18 VGSHGVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSR 77
+G G Q+W+ + + + D++ +F G + VF+ + + G
Sbjct: 321 LGGEGSKAQKWK-------LIIRNLPFQAKPSDIKVVFSAVGFVWDVFIPKNFETGLPKG 373
Query: 78 FAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRARYDYTNRRNVTNRHSKVWKQKFVGEK 137
FAF++F ++DA AI+ NG F + + V D+ +N+ N + G+K
Sbjct: 374 FAFVKFTCKKDAANAIKKFNGHMFGKRPIAV-----DWAVPKNIYNGAADATTASADGDK 428
Query: 138 Q 138
+
Sbjct: 429 E 429
Score = 34.7 bits (78), Expect = 0.070
Identities = 18/64 (28%), Positives = 29/64 (45%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRS 95
T+ V G+ +++ L F G + FL + FAF++F ++D AI
Sbjct: 21 TVCVSGLPYSITNAQLEEAFSEVGPVRRCFLVTNKGSDEHRGFAFVKFALQEDVNRAIEL 80
Query: 96 LNGS 99
NGS
Sbjct: 81 KNGS 84
>At4g25500 splicing factor At-SRp40
Length = 350
Score = 41.2 bits (95), Expect = 7e-04
Identities = 29/92 (31%), Positives = 44/92 (47%), Gaps = 10/92 (10%)
Query: 29 RTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQD 88
R K +F I D +DN DL + FE YG + V ++R FAFI+++ ++D
Sbjct: 94 RPSKTLFVINFD--ADNTRTRDLEKHFEPYGKIVNVRIRRN--------FAFIQYEAQED 143
Query: 89 ACEAIRSLNGSWFFESFLMVNRARYDYTNRRN 120
A A+ + N S + + V A D R N
Sbjct: 144 ATRALDASNNSKLMDKVISVEYAVKDDDARGN 175
Score = 32.7 bits (73), Expect = 0.26
Identities = 24/77 (31%), Positives = 39/77 (50%), Gaps = 11/77 (14%)
Query: 50 DLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVN 109
DL R+F +YG + V + K+ FAF+ + E+DA +AIR+L+ FE
Sbjct: 17 DLERLFRKYGKVERVDM--------KAGFAFVYMEDERDAEDAIRALDR---FEFGRKGR 65
Query: 110 RARYDYTNRRNVTNRHS 126
R R ++T ++ S
Sbjct: 66 RLRVEWTKSERGGDKRS 82
>At1g22760 putative polyA-binding protein, PAB3
Length = 660
Score = 41.2 bits (95), Expect = 7e-04
Identities = 25/102 (24%), Positives = 47/102 (45%), Gaps = 6/102 (5%)
Query: 16 EVVGSHGVDRQQWRTDKKVFT-----IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRR 70
+V H + RQ+ D+ T ++V + + +LR+ F ++GV+ + R +
Sbjct: 205 QVFVGHFIRRQERARDENTPTPRFTNVYVKNLPKEIGEDELRKTFGKFGVISSAVVMRDQ 264
Query: 71 KQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRAR 112
G F F+ F+ + A A+ +NG + L V RA+
Sbjct: 265 S-GNSRCFGFVNFECTEAAASAVEKMNGISLGDDVLYVGRAQ 305
Score = 30.8 bits (68), Expect = 1.0
Identities = 19/92 (20%), Positives = 39/92 (41%), Gaps = 1/92 (1%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
+++ + D+V L+ MF YG + + QG F F+ + ++A A+ +
Sbjct: 334 LYLKNLDDSVDDEKLKEMFSEYGNVTSSKVMLN-PQGMSRGFGFVAYSNPEEALRALSEM 392
Query: 97 NGSWFFESFLMVNRARYDYTNRRNVTNRHSKV 128
NG L + A+ R ++ S++
Sbjct: 393 NGKMIGRKPLYIALAQRKEDRRAHLQALFSQI 424
Score = 30.0 bits (66), Expect = 1.7
Identities = 31/121 (25%), Positives = 52/121 (42%), Gaps = 12/121 (9%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
IF+ + ++ L F +G + + GR + F++F+ E+ A AI L
Sbjct: 138 IFIKNLDASIDNKALFETFSSFGTILSCKVAMD-VTGRSKGYGFVQFEKEESAQAAIDKL 196
Query: 97 NGSWFFESFLMV-------NRARYDYTNRRNVTNRHSKVWKQKFVGE---KQAFGQETVV 146
NG + + V RAR + T TN + K K +GE ++ FG+ V+
Sbjct: 197 NGMLMNDKQVFVGHFIRRQERARDENTPTPRFTNVYVKN-LPKEIGEDELRKTFGKFGVI 255
Query: 147 S 147
S
Sbjct: 256 S 256
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/78 (29%), Positives = 39/78 (49%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIR 94
+ FV G++ + DL+R F ++G + + R+ GR F F+ FK E+ +AI
Sbjct: 6 YRCFVGGLAWATNDEDLQRTFSQFGDVIDSKIINDRESGRSRGFGFVTFKDEKAMRDAIE 65
Query: 95 SLNGSWFFESFLMVNRAR 112
+NG + VN A+
Sbjct: 66 EMNGKELDGRVITVNEAQ 83
>At1g54080 unknown protein
Length = 426
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/66 (33%), Positives = 31/66 (46%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIR 94
F IFV +S V+ L F + + +K GR F F+ F+ +QDA AI
Sbjct: 148 FNIFVGDLSPEVTDAALFDSFSAFNSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQTAIN 207
Query: 95 SLNGSW 100
+NG W
Sbjct: 208 EMNGKW 213
Score = 30.8 bits (68), Expect = 1.0
Identities = 24/83 (28%), Positives = 39/83 (46%), Gaps = 8/83 (9%)
Query: 36 TIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRS 95
T++V +S V+ DL R+F G GV + R + R F F+R+ +A AI+
Sbjct: 270 TVYVGNLSPEVTQLDLHRLFYTLGA--GVIEEVRVQ--RDKGFGFVRYNTHDEAALAIQM 325
Query: 96 LNGSWFFESFLMVNRARYDYTNR 118
N + FL + R + N+
Sbjct: 326 GNA----QPFLFSRQIRCSWGNK 344
>At2g46780 putative RNA-binding protein
Length = 304
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/68 (29%), Positives = 35/68 (51%)
Query: 30 TDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDA 89
TD K+ IFV G++ +RR FE++G + + + GR + F+ FK + A
Sbjct: 17 TDTKLTKIFVGGLAWETQRDTMRRYFEQFGEIVEAVVITDKNTGRSKGYGFVTFKEAEAA 76
Query: 90 CEAIRSLN 97
A +++N
Sbjct: 77 MRACQNMN 84
>At2g18510 putative spliceosome associated protein
Length = 363
Score = 39.7 bits (91), Expect = 0.002
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 6/109 (5%)
Query: 6 SARAWPVSRPEVVGSHGVDRQQWRTDKKVFTIFVDGVSDNVSYHDLRRMFERYGVLHGVF 65
+ R P ++G H +R Q T ++V G+ +S L +F + G + V+
Sbjct: 2 TTRIAPGVGANLLGQHSAERNQDAT------VYVGGLDAQLSEELLWELFVQAGPVVNVY 55
Query: 66 LQRRRKQGRKSRFAFIRFKYEQDACEAIRSLNGSWFFESFLMVNRARYD 114
+ + R + FI ++ E+DA AI+ LN + VN+A D
Sbjct: 56 VPKDRVTNLHQNYGFIEYRSEEDADYAIKVLNMIKLHGKPIRVNKASQD 104
>At1g58470 unknown protein
Length = 360
Score = 39.7 bits (91), Expect = 0.002
Identities = 17/60 (28%), Positives = 32/60 (53%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIR 94
+ +FV G++ S L++ F RYG + + + + G+ F F+RF + D +A+R
Sbjct: 6 YKLFVGGIAKETSEEALKQYFSRYGAVLEAVVAKEKVTGKPRGFGFVRFANDCDVVKALR 65
Score = 35.0 bits (79), Expect = 0.053
Identities = 27/111 (24%), Positives = 46/111 (41%), Gaps = 7/111 (6%)
Query: 37 IFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIRSL 96
IFV G+S N + + + FER+G V + R F F+ + E D+ E +
Sbjct: 122 IFVGGLSSNTTEEEFKSYFERFGRTTDVVVMHDGVTNRPRGFGFVTYDSE-DSVEVVMQS 180
Query: 97 NGSWFFESFLMVNRA------RYDYTNRRNVTNRHSKVWKQKFVGEKQAFG 141
N + + V RA + + N N+ +S +V E+ +G
Sbjct: 181 NFHELSDKRVEVKRAIPKEGIQSNNGNAVNIPPSYSSFQATPYVPEQNGYG 231
>At1g17370 oligouridylate binding protein, putative
Length = 419
Score = 39.7 bits (91), Expect = 0.002
Identities = 27/93 (29%), Positives = 39/93 (41%)
Query: 35 FTIFVDGVSDNVSYHDLRRMFERYGVLHGVFLQRRRKQGRKSRFAFIRFKYEQDACEAIR 94
F IFV +S V+ L F Y + +K GR F F+ F+ +QDA AI
Sbjct: 139 FNIFVGDLSPEVTDAMLFTCFSVYPTCSDARVMWDQKTGRSRGFGFVSFRNQQDAQTAID 198
Query: 95 SLNGSWFFESFLMVNRARYDYTNRRNVTNRHSK 127
+ G W + N A T+ + + SK
Sbjct: 199 EITGKWLGSRQIRCNWATKGATSGEDKQSSDSK 231
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,996,105
Number of Sequences: 26719
Number of extensions: 283488
Number of successful extensions: 990
Number of sequences better than 10.0: 135
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 828
Number of HSP's gapped (non-prelim): 191
length of query: 316
length of database: 11,318,596
effective HSP length: 99
effective length of query: 217
effective length of database: 8,673,415
effective search space: 1882131055
effective search space used: 1882131055
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0383.7


