

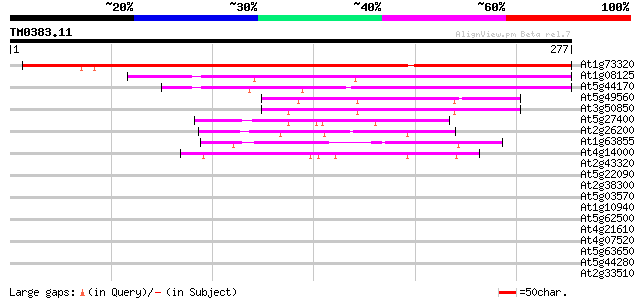
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0383.11
(277 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g73320 unknown protein 344 3e-95
At1g08125 DNA ligase 129 2e-30
At5g44170 unknown protein 72 3e-13
At5g49560 putative protein 64 9e-11
At3g50850 unknown protein 64 9e-11
At5g27400 unknown protein 58 6e-09
At2g26200 unknown protein 57 8e-09
At1g63855 putative protein 49 4e-06
At4g14000 unknown protein 42 3e-04
At2g43320 unknown protein 35 0.034
At5g22090 unknown protein 30 1.1
At2g38300 unknown protein 30 1.1
At5g03570 transporter like protein 30 1.4
At1g10940 Ser/Thr kinase 30 1.4
At5g62500 microtubule-associated protein EB1-like protein 30 1.9
At4g21610 Lsd1 like protein 30 1.9
At4g07520 hypothetical protein 30 1.9
At5g63650 serine/threonine-protein kinase 29 2.4
At5g44280 putative protein 29 2.4
At2g33510 unknown protein 29 2.4
>At1g73320 unknown protein
Length = 314
Score = 344 bits (883), Expect = 3e-95
Identities = 180/281 (64%), Positives = 213/281 (75%), Gaps = 12/281 (4%)
Query: 7 EEEEEEEEEDAPPPMVKLGSYGGEVRLV--VPGEES--------AAEETMLLWGIQQPTL 56
E E+ +E++A + ++ YGG+V +V P ES AA E M++W IQ PT
Sbjct: 21 EAEKMGKEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTS 80
Query: 57 SKPNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLG 116
PN V+QSSL+LRLD+CGHSLSILQSP SL PGVTG+VMWDSGVVLGKFLEHSVD
Sbjct: 81 FAPNTLVAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSK 140
Query: 117 TLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHVSLRGSAT 176
L L+ KKIVELGSGCGLVGCIAALLGG +LTDL DRLRLL+KNI+TN+ + RGSA
Sbjct: 141 VLSLEGKKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAI 200
Query: 177 ATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRND 236
EL WG+DPD +LI+P PD+ GSDV+YSE AV LV+TL QL TTIFL+GELRND
Sbjct: 201 VQELVWGDDPDPDLIEPFPDY--GSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRND 258
Query: 237 AILEYFLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
A+LEYFLE A+ DF IGRV+QT WHP+YRS+RVVLYVL KK
Sbjct: 259 AVLEYFLETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 299
>At1g08125 DNA ligase
Length = 315
Score = 129 bits (324), Expect = 2e-30
Identities = 73/228 (32%), Positives = 122/228 (53%), Gaps = 13/228 (5%)
Query: 59 PNAFVSQSSLQLRLDSCGHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGTL 118
P+ S S+ + ++ GH L Q P+S G +WD+ +V K+L + G
Sbjct: 3 PDRLNSPSTCTVTIEVLGHELDFAQDPNSKHL----GTTVWDASMVFAKYLGKNSRKGRF 58
Query: 119 V---LQEKKIVELGSGCGLVGCIAALLGGEVILTDLLDRLRLLRKNIETNMKHV------ 169
L+ K+ +ELG+GCG+ G A+LG +V+ TD + L LL++N+E N +
Sbjct: 59 SSSKLKGKRAIELGAGCGVAGFALAMLGCDVVTTDQKEVLPLLKRNVEWNTSRIVQMNPG 118
Query: 170 SLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFL 229
S GS EL WG + ++P D+V+G+DVVYSE + L+ T+ LSGP TT+ L
Sbjct: 119 SAFGSLRVAELDWGNEDHITAVEPPFDYVIGTDVVYSEQLLEPLLRTILALSGPKTTVML 178
Query: 230 AGELRNDAILEYFLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
E+R+ + E L+ ++F + + ++ EY+ + LY++ +K
Sbjct: 179 GYEIRSTVVHEKMLQMWKDNFEVKTIPRSKMDGEYQDPSIHLYIMAQK 226
>At5g44170 unknown protein
Length = 234
Score = 72.0 bits (175), Expect = 3e-13
Identities = 57/216 (26%), Positives = 93/216 (42%), Gaps = 20/216 (9%)
Query: 76 GHSLSILQSPSSLGAPGVTGAVMWDSGVVLGKFLEHSVDLGT-------------LVLQE 122
G LSI Q S+ G +W ++L KF E L + +
Sbjct: 15 GTKLSIQQDNGSMHV----GTSVWPCSLILSKFAERWSTLDSSSSTTSPNPYAELFDFRR 70
Query: 123 KKIVELGSGCGLVGCIAALLG-GEVILTDLLDRLRLLRKNIETNMKHVSLRGSATATELT 181
++ +ELG+GCG+ G LLG E++LTD+ + L+ N++ N +L S + +
Sbjct: 71 RRGIELGTGCGVAGMAFYLLGLTEIVLTDIAPVMPALKHNLKRNK--TALGKSLKTSIVY 128
Query: 182 WGEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLGQLSGPNTTIFLAGELRNDAILEY 241
W + P D V+ +DVVY E +V LV + L + + L ++R+ +
Sbjct: 129 WNNRDQISALKPPFDLVIAADVVYIEESVGQLVTAMELLVADDGAVLLGYQIRSPEADKL 188
Query: 242 FLEAAMNDFTIGRVDQTLWHPEYRSNRVVLYVLVKK 277
F E F I +V H +Y +Y+ KK
Sbjct: 189 FWELCDIVFKIEKVPHEHLHSDYAYEETDVYIFRKK 224
>At5g49560 putative protein
Length = 274
Score = 63.9 bits (154), Expect = 9e-11
Identities = 50/132 (37%), Positives = 64/132 (47%), Gaps = 5/132 (3%)
Query: 125 IVELGSGCGLVGCIAAL-LGGEVILTDLLDRLRLLRKNIETNMKHVS-LRGSATATELTW 182
I+ELGSG GLVG AA+ L V +TDL L L N E N + V G L W
Sbjct: 109 ILELGSGTGLVGIAAAITLSANVTVTDLPHVLDNLNFNAEANAEIVERFGGKVNVAPLRW 168
Query: 183 GEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLG--QLSGPNTTIFLAGELRNDAILE 240
GE D E++ D ++ SDVVY ++ L++TL QL G IFL LR
Sbjct: 169 GEADDVEVLGQNVDLILASDVVYHDHLYEPLLKTLRLMQLEG-KRLIFLMAHLRRWKKES 227
Query: 241 YFLEAAMNDFTI 252
F + A F +
Sbjct: 228 VFFKKARKLFDV 239
>At3g50850 unknown protein
Length = 251
Score = 63.9 bits (154), Expect = 9e-11
Identities = 46/132 (34%), Positives = 65/132 (48%), Gaps = 4/132 (3%)
Query: 125 IVELGSGCGLVG-CIAALLGGEVILTDLLDRLRLLRKNIETNMKHVS-LRGSATATELTW 182
IVELGSG G+VG AA LG V +TDL + + L+ N + N + V+ G L W
Sbjct: 92 IVELGSGTGIVGIAAAATLGANVTVTDLPNVIENLKFNADANAQVVAKFGGKVHVASLRW 151
Query: 183 GEDPDRELIDPTPDFVVGSDVVYSENAVVDLVETLG--QLSGPNTTIFLAGELRNDAILE 240
GE D E + D ++ SDVVY + L++TL L G + +FL L+
Sbjct: 152 GEIDDVESLGQNVDLILASDVVYHVHLYEPLLKTLRFLLLEGSSERVFLMAHLKRWKKES 211
Query: 241 YFLEAAMNDFTI 252
F + A F +
Sbjct: 212 IFFKKARRFFDV 223
>At5g27400 unknown protein
Length = 369
Score = 57.8 bits (138), Expect = 6e-09
Identities = 45/138 (32%), Positives = 68/138 (48%), Gaps = 16/138 (11%)
Query: 92 GVTGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVG-CIAALLGGEVILTD 150
G TG +W S + L +F+ +L K E+GSG G+VG C+A + EVILTD
Sbjct: 145 GDTGCSIWPSSLFLSEFVLSFPEL----FANKACFEVGSGVGMVGICLAHVKAKEVILTD 200
Query: 151 --LLD----RLRLLRKNIETNMKHVSLRGSATATE-----LTWGEDPDRELIDPTPDFVV 199
LL +L L R ++ + + + G A +T L W + EL PD V+
Sbjct: 201 GDLLTLSNMKLNLERNHLNYDDEFLKQPGEAQSTRVKCTHLPWETASESELSQYRPDIVL 260
Query: 200 GSDVVYSENAVVDLVETL 217
G+DV+Y + + L+ L
Sbjct: 261 GADVIYDPSCLPHLLRVL 278
>At2g26200 unknown protein
Length = 565
Score = 57.4 bits (137), Expect = 8e-09
Identities = 40/133 (30%), Positives = 68/133 (51%), Gaps = 11/133 (8%)
Query: 94 TGAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGC-GLVGCIAALLGGEVILTDLL 152
TG ++W+S ++ L+ + + ++ K+++ELG GC G+ +AA V+ TD
Sbjct: 353 TGLMLWESARLMASVLDRNPN----IVSGKRVLELGCGCTGICSMVAARSANLVVATDAD 408
Query: 153 DR-LRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTP----DFVVGSDVVYSE 207
+ L LL +NI N++ SL G + L WG E I + ++G+DV Y
Sbjct: 409 TKALTLLTENITMNLQS-SLLGKLKTSVLEWGNKEHIESIKRLACEGFEVIMGTDVTYVA 467
Query: 208 NAVVDLVETLGQL 220
A++ L ET +L
Sbjct: 468 EAIIPLFETAKEL 480
>At1g63855 putative protein
Length = 205
Score = 48.5 bits (114), Expect = 4e-06
Identities = 42/152 (27%), Positives = 67/152 (43%), Gaps = 29/152 (19%)
Query: 95 GAVMWDSGVVLGKFL-EHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVILTDLLD 153
G +W V+L +++ +H ++ I+ELG+G L G +AA +G V LTD
Sbjct: 34 GLFVWPCSVILAEYVWQHRSRF-----RDSSILELGAGTSLPGLVAAKVGANVTLTDDAT 88
Query: 154 RLRLLRKNIETNMKHVSLRGSATATELTWGEDPDRELIDPTPDFVVGSDVVYSENAVVDL 213
+ +L LTWG D ++D P+ ++G+DV+Y +A DL
Sbjct: 89 KPEVL--------------------GLTWGV-WDAPILDLRPNIILGADVLYDSSAFDDL 127
Query: 214 VETLGQL--SGPNTTIFLAGELRNDAILEYFL 243
T+ L S P+ R+ L FL
Sbjct: 128 FATVSFLLQSSPDAVFITTYHNRSGHHLIEFL 159
>At4g14000 unknown protein
Length = 290
Score = 42.4 bits (98), Expect = 3e-04
Identities = 43/168 (25%), Positives = 73/168 (42%), Gaps = 20/168 (11%)
Query: 85 PSSLGAPGVT--GAVMWDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALL 142
P+S PGV G +W+ + L K LE G L K+++ELG G L G A L
Sbjct: 76 PNSDLVPGVYEGGLKLWEGSIDLVKALEKESQTGNLSFSGKRVLELGCGHALPGIYACLK 135
Query: 143 GGEVI-LTDL-LDRLRLLR-KNIETNMKHVSLRGSATATELTWGEDPDRELIDPTP---- 195
G + + D + LR L N+ N+ S S + TE+ + E+ P
Sbjct: 136 GSDAVHFQDFNAEVLRCLTIPNLNANLSEKSSSVSVSETEVRFFAGEWSEVHQVLPLVNS 195
Query: 196 ----------DFVVGSDVVYSENAVVDLVETLGQ-LSGPNTTIFLAGE 232
D ++ ++ +YS +A E + + L+ P+ +++A +
Sbjct: 196 DGETNKKGGYDIILMAETIYSISAQKSQYELIKRCLAYPDGAVYMAAK 243
>At2g43320 unknown protein
Length = 351
Score = 35.4 bits (80), Expect = 0.034
Identities = 17/49 (34%), Positives = 27/49 (54%)
Query: 99 WDSGVVLGKFLEHSVDLGTLVLQEKKIVELGSGCGLVGCIAALLGGEVI 147
W+S VVL L++ + G L + K+++ELG G+ G A L G +
Sbjct: 92 WESSVVLVNVLKNEIRDGQLSFRGKRVLELGCNFGVPGIFACLKGASSV 140
>At5g22090 unknown protein
Length = 463
Score = 30.4 bits (67), Expect = 1.1
Identities = 12/17 (70%), Positives = 15/17 (87%)
Query: 2 ENRQEEEEEEEEEEDAP 18
E +EEEEEE+EEE+AP
Sbjct: 301 EEEEEEEEEEDEEEEAP 317
>At2g38300 unknown protein
Length = 340
Score = 30.4 bits (67), Expect = 1.1
Identities = 14/24 (58%), Positives = 16/24 (66%)
Query: 3 NRQEEEEEEEEEEDAPPPMVKLGS 26
NR+EEEEEEEEEE+ V S
Sbjct: 12 NREEEEEEEEEEEEGEESKVSSNS 35
>At5g03570 transporter like protein
Length = 498
Score = 30.0 bits (66), Expect = 1.4
Identities = 14/21 (66%), Positives = 14/21 (66%)
Query: 2 ENRQEEEEEEEEEEDAPPPMV 22
E QEEEEEEEEE P MV
Sbjct: 14 EQHQEEEEEEEEEPSLPRSMV 34
Score = 27.3 bits (59), Expect = 9.2
Identities = 11/20 (55%), Positives = 14/20 (70%)
Query: 1 MENRQEEEEEEEEEEDAPPP 20
+ N Q +EEEEEEEE+ P
Sbjct: 11 LSNEQHQEEEEEEEEEPSLP 30
>At1g10940 Ser/Thr kinase
Length = 363
Score = 30.0 bits (66), Expect = 1.4
Identities = 15/33 (45%), Positives = 20/33 (60%)
Query: 1 MENRQEEEEEEEEEEDAPPPMVKLGSYGGEVRL 33
+E +EE EEEE++ED VK GEVR+
Sbjct: 330 VEEEEEEVEEEEDDEDEYDKTVKEVHASGEVRI 362
>At5g62500 microtubule-associated protein EB1-like protein
Length = 293
Score = 29.6 bits (65), Expect = 1.9
Identities = 12/16 (75%), Positives = 15/16 (93%)
Query: 2 ENRQEEEEEEEEEEDA 17
E ++EEEEEEEEEE+A
Sbjct: 270 EGKEEEEEEEEEEEEA 285
>At4g21610 Lsd1 like protein
Length = 155
Score = 29.6 bits (65), Expect = 1.9
Identities = 11/19 (57%), Positives = 14/19 (72%)
Query: 2 ENRQEEEEEEEEEEDAPPP 20
+ Q+ EEEEEEE+ PPP
Sbjct: 10 KEEQKHREEEEEEEEGPPP 28
>At4g07520 hypothetical protein
Length = 734
Score = 29.6 bits (65), Expect = 1.9
Identities = 17/43 (39%), Positives = 20/43 (45%)
Query: 2 ENRQEEEEEEEEEEDAPPPMVKLGSYGGEVRLVVPGEESAAEE 44
E ++ EEEEE EE D PM G GEE +EE
Sbjct: 82 EEKEGEEEEESEEGDDVEPMQSQGMEENPKEEEKEGEEEESEE 124
>At5g63650 serine/threonine-protein kinase
Length = 360
Score = 29.3 bits (64), Expect = 2.4
Identities = 14/22 (63%), Positives = 15/22 (67%)
Query: 2 ENRQEEEEEEEEEEDAPPPMVK 23
E +EEEEEEEEEED VK
Sbjct: 328 EEEEEEEEEEEEEEDEYEKHVK 349
Score = 27.7 bits (60), Expect = 7.0
Identities = 11/16 (68%), Positives = 14/16 (86%)
Query: 1 MENRQEEEEEEEEEED 16
+E +EEEEEEEEEE+
Sbjct: 323 VEEEEEEEEEEEEEEE 338
Score = 27.3 bits (59), Expect = 9.2
Identities = 11/15 (73%), Positives = 13/15 (86%)
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEE+
Sbjct: 326 EEEEEEEEEEEEEEE 340
Score = 27.3 bits (59), Expect = 9.2
Identities = 11/15 (73%), Positives = 13/15 (86%)
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEE+
Sbjct: 325 EEEEEEEEEEEEEEE 339
Score = 27.3 bits (59), Expect = 9.2
Identities = 11/15 (73%), Positives = 13/15 (86%)
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEE+
Sbjct: 327 EEEEEEEEEEEEEEE 341
>At5g44280 putative protein
Length = 486
Score = 29.3 bits (64), Expect = 2.4
Identities = 12/16 (75%), Positives = 14/16 (87%)
Query: 2 ENRQEEEEEEEEEEDA 17
E +EEEEEEEEEED+
Sbjct: 92 EEEEEEEEEEEEEEDS 107
Score = 27.3 bits (59), Expect = 9.2
Identities = 11/15 (73%), Positives = 13/15 (86%)
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEE+
Sbjct: 91 EEEEEEEEEEEEEEE 105
>At2g33510 unknown protein
Length = 189
Score = 29.3 bits (64), Expect = 2.4
Identities = 12/15 (80%), Positives = 14/15 (93%)
Query: 2 ENRQEEEEEEEEEED 16
EN +EEEEEEEEEE+
Sbjct: 130 ENHKEEEEEEEEEEE 144
Score = 28.9 bits (63), Expect = 3.2
Identities = 12/15 (80%), Positives = 13/15 (86%)
Query: 2 ENRQEEEEEEEEEED 16
E +EEEEEEEEEED
Sbjct: 134 EEEEEEEEEEEEEED 148
Score = 28.9 bits (63), Expect = 3.2
Identities = 14/34 (41%), Positives = 21/34 (61%)
Query: 3 NRQEEEEEEEEEEDAPPPMVKLGSYGGEVRLVVP 36
+++EEEEEEEEEE+ +V G + +VP
Sbjct: 132 HKEEEEEEEEEEEEEEDVLVVAGCKACFMYFMVP 165
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.135 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,464,169
Number of Sequences: 26719
Number of extensions: 279635
Number of successful extensions: 3186
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2457
Number of HSP's gapped (non-prelim): 478
length of query: 277
length of database: 11,318,596
effective HSP length: 98
effective length of query: 179
effective length of database: 8,700,134
effective search space: 1557323986
effective search space used: 1557323986
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0383.11


