

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
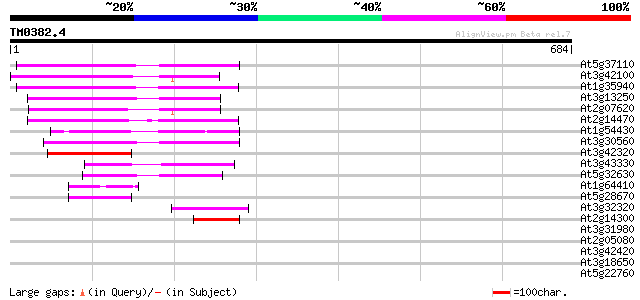
Query= TM0382.4
(684 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g37110 putative helicase 150 3e-36
At3g42100 putative protein 145 6e-35
At1g35940 hypothetical protein 139 5e-33
At3g13250 hypothetical protein 132 6e-31
At2g07620 putative helicase 129 4e-30
At2g14470 pseudogene 129 5e-30
At1g54430 hypothetical protein 117 2e-26
At3g30560 hypothetical protein 113 3e-25
At3g42320 putative protein 93 4e-19
At3g43330 putative protein 82 8e-16
At5g32630 putative protein 78 1e-14
At1g64410 unknown protein 70 5e-12
At5g28670 putative protein 68 1e-11
At3g32320 hypothetical protein 43 7e-04
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 43 7e-04
At3g31980 hypothetical protein 42 0.001
At2g05080 putative helicase 35 0.11
At3g42420 putative protein 35 0.14
At3g18650 hypothetical protein 33 0.40
At5g22760 putative protein 33 0.53
>At5g37110 putative helicase
Length = 1307
Score = 150 bits (378), Expect = 3e-36
Identities = 87/273 (31%), Positives = 131/273 (47%), Gaps = 28/273 (10%)
Query: 9 GDASYVCSNCQSIMWYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQLLYNLMNEIN 68
GD Y C C +IMW+ E K ++ KF CC++ ++ LPFL+ P+LLY L +
Sbjct: 99 GDPIYACEYCGAIMWHGERIEKTVKNKKSKFTSCCLQGQVKLPFLKNSPELLYALPTNDD 158
Query: 69 DRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPA 128
+ ++F+ENIRA N +F FTSLGG+ E+ + P F + +N HRIGSL P P
Sbjct: 159 EISRHFRENIRAYNMIFYFTSLGGDTENSVRASGGPQMFQIHRENYHRIGSLKPDNDIPP 218
Query: 129 KFTQLYIYDTHNEFSNRMKIFRYFLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKS 188
KF QLYI DT NE NR + +N + +K
Sbjct: 219 KFMQLYIVDTENEVDNRSEALSNGVN---------------------------ADKYKKK 251
Query: 189 PLDPSLMKDLTALIDKDNELAKMFQRVKDFVETDGSSNFYLKLYCNKTKDPRTYNMPTAD 248
L ++ + L++ N + +D + ++++ + KD R YN+PT
Sbjct: 252 ELRKEIIDAVIKLLNSINPYVSHLRTARDRFNANPEETLHMRIVSKREKDGRVYNVPTTS 311
Query: 249 EVAALIIGDLD-TVNEGNIIVKGTDGGFQRLPE 280
EVA LI GD + +I+V+ G QR+ E
Sbjct: 312 EVAMLIPGDFTIDMPSRDIVVQEKSGKLQRISE 344
>At3g42100 putative protein
Length = 1752
Score = 145 bits (367), Expect = 6e-35
Identities = 89/262 (33%), Positives = 130/262 (48%), Gaps = 37/262 (14%)
Query: 2 AAKVFYL--GDASYVCSNCQSIMWYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQL 59
A K YL GDA+Y C+ C ++MW+ E K ++ P F +CC K + LP L+ P L
Sbjct: 310 ATKSAYLDHGDATYKCNYCGALMWFAERINKKQQNKSPTFTLCCGKGNVKLPLLKDSPAL 369
Query: 60 LYNLMNEINDRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGS 119
+ NL+ + +NF+ENIR N +F+ TSLGG V++ + G P F L N H IGS
Sbjct: 370 INNLLTGDDALSRNFRENIRIYNMIFAMTSLGGRVDNSMPKGKGPNMFRLQGGNYHLIGS 429
Query: 120 LIPPKGAPAKFTQLYIYDTHNEFSNRMKIFRYFLNYLQIYEQNLYLDVMVADAKFLLSFF 179
L P G AK++QLYI DT NE NR +
Sbjct: 430 LKPNPGDYAKYSQLYIVDTENEVDNRATVINK---------------------------- 461
Query: 180 YISSSSRKSPLDPSLMKD----LTALIDKDNELAKMFQRVKDFVETDGSSNFYLKLYCN- 234
R +P L K+ L +++K N F++ ++ ++ D F++++ +
Sbjct: 462 --GKGRRNTPAKQKLKKEVIEALIEMLNKVNPYVDKFRQARERIQDDNDEPFHMRIVADR 519
Query: 235 KTKDPRTYNMPTADEVAALIIG 256
K D RTY+MPT+ EVAALI G
Sbjct: 520 KGVDRRTYSMPTSSEVAALIPG 541
>At1g35940 hypothetical protein
Length = 1678
Score = 139 bits (350), Expect = 5e-33
Identities = 83/273 (30%), Positives = 139/273 (50%), Gaps = 29/273 (10%)
Query: 9 GDASYVCSNCQSIMWYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQLLYNLMNEIN 68
GD +Y C C ++MWY+E K + ++ F +CC + + LPFL++ P LL NL++ +
Sbjct: 329 GDPTYKCKYCGAMMWYDERIRKKETNKESVFTLCCGEGSVKLPFLKESPHLLKNLLSGNH 388
Query: 69 DRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPA 128
K++++N R N +F+ TS GG V+ + G P F L N H IGSL G A
Sbjct: 389 PLSKHYRDNARIFNMVFAMTSFGGKVDKSMPKGRGPAMFRLQGGNYHLIGSLKLTPGDYA 448
Query: 129 KFTQLYIYDTHNEFSNRMKIFRYFLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKS 188
K++QLYI DT NE NR + N +S K
Sbjct: 449 KYSQLYIIDTENEVENRATVINKGKN--------------------------AKPASGKP 482
Query: 189 PLDPSLMKDLTALIDKDNELAKMFQRVKDFVETDGSSNFYLKLYCNKT-KDPRTYNMPTA 247
LD +L++ + ++++ N + F+ ++ ++T+ F++++ ++ + RTY+MPT
Sbjct: 483 NLDKNLIEAIIKMLNRCNPYVRRFRTARERIQTNDEEPFHMRIIADRQGVEGRTYSMPTT 542
Query: 248 DEVAALIIGDL--DTVNEGNIIVKGTDGGFQRL 278
EVAALI GD + +I K ++G +R+
Sbjct: 543 SEVAALIPGDFRHGMPDRDIVIGKKSNGHLKRI 575
>At3g13250 hypothetical protein
Length = 1419
Score = 132 bits (332), Expect = 6e-31
Identities = 78/237 (32%), Positives = 122/237 (50%), Gaps = 27/237 (11%)
Query: 22 MWYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQLLYNLMNEINDRYKNFKENIRA* 81
MW+ E K +SE PKF +CC + + LPFL++ P+L+ L+ + +++++ IR
Sbjct: 1 MWFNERINKKSNSENPKFSLCCGQGSVKLPFLKESPELIKKLLKGNDALSRHYRQFIRIY 60
Query: 82 NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNE 141
N +F+ TSLGG V+ + G P F L N H+IGSL P G AK++QLYI DT NE
Sbjct: 61 NMIFAMTSLGGKVDKSMPKGRGPAMFRLQGGNYHQIGSLKPKDGDYAKYSQLYIVDTENE 120
Query: 142 FSNRMKIFRYFLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKSPLDPSLMKDLTAL 201
NR + N SS+ K L+ L+ + +
Sbjct: 121 VENRANVIGKGNNG--------------------------SSTKGKKNLNKQLIDAIIKM 154
Query: 202 IDKDNELAKMFQRVKDFVETDGSSNFYLKLYCN-KTKDPRTYNMPTADEVAALIIGD 257
+++ N + F+ ++ +++ F++++ + K D R YNMPTA EVAALI GD
Sbjct: 155 LNQVNPYVEKFRSARERIDSTNDEPFHMRIVSDRKGTDGRLYNMPTAGEVAALIPGD 211
>At2g07620 putative helicase
Length = 1241
Score = 129 bits (325), Expect = 4e-30
Identities = 82/240 (34%), Positives = 121/240 (50%), Gaps = 41/240 (17%)
Query: 23 WYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQLLYNLMNEINDRYKNFKENIRA*N 82
WY+E K S+ PKF +CC++ + LPFL + P+L+ L++ + ++F+ENIRA N
Sbjct: 3 WYDERVNKRRKSKKPKFSLCCLQGSVKLPFLTESPELIRELLSCDDALRRHFRENIRAYN 62
Query: 83 NLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNEF 142
LFS TSLGG V+ G P +F L N H IGS++P +G AKF+QLYI DT NE
Sbjct: 63 MLFSMTSLGGKVDRSNPQGKGPNQFQLHGANYHLIGSMLPGEGDYAKFSQLYIVDTGNEV 122
Query: 143 SNRMKIFRYFLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKSPLDPSLMKD----L 198
NR S++ +K+ + KD L
Sbjct: 123 ENR------------------------------------STNPKKATFKNQVRKDLIEKL 146
Query: 199 TALIDKDNELAKMFQRVKDFVETDGSSNFYLKLYCNKT-KDPRTYNMPTADEVAALIIGD 257
++D N + F+ K ++++ FY+++ ++ KD RTY P EVAALI GD
Sbjct: 147 IRMLDACNPYIENFRLAKYKLDSNNGEPFYMQIVSDRVGKDGRTYCNPRTSEVAALIPGD 206
>At2g14470 pseudogene
Length = 1265
Score = 129 bits (324), Expect = 5e-30
Identities = 80/260 (30%), Positives = 133/260 (50%), Gaps = 29/260 (11%)
Query: 22 MWYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQLLYNLMNEINDRYKNFKENIRA* 81
MWY+E K + + F +CC + + LPFL++ P LL NL++ + K++++N R
Sbjct: 1 MWYDERIRKKETKKESGFTLCCGEGSVKLPFLKESPDLLKNLLSGNHPLSKHYRDNARTF 60
Query: 82 NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNE 141
N +F+ TSLGG V+ + G P F L N H IGSL P G AK++QLYI DT NE
Sbjct: 61 NMVFAVTSLGGKVDKSMPKGRGPAMFRLQGGNYHLIGSLKPTPGDYAKYSQLYIVDTENE 120
Query: 142 FSNRMKIFRYFLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKSPLDPSLMKDLTAL 201
NR +++ K +S K LD +L++ + +
Sbjct: 121 VENRA--------------------IVIGKGKI------AKPASGKPNLDKNLIEVIIKM 154
Query: 202 IDKDNELAKMFQRVKDFVETDGSSNFYLKLYCNKT-KDPRTYNMPTADEVAALIIGDL-- 258
+++ N + F+ ++ ++T+ F++++ ++ D RTY+M T EVAALI GD
Sbjct: 155 LNRCNPYVRKFRTARERIQTNDEEPFHMRIIADRQGVDGRTYSMHTTSEVAALIPGDFRH 214
Query: 259 DTVNEGNIIVKGTDGGFQRL 278
+ +I K ++G +R+
Sbjct: 215 GMPDRDIVIEKKSNGHLKRI 234
>At1g54430 hypothetical protein
Length = 1639
Score = 117 bits (293), Expect = 2e-26
Identities = 71/232 (30%), Positives = 114/232 (48%), Gaps = 38/232 (16%)
Query: 50 LPFLRKPPQLLYNLMNEINDRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFIL 109
LP +PPQ+L L+ E +++ NIR N++ +FTS+G ++ + P F +
Sbjct: 320 LPPEPQPPQMLKKLLTES----PHYQRNIRTYNSILAFTSMGAQIDKNVMHKGGPFTFRI 375
Query: 110 SAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNEFSNRMKIFRYFLNYLQIYEQNLYLDVMV 169
QN H++GSL+P +G P K QLYI+DT NE NR+
Sbjct: 376 HGQNHHKLGSLVPEEGKPPKILQLYIFDTANEVHNRISA--------------------- 414
Query: 170 ADAKFLLSFFYISSSSRKSPLDPSLMKDLTALIDKDNELAKMFQRVKDFVETDGSSNFYL 229
+ +++ L+ ++KDL +D N LAK+F++ +D E F +
Sbjct: 415 -----------VKRTTKVGELNEKIVKDLITTVDTFNCLAKVFRKARDRYEAGDCPEFSI 463
Query: 230 KLYCNKTKDPRTYNMPTADEVAALIIGDLD-TVNEGNIIVKGTDGGFQRLPE 280
KL K K + Y+MPT DE+A LI+GD + E ++IV G Q++ +
Sbjct: 464 KLIGQKKKG-KQYDMPTTDEIAGLIVGDFSKNIGERDVIVHHKSSGLQQISD 514
>At3g30560 hypothetical protein
Length = 1473
Score = 113 bits (283), Expect = 3e-25
Identities = 74/241 (30%), Positives = 116/241 (47%), Gaps = 29/241 (12%)
Query: 42 CCMKEKITLPFLRKPPQLLYNLMNEINDRYKNFKENIRA*NNLFSFTSLGGNVESGINDG 101
CCM+ +I LP L++ P+ ++ L + KNF+ N+R N LFSFTSLGG V+ + G
Sbjct: 128 CCMQGQIVLPMLKESPEYMWWLFTSDHPDAKNFRANVRPYNMLFSFTSLGGKVDRSVKKG 187
Query: 102 NDPPRFILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNEFSNRMKIFRYFLNYLQIYEQ 161
P F L +N H I +L P G AKF QLY+ DT NE NR+++ N
Sbjct: 188 RGPSMFALQGENYHLIDALKPKPGDYAKFQQLYVMDTDNEVDNRIEVMSKGNNG------ 241
Query: 162 NLYLDVMVADAKFLLSFFYISSSSRKSPLDPSLMKDLTALIDKDNELAKMFQRVKD-FVE 220
+ K + L ++D+ N F+ +D F
Sbjct: 242 --------------------KTEGVKQKFKKETVSALLKMLDEINPHVANFRIARDRFNI 281
Query: 221 TDGSSNFYLKLYCNKTKDPRTYNMPTADEVAALIIGDL-DTVNEGNIIVKGTDGGFQRLP 279
+NF++++ N+ D R YN+P+ EV ALI GD D +++ +I+++ +R+
Sbjct: 282 EKEDANFHMRIISNRDTDGRVYNLPSVGEV-ALIPGDFDDNLDKRDIVLQIKSEKLRRIH 340
Query: 280 E 280
E
Sbjct: 341 E 341
>At3g42320 putative protein
Length = 541
Score = 93.2 bits (230), Expect = 4e-19
Identities = 47/102 (46%), Positives = 64/102 (62%)
Query: 47 KITLPFLRKPPQLLYNLMNEINDRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPR 106
+I LP L+ P+ L++L+ ++ K+F+ENIRA N LFSFTS+G V+ + G P
Sbjct: 6 QIVLPSLKDSPEFLWHLLTSDDELSKHFRENIRAYNTLFSFTSIGSKVDHFLPKGPQPNM 65
Query: 107 FILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNEFSNRMKI 148
F + +N H IG+L P A AKF QLYI DT NE +NR I
Sbjct: 66 FAIQGENYHLIGALKPKSSAKAKFQQLYIADTENEVNNRYNI 107
Score = 33.5 bits (75), Expect = 0.40
Identities = 16/23 (69%), Positives = 17/23 (73%)
Query: 234 NKTKDPRTYNMPTADEVAALIIG 256
N DPR YN+PTA EVAALI G
Sbjct: 106 NIMSDPRIYNLPTAYEVAALIPG 128
>At3g43330 putative protein
Length = 489
Score = 82.4 bits (202), Expect = 8e-16
Identities = 53/184 (28%), Positives = 83/184 (44%), Gaps = 35/184 (19%)
Query: 92 GNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNEFSNRMKIFRY 151
G V+ + P F + +N + +G+L P GA AKF QLYI DT NE NR I
Sbjct: 163 GKVDHCLPKDRGPNMFAIQGENYYLMGALKPKSGAKAKFQQLYIVDTENEVKNRYNIM-- 220
Query: 152 FLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKSPLDPSLMKDLTALIDKDNELAKM 211
S+ + K + L+++ N K
Sbjct: 221 --------------------------------SNENESENGQKKKKIMKLLNRVNPHVKA 248
Query: 212 FQRVKDFVETDGSSNFYLKLYCNKTKDPRTYNMPTADEVAALIIGDL-DTVNEGNIIVKG 270
F+ +D TD +F++++ + DPR YN+PTA EVAALI GD + +++ +I+++
Sbjct: 249 FRYARDRFNTDAQESFHMRIIAARKTDPRIYNLPTASEVAALIPGDFHEDMDKLDIVLQE 308
Query: 271 TDGG 274
T G
Sbjct: 309 TTSG 312
>At5g32630 putative protein
Length = 856
Score = 78.2 bits (191), Expect = 1e-14
Identities = 52/172 (30%), Positives = 80/172 (46%), Gaps = 27/172 (15%)
Query: 89 SLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFTQLYIYDTHNEFSNRMKI 148
SLGG V++ + G P F L N H IGSL P G AK++QLYI DT N+ NR +
Sbjct: 3 SLGGRVDNSMPKGKGPNMFRLQEGNYHLIGSLKPKPGDYAKYSQLYIVDTENKVENRATV 62
Query: 149 FRYFLNYLQIYEQNLYLDVMVADAKFLLSFFYISSSSRKSPLDPSLMKDLTALIDKDNEL 208
++ K L +++ L +++K N
Sbjct: 63 TNKGKGG--------------------------QNTVAKQKLKKEVIEALIEMLNKVNPY 96
Query: 209 AKMFQRVKDFVETDGSSNFYLKLYC-NKTKDPRTYNMPTADEVAALIIGDLD 259
+ F++ ++ ++ D F++++ K D RTYNMPT+ +VAALI G D
Sbjct: 97 VEKFKQSRERIQADNDELFHMRIVAYRKGVDRRTYNMPTSSKVAALIPGGFD 148
>At1g64410 unknown protein
Length = 753
Score = 69.7 bits (169), Expect = 5e-12
Identities = 41/86 (47%), Positives = 51/86 (58%), Gaps = 9/86 (10%)
Query: 72 KNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFT 131
K+F+ENIRA N LFSFTS+GG V+ + G P F + G+L P A AKF
Sbjct: 73 KHFRENIRAYNMLFSFTSIGGKVDHCLPKGRGPNMFAIQ-------GALKPKSVAKAKFQ 125
Query: 132 QLYIYDTHNEFSNRMKIFRYFLNYLQ 157
QLYI DT NE +NR I R L Y++
Sbjct: 126 QLYIVDTENEVNNRYNIMR--LRYIK 149
>At5g28670 putative protein
Length = 647
Score = 68.2 bits (165), Expect = 1e-11
Identities = 35/77 (45%), Positives = 44/77 (56%)
Query: 72 KNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFILSAQNCHRIGSLIPPKGAPAKFT 131
K F+ NIR N LFS TS+GG V + P F L +N H IG+L P AKF
Sbjct: 178 KTFRANIRPYNMLFSVTSIGGKVYRSVKKRRGPSMFALQGENYHLIGALKPKADDYAKFQ 237
Query: 132 QLYIYDTHNEFSNRMKI 148
QLYI DT NE N++++
Sbjct: 238 QLYIMDTENEVDNQIEV 254
>At3g32320 hypothetical protein
Length = 494
Score = 42.7 bits (99), Expect = 7e-04
Identities = 28/96 (29%), Positives = 48/96 (49%), Gaps = 2/96 (2%)
Query: 198 LTALIDKDNELAKMFQRVKD-FVETDGSSNFYLKLYCNKTKDPRTYNMPTADEVAALIIG 256
L ++DK N F+ +D F NF++++ + D R YN+P+ EVAALI G
Sbjct: 37 LLKMLDKINPHVANFRIARDRFNIEKEEENFHMRIISRRETDGRVYNLPSVAEVAALIPG 96
Query: 257 DL-DTVNEGNIIVKGTDGGFQRLPERKKKPFLHSSS 291
D D +++ +I+++ G Q K + +S
Sbjct: 97 DFDDNLDKRDIVLQMKSGKNQTKLRNTNKQAVQDTS 132
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 42.7 bits (99), Expect = 7e-04
Identities = 20/57 (35%), Positives = 37/57 (64%), Gaps = 1/57 (1%)
Query: 225 SNFYLKLYCNKTKDPRTYNMPTADEVAALIIGDL-DTVNEGNIIVKGTDGGFQRLPE 280
+NF++++ + D R YN+P+ EVAALI GD D +++ +I+++ G +R+ E
Sbjct: 173 ANFHMRIISKRETDGRVYNLPSVAEVAALIPGDFDDNLDKKDIVLQMKSGKLRRIHE 229
Score = 38.1 bits (87), Expect = 0.016
Identities = 17/57 (29%), Positives = 31/57 (53%)
Query: 20 SIMWYEEEAVKFDHSEIPKFKICCMKEKITLPFLRKPPQLLYNLMNEINDRYKNFKE 76
+I WY E + S P + CCM+ +I LP L++ P+ ++ L+ + KN ++
Sbjct: 73 AIFWYGERLNRRKKSANPVYTGCCMQGQIVLPMLKESPEYMWWLLTSDHPDAKNLEQ 129
>At3g31980 hypothetical protein
Length = 1099
Score = 41.6 bits (96), Expect = 0.001
Identities = 20/58 (34%), Positives = 34/58 (58%), Gaps = 1/58 (1%)
Query: 201 LIDKDNELAKMFQRVKDFVETDGSSNFYLKLYCNKT-KDPRTYNMPTADEVAALIIGD 257
++D N + F+ KD ++++ FY+++ ++ KD +TY PT EV ALI GD
Sbjct: 1 MLDASNPYVEKFRLAKDKLDSNNGEPFYMQIVSDRVGKDGKTYCNPTTSEVTALIPGD 58
>At2g05080 putative helicase
Length = 1219
Score = 35.4 bits (80), Expect = 0.11
Identities = 20/53 (37%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Query: 229 LKLYCNKTKDPRTYNMPTADEVAALIIGDLD-TVNEGNIIVKGTDGGFQRLPE 280
+++ + D R YN+PT EVA LI GD + +IIV+ G QR+ E
Sbjct: 1 MRIVSKRETDGRVYNVPTTSEVAMLIPGDFTIDIPCRDIIVEEKSGKLQRISE 53
>At3g42420 putative protein
Length = 1018
Score = 35.0 bits (79), Expect = 0.14
Identities = 24/62 (38%), Positives = 31/62 (49%), Gaps = 8/62 (12%)
Query: 235 KTKDPRTYNMPTADEVAALIIG-------DLDTVNEGNIIVKGTDGGFQ-RLPERKKKPF 286
K KD RTY PT EVA LI G DLD V + I D +P+++K+P
Sbjct: 379 KEKDQRTYIRPTTSEVAELIPGDFENGMPDLDIVIQEKITADDIDKMISAEIPDKEKEPE 438
Query: 287 LH 288
L+
Sbjct: 439 LY 440
Score = 30.4 bits (67), Expect = 3.4
Identities = 14/52 (26%), Positives = 30/52 (56%)
Query: 58 QLLYNLMNEINDRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFIL 109
+L+ L+ + + + +N++++ N +F+ TS+GG V+ + G P F L
Sbjct: 321 ELVKELLTDDDAQGRNYRKHAWIYNMVFAMTSIGGKVDKSMPKGQGPIMFRL 372
>At3g18650 hypothetical protein
Length = 386
Score = 33.5 bits (75), Expect = 0.40
Identities = 27/95 (28%), Positives = 43/95 (44%), Gaps = 7/95 (7%)
Query: 53 LRKPPQLLYNLMNEINDRYKNFKENIRA*NNLFSFTSLGGNVESGINDGNDPPRFILSAQ 112
L+ L YN +N +ND K F +N RA S++SL G E G N+ +P +
Sbjct: 262 LQNMNMLTYNNINSVNDFSKQFDQNSRA----ESYSSLLGVHEDGNNEFENPNMSSRNNF 317
Query: 113 NCHRIGSLIPPKGAPAKFTQ---LYIYDTHNEFSN 144
N L+ +GA Q ++ Y +N ++
Sbjct: 318 NVQDCAGLLGMQGAGTNGLQSMNMHDYSNNNSINS 352
>At5g22760 putative protein
Length = 1516
Score = 33.1 bits (74), Expect = 0.53
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 7/52 (13%)
Query: 4 KVFYLGDASYV-CSNCQSIMWYEEEAVKFDHSEIPK---FKICCMKEKITLP 51
K+ Y +Y+ C++C MWY EAVK + S+IP+ FK CC +I P
Sbjct: 1199 KLPYNPGLTYIHCTSCD--MWYHIEAVKLEESKIPEVVGFK-CCRCRRIRSP 1247
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.345 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,855,384
Number of Sequences: 26719
Number of extensions: 560806
Number of successful extensions: 2206
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2160
Number of HSP's gapped (non-prelim): 47
length of query: 684
length of database: 11,318,596
effective HSP length: 106
effective length of query: 578
effective length of database: 8,486,382
effective search space: 4905128796
effective search space used: 4905128796
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0382.4


