

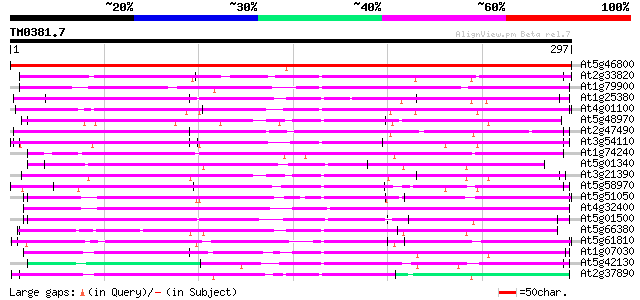
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0381.7
(297 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g46800 carnitine/acylcarnitine translocase-like protein 475 e-134
At2g33820 mitochondrial basic amino acid carrier (BAC1) 160 6e-40
At1g79900 putative carnitine/acylcarnitine translocase 152 2e-37
At1g25380 unknown protein 125 2e-29
At4g01100 putative carrier protein 111 4e-25
At5g48970 mitochondrial carrier protein-like 107 1e-23
At2g47490 mitochondrial carrier like protein 106 2e-23
At3g54110 uncoupling protein (ucp/PUMP) 101 6e-22
At1g74240 mitochondrial carrier like protein 100 1e-21
At5g01340 unknown protein 99 4e-21
At3g21390 unknown protein 97 1e-20
At5g58970 uncoupling protein AtUCP2 96 3e-20
At5g51050 calcium-binding transporter-like protein 92 3e-19
At4g32400 adenylate translocator (brittle-1) - like protein 90 1e-18
At5g01500 unknown protein 89 2e-18
At5g66380 unknown protein 89 4e-18
At5g61810 peroxisomal Ca-dependent solute carrier - like protein 87 8e-18
At1g07030 unknown protein 87 8e-18
At5g42130 mitochondrial carrier protein-like 87 1e-17
At2g37890 mitochondrial carrier like protein 85 4e-17
>At5g46800 carnitine/acylcarnitine translocase-like protein
Length = 300
Score = 475 bits (1223), Expect = e-134
Identities = 238/300 (79%), Positives = 259/300 (86%), Gaps = 3/300 (1%)
Query: 1 MGDVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAE 60
M D KDL +GT GGAAQL+VGHPFDTIKVKLQSQPTP PGQLPRY+GAIDAVKQT+A+E
Sbjct: 1 MADAWKDLASGTVGGAAQLVVGHPFDTIKVKLQSQPTPAPGQLPRYTGAIDAVKQTVASE 60
Query: 61 GPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSF 120
G +GLYKGMGAPLATVAA NAVLFTVRGQME L RS G LT+ QQ GAGAG AVSF
Sbjct: 61 GTKGLYKGMGAPLATVAAFNAVLFTVRGQMEGLLRSEAGVPLTISQQFVAGAGAGFAVSF 120
Query: 121 LVCPTELIKCRLQAQSTLAGSGTAA---VAVKYGGPMDVARHVLRSEGGARGLFKGLVPT 177
L CPTELIKCRLQAQ LAG+ T + AVKYGGPMDVARHVLRSEGGARGLFKGL PT
Sbjct: 121 LACPTELIKCRLQAQGALAGASTTSSVVAAVKYGGPMDVARHVLRSEGGARGLFKGLFPT 180
Query: 178 MAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSV 237
AREVPGNA MF AYEA K+ LAGG DTS LG+GSLI+AGG+AGASFW VYPTDVVKSV
Sbjct: 181 FAREVPGNATMFAAYEAFKRFLAGGSDTSSLGQGSLIMAGGVAGASFWGIVYPTDVVKSV 240
Query: 238 IQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSALG 297
+QVDDYK P+++GS+DAFR+IL SEG+KGLYKGFGPAM RSVPANAACFLAYEMTRS+LG
Sbjct: 241 LQVDDYKNPRYTGSMDAFRKILKSEGVKGLYKGFGPAMARSVPANAACFLAYEMTRSSLG 300
>At2g33820 mitochondrial basic amino acid carrier (BAC1)
Length = 311
Score = 160 bits (406), Expect = 6e-40
Identities = 102/303 (33%), Positives = 149/303 (48%), Gaps = 22/303 (7%)
Query: 6 KDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGL 65
K+ AG G A + VGHPFDT+KVKLQ T + G RY + + L EG +GL
Sbjct: 16 KEYVAGMMAGLATVAVGHPFDTVKVKLQKHNTDVQGL--RYKNGLHCASRILQTEGVKGL 73
Query: 66 YKGMGAPLATVAALNAVLFTVRGQMEALFR-----SHPGASLTVHQQVFCGAGAGLAVSF 120
Y+G + +A ++++F + Q + R P + V +F GA +SF
Sbjct: 74 YRGATSSFMGMAFESSLMFGIYSQAKLFLRGTLPDDGPRPEIIVPSAMFGGA----IISF 129
Query: 121 LVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAR 180
++CPTEL+KCR+Q Q T +Y P+D A ++++ G G+F+G T+ R
Sbjct: 130 VLCPTELVKCRMQIQGT---DSLVPNFRRYNSPLDCAVQTVKND-GVTGIFRGGSATLLR 185
Query: 181 EVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSL------IVAGGLAGASFWFFVYPTDVV 234
E GNA F YE + + + S L G L ++ GGL G + W V P DV
Sbjct: 186 ECTGNAVFFTVYEYLRYHIHSRLEDSKLKDGYLVDMGIGVLTGGLGGIACWSAVLPFDVA 245
Query: 235 KSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRS 294
K++IQ K + I G+KG Y G GP + R+ PANAA +A+E +
Sbjct: 246 KTIIQTSSEKATE-RNPFKVLSSIHKRAGLKGCYAGLGPTIVRAFPANAAAIVAWEFSMK 304
Query: 295 ALG 297
LG
Sbjct: 305 MLG 307
Score = 67.8 bits (164), Expect = 7e-12
Identities = 46/200 (23%), Positives = 89/200 (44%), Gaps = 12/200 (6%)
Query: 99 GASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVAR 158
G +++ G AGLA + P + +K +LQ + T ++Y + A
Sbjct: 9 GEGFGFYKEYVAGMMAGLATVAVGHPFDTVKVKLQKHN------TDVQGLRYKNGLHCAS 62
Query: 159 HVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGG 218
+L++E G +GL++G + ++ MFG Y +K L G G ++ +
Sbjct: 63 RILQTE-GVKGLYRGATSSFMGMAFESSLMFGIYSQAKLFLRGTLPDDGPRPEIIVPSAM 121
Query: 219 LAGASFWFFVYPTDVVKSVIQVDDY-----KYPKFSGSIDAFRRILASEGIKGLYKGFGP 273
GA F + PT++VK +Q+ + +++ +D + + ++G+ G+++G
Sbjct: 122 FGGAIISFVLCPTELVKCRMQIQGTDSLVPNFRRYNSPLDCAVQTVKNDGVTGIFRGGSA 181
Query: 274 AMFRSVPANAACFLAYEMTR 293
+ R NA F YE R
Sbjct: 182 TLLRECTGNAVFFTVYEYLR 201
>At1g79900 putative carnitine/acylcarnitine translocase
Length = 296
Score = 152 bits (385), Expect = 2e-37
Identities = 101/298 (33%), Positives = 154/298 (50%), Gaps = 35/298 (11%)
Query: 6 KDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGL 65
++ AG FGG A +I G+P DT++++ Q Q + A +++ LA EGP L
Sbjct: 14 REFVAGGFGGVAGIISGYPLDTLRIRQQ--------QSSKSGSAFSILRRMLAIEGPSSL 65
Query: 66 YKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQ------VFCGAGAGLAVS 119
Y+GM APLA+V NA++F Q+ A+F +S+ + + G G S
Sbjct: 66 YRGMAAPLASVTFQNAMVF----QIYAIFSRSFDSSVPLVEPPSYRGVALGGVATGAVQS 121
Query: 120 FLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMA 179
L+ P ELIK RLQ Q T + GP+ +A+ +LR + G +GL++GL T+
Sbjct: 122 LLLTPVELIKIRLQLQQTKS------------GPITLAKSILRRQ-GLQGLYRGLTITVL 168
Query: 180 REVPGNAAMFGAYEASKQLLAGGPDTSGL-GRGSLIVAGGLAGASFWFFVYPTDVVKSVI 238
R+ P + F YE ++ L G +G +++VAGGLAG + W YP DVVK+ +
Sbjct: 169 RDAPAHGLYFWTYEYVRERLHPGCRKTGQENLRTMLVAGGLAGVASWVACYPLDVVKTRL 228
Query: 239 QVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
Q + + G D FR+ + EG L++G G A+ R+ N A F AYE+ L
Sbjct: 229 Q---QGHGAYEGIADCFRKSVKQEGYTVLWRGLGTAVARAFVVNGAIFAAYEVALRCL 283
Score = 35.4 bits (80), Expect = 0.037
Identities = 24/87 (27%), Positives = 34/87 (38%), Gaps = 4/87 (4%)
Query: 203 PDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASE 262
P+ G VAGG G + YP D +++ + K + RR+LA E
Sbjct: 5 PEFMATSWGREFVAGGFGGVAGIISGYPLD----TLRIRQQQSSKSGSAFSILRRMLAIE 60
Query: 263 GIKGLYKGFGPAMFRSVPANAACFLAY 289
G LY+G + NA F Y
Sbjct: 61 GPSSLYRGMAAPLASVTFQNAMVFQIY 87
>At1g25380 unknown protein
Length = 363
Score = 125 bits (315), Expect = 2e-29
Identities = 92/295 (31%), Positives = 138/295 (46%), Gaps = 15/295 (5%)
Query: 3 DVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTP-IPGQLPRYSGAIDAVKQTLAAEG 61
+VA + AG GA P D IK +LQ P P R I ++K + EG
Sbjct: 16 EVAANAGAGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQRGGVIITSLKNIIKEEG 75
Query: 62 PRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFL 121
RG+Y+G+ + + AV F+V G+++ + +S G L++ + AGAG A S
Sbjct: 76 YRGMYRGLSPTIIALLPNWAVYFSVYGKLKDVLQSSDG-KLSIGSNMIAAAGAGAATSIA 134
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
P ++K RL Q G V Y M + EG RGL+ G++P++A
Sbjct: 135 TNPLWVVKTRLMTQGIRPG------VVPYKSVMSAFSRICHEEG-VRGLYSGILPSLAG- 186
Query: 182 VPGNAAMFGAYEASKQLLAGGPDTS--GLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQ 239
V A F AYE KQ +A +TS L G++ +A +A YP +V+++ +Q
Sbjct: 187 VSHVAIQFPAYEKIKQYMAKMDNTSVENLSPGNVAIASSIAKVIASILTYPHEVIRAKLQ 246
Query: 240 VDDY---KYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEM 291
K+SG ID ++ SEGI GLY+G + R+ P+ F YEM
Sbjct: 247 EQGQIRNAETKYSGVIDCITKVFRSEGIPGLYRGCATNLLRTTPSAVITFTTYEM 301
Score = 44.7 bits (104), Expect = 6e-05
Identities = 28/86 (32%), Positives = 40/86 (45%), Gaps = 5/86 (5%)
Query: 216 AGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGS-----IDAFRRILASEGIKGLYKG 270
AG AGA FV P DV+K+ +QV SG I + + I+ EG +G+Y+G
Sbjct: 23 AGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQRGGVIITSLKNIIKEEGYRGMYRG 82
Query: 271 FGPAMFRSVPANAACFLAYEMTRSAL 296
P + +P A F Y + L
Sbjct: 83 LSPTIIALLPNWAVYFSVYGKLKDVL 108
Score = 42.4 bits (98), Expect = 3e-04
Identities = 25/76 (32%), Positives = 35/76 (45%), Gaps = 1/76 (1%)
Query: 20 IVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAAL 79
I+ +P + I+ KLQ Q I +YSG ID + + +EG GLY+G L
Sbjct: 233 ILTYPHEVIRAKLQEQGQ-IRNAETKYSGVIDCITKVFRSEGIPGLYRGCATNLLRTTPS 291
Query: 80 NAVLFTVRGQMEALFR 95
+ FT M FR
Sbjct: 292 AVITFTTYEMMLRFFR 307
>At4g01100 putative carrier protein
Length = 352
Score = 111 bits (278), Expect = 4e-25
Identities = 93/320 (29%), Positives = 140/320 (43%), Gaps = 38/320 (11%)
Query: 4 VAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPR 63
+ K L AG G P + +K+ LQ Q P + +YSG + +K EG R
Sbjct: 38 ICKSLFAGGVAGGVSRTAVAPLERMKILLQVQN---PHNI-KYSGTVQGLKHIWRTEGLR 93
Query: 64 GLYKGMGAPLATVAALNAVLFTVRGQMEA----LFRSHPG---ASLTVHQQVFCGAGAGL 116
GL+KG G A + +AV F Q ++R G A LT ++ GA AG+
Sbjct: 94 GLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENAQLTPLLRLGAGATAGI 153
Query: 117 AVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVP 176
P ++++ RL Q TA +Y G VLR EG R L++G +P
Sbjct: 154 IAMSATYPMDMVRGRLTVQ-------TANSPYQYRGIAHALATVLREEG-PRALYRGWLP 205
Query: 177 TMAREVPGNAAMFGAYEASKQLLA-----GGPDTSGLGRGSLIVAGGLAGASFWFFVYPT 231
++ VP F YE+ K L G + + L + + G +AG YP
Sbjct: 206 SVIGVVPYVGLNFSVYESLKDWLVKENPYGLVENNELTVVTRLTCGAIAGTVGQTIAYPL 265
Query: 232 DVVKSVIQVDDYKYP--------------KFSGSIDAFRRILASEGIKGLYKGFGPAMFR 277
DV++ +Q+ +K +++G +DAFR+ + EG LYKG P +
Sbjct: 266 DVIRRRMQMVGWKDASAIVTGEGRSTASLEYTGMVDAFRKTVRHEGFGALYKGLVPNSVK 325
Query: 278 SVPANAACFLAYEMTRSALG 297
VP+ A F+ YEM + LG
Sbjct: 326 VVPSIAIAFVTYEMVKDVLG 345
Score = 73.6 bits (179), Expect = 1e-13
Identities = 62/201 (30%), Positives = 90/201 (43%), Gaps = 16/201 (7%)
Query: 103 TVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLR 162
++ + +F G AG V P E +K LQ Q+ +KY G + +H+ R
Sbjct: 37 SICKSLFAGGVAGGVSRTAVAPLERMKILLQVQN--------PHNIKYSGTVQGLKHIWR 88
Query: 163 SEGGARGLFKGLVPTMAREVPGNAAMFGAYE-ASKQLLAGGPDTSGLGRGSL-----IVA 216
+E G RGLFKG AR VP +A F +YE AS +L +G L + A
Sbjct: 89 TE-GLRGLFKGNGTNCARIVPNSAVKFFSYEQASNGILYMYRQRTGNENAQLTPLLRLGA 147
Query: 217 GGLAGASFWFFVYPTDVVKSVIQVDDYKYP-KFSGSIDAFRRILASEGIKGLYKGFGPAM 275
G AG YP D+V+ + V P ++ G A +L EG + LY+G+ P++
Sbjct: 148 GATAGIIAMSATYPMDMVRGRLTVQTANSPYQYRGIAHALATVLREEGPRALYRGWLPSV 207
Query: 276 FRSVPANAACFLAYEMTRSAL 296
VP F YE + L
Sbjct: 208 IGVVPYVGLNFSVYESLKDWL 228
>At5g48970 mitochondrial carrier protein-like
Length = 339
Score = 107 bits (266), Expect = 1e-23
Identities = 91/321 (28%), Positives = 137/321 (42%), Gaps = 46/321 (14%)
Query: 7 DLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTP------IPGQLP---RYSGAIDAVKQTL 57
D +AG G V P D IK++ Q Q P + G L +Y+G + A K
Sbjct: 21 DASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIF 80
Query: 58 AAEGPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFC----GAG 113
EG RG ++G L V ++ FTV ++++ +H + GA
Sbjct: 81 REEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFVSGAL 140
Query: 114 AGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKG 173
AG A + P +L++ + LA G V Y +++S G RGL+ G
Sbjct: 141 AGCAATLGSYPFDLLR------TILASQGEPKV---YPTMRSAFVDIIQSRG-IRGLYNG 190
Query: 174 LVPTMAREVPGNAAMFGAYEASKQLLAG------------GPDTSGLGRGSLIVAGGLAG 221
L PT+ VP FG Y+ K+ + DT+ L L + G AG
Sbjct: 191 LTPTLVEIVPYAGLQFGTYDMFKRWMMDWNRYKLSSKIPINVDTN-LSSFQLFICGLGAG 249
Query: 222 ASFWFFVYPTDVVKSVIQVDDY-KYPKFSGSI---------DAFRRILASEGIKGLYKGF 271
S +P DVVK Q++ ++P++ + D R+I+ SEG GLYKG
Sbjct: 250 TSAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYRNMLDGLRQIMISEGWHGLYKGI 309
Query: 272 GPAMFRSVPANAACFLAYEMT 292
P+ ++ PA A F+AYE T
Sbjct: 310 VPSTVKAAPAGAVTFVAYEFT 330
Score = 66.6 bits (161), Expect = 2e-11
Identities = 52/204 (25%), Positives = 83/204 (40%), Gaps = 20/204 (9%)
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGM 69
+G G A + +PFD ++ L SQ G+ Y A + + G RGLY G+
Sbjct: 137 SGALAGCAATLGSYPFDLLRTILASQ-----GEPKVYPTMRSAFVDIIQSRGIRGLYNGL 191
Query: 70 GAPLATVAALNAVLFTV------------RGQMEALFRSHPGASLTVHQQVFCGAGAGLA 117
L + + F R ++ + + +L+ Q CG GAG +
Sbjct: 192 TPTLVEIVPYAGLQFGTYDMFKRWMMDWNRYKLSSKIPINVDTNLSSFQLFICGLGAGTS 251
Query: 118 VSFLVCPTELIKCRLQAQSTLAGS--GTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLV 175
+ P +++K R Q + G Y +D R ++ SEG GL+KG+V
Sbjct: 252 AKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYRNMLDGLRQIMISEGW-HGLYKGIV 310
Query: 176 PTMAREVPGNAAMFGAYEASKQLL 199
P+ + P A F AYE + L
Sbjct: 311 PSTVKAAPAGAVTFVAYEFTSDWL 334
Score = 30.4 bits (67), Expect = 1.2
Identities = 22/92 (23%), Positives = 36/92 (38%), Gaps = 13/92 (14%)
Query: 216 AGGLAGASFWFFVYPTDVVKSVIQVDDYKYP-------------KFSGSIDAFRRILASE 262
AG ++G P DV+K QV K++G + A + I E
Sbjct: 24 AGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIFREE 83
Query: 263 GIKGLYKGFGPAMFRSVPANAACFLAYEMTRS 294
G +G ++G PA+ +P + F +S
Sbjct: 84 GFRGFWRGNVPALLMVMPYTSIQFTVLHKLKS 115
>At2g47490 mitochondrial carrier like protein
Length = 312
Score = 106 bits (264), Expect = 2e-23
Identities = 84/298 (28%), Positives = 129/298 (43%), Gaps = 13/298 (4%)
Query: 3 DVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLP-RYSGAIDAVKQTLAAEG 61
+V + AG G P D IK + Q P G + S + +++Q EG
Sbjct: 12 NVLCNAAAGAAAGVVAATFVCPLDVIKTRFQVHGLPKLGDANIKGSLIVGSLEQIFKREG 71
Query: 62 PRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFL 121
RGLY+G+ + + + A+ FT+ Q+++ S+ L+V V +GAG A +
Sbjct: 72 MRGLYRGLSPTVMALLSNWAIYFTMYDQLKSFLCSNDH-KLSVGANVLAASGAGAATTIA 130
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
P ++K RLQ Q G V Y R + EG RGL+ GLVP +A
Sbjct: 131 TNPLWVVKTRLQTQGMRVG------IVPYKSTFSALRRIAYEEG-IRGLYSGLVPALAG- 182
Query: 182 VPGNAAMFGAYEASKQLLA--GGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQ 239
+ A F YE K LA G L + VA +A YP +VV++ +Q
Sbjct: 183 ISHVAIQFPTYEMIKVYLAKKGDKSVDNLNARDVAVASSIAKIFASTLTYPHEVVRARLQ 242
Query: 240 VDDYKYPK-FSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
+ K +SG D +++ +G G Y+G + R+ PA F ++EM L
Sbjct: 243 EQGHHSEKRYSGVRDCIKKVFEKDGFPGFYRGCATNLLRTTPAAVITFTSFEMVHRFL 300
Score = 75.5 bits (184), Expect = 3e-14
Identities = 62/200 (31%), Positives = 87/200 (43%), Gaps = 9/200 (4%)
Query: 96 SHPGASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMD 155
SHP S V GA AG+ + VCP ++IK R Q L G A + G +
Sbjct: 5 SHPPNSKNVLCNAAAGAAAGVVAATFVCPLDVIKTRFQVHG-LPKLGDANIK---GSLIV 60
Query: 156 VARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIV 215
+ + G RGL++GL PT+ + A F Y+ K L +G L
Sbjct: 61 GSLEQIFKREGMRGLYRGLSPTVMALLSNWAIYFTMYDQLKSFLCSNDHKLSVGANVLAA 120
Query: 216 AGGLAGASFWFFVYPTDVVKSVIQVDDYK--YPKFSGSIDAFRRILASEGIKGLYKGFGP 273
+G AGA+ P VVK+ +Q + + + A RRI EGI+GLY G P
Sbjct: 121 SG--AGAATTIATNPLWVVKTRLQTQGMRVGIVPYKSTFSALRRIAYEEGIRGLYSGLVP 178
Query: 274 AMFRSVPANAACFLAYEMTR 293
A+ + A F YEM +
Sbjct: 179 AL-AGISHVAIQFPTYEMIK 197
>At3g54110 uncoupling protein (ucp/PUMP)
Length = 306
Score = 101 bits (251), Expect = 6e-22
Identities = 88/298 (29%), Positives = 126/298 (41%), Gaps = 26/298 (8%)
Query: 6 KDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQ--LPRYSGAIDAVKQTLAAEGPR 63
K F + P DT KV+LQ Q + + G LP+Y G + V EG R
Sbjct: 13 KTFACSAFAACVGEVCTIPLDTAKVRLQLQKSALAGDVTLPKYRGLLGTVGTIAREEGLR 72
Query: 64 GLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPG----ASLTVHQQVFCGAGAGLAVS 119
L+KG+ L + G E + + G + + +++ G G
Sbjct: 73 SLWKGVVPGLHRQCLFGGLRI---GMYEPVKNLYVGKDFVGDVPLSKKILAGLTTGALGI 129
Query: 120 FLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMA 179
+ PT+L+K RLQA+ LA A +Y G ++ ++R EG R L+ GL P +A
Sbjct: 130 MVANPTDLVKVRLQAEGKLA----AGAPRRYSGALNAYSTIVRQEG-VRALWTGLGPNVA 184
Query: 180 REVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVY----PTDVVK 235
R NAA +Y+ K+ + P G +V L+G FF P DVVK
Sbjct: 185 RNAIINAAELASYDQVKETILKIP-----GFTDNVVTHILSGLGAGFFAVCIGSPVDVVK 239
Query: 236 SVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTR 293
S + D Y G+ID F + L S+G YKGF P R N FL E +
Sbjct: 240 SRMMGDSGAY---KGTIDCFVKTLKSDGPMAFYKGFIPNFGRLGSWNVIMFLTLEQAK 294
Score = 84.3 bits (207), Expect = 7e-17
Identities = 59/199 (29%), Positives = 92/199 (45%), Gaps = 14/199 (7%)
Query: 1 MGDV--AKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLA 58
+GDV +K + AG GA ++V +P D +KV+LQ++ G RYSGA++A +
Sbjct: 109 VGDVPLSKKILAGLTTGALGIMVANPTDLVKVRLQAEGKLAAGAPRRYSGALNAYSTIVR 168
Query: 59 AEGPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAV 118
EG R L+ G+G +A A +NA Q++ PG + V + G GAG
Sbjct: 169 QEGVRALWTGLGPNVARNAIINAAELASYDQVKETILKIPGFTDNVVTHILSGLGAGFFA 228
Query: 119 SFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTM 178
+ P +++K R+ S Y G +D L+S+ G +KG +P
Sbjct: 229 VCIGSPVDVVKSRMMGDSG-----------AYKGTIDCFVKTLKSD-GPMAFYKGFIPNF 276
Query: 179 AREVPGNAAMFGAYEASKQ 197
R N MF E +K+
Sbjct: 277 GRLGSWNVIMFLTLEQAKK 295
Score = 80.5 bits (197), Expect = 1e-15
Identities = 60/202 (29%), Positives = 92/202 (44%), Gaps = 9/202 (4%)
Query: 100 ASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQ-STLAGSGTAAVAVKYGGPMDVAR 158
+ L++ + C A A P + K RLQ Q S LAG T KY G +
Sbjct: 7 SDLSLPKTFACSAFAACVGEVCTIPLDTAKVRLQLQKSALAGDVTLP---KYRGLLGTVG 63
Query: 159 HVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGG 218
+ R EG R L+KG+VP + R+ G YE K L G + I+AG
Sbjct: 64 TIAREEG-LRSLWKGVVPGLHRQCLFGGLRIGMYEPVKNLYVGKDFVGDVPLSKKILAGL 122
Query: 219 LAGASFWFFVYPTDVVKSVIQVDDYKYP----KFSGSIDAFRRILASEGIKGLYKGFGPA 274
GA PTD+VK +Q + ++SG+++A+ I+ EG++ L+ G GP
Sbjct: 123 TTGALGIMVANPTDLVKVRLQAEGKLAAGAPRRYSGALNAYSTIVRQEGVRALWTGLGPN 182
Query: 275 MFRSVPANAACFLAYEMTRSAL 296
+ R+ NAA +Y+ + +
Sbjct: 183 VARNAIINAAELASYDQVKETI 204
Score = 40.8 bits (94), Expect = 9e-04
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 7/93 (7%)
Query: 3 DVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGP 62
+V + +G G + +G P D +K ++ G Y G ID +TL ++GP
Sbjct: 213 NVVTHILSGLGAGFFAVCIGSPVDVVKSRMM-------GDSGAYKGTIDCFVKTLKSDGP 265
Query: 63 RGLYKGMGAPLATVAALNAVLFTVRGQMEALFR 95
YKG + + N ++F Q + R
Sbjct: 266 MAFYKGFIPNFGRLGSWNVIMFLTLEQAKKYVR 298
>At1g74240 mitochondrial carrier like protein
Length = 364
Score = 100 bits (248), Expect = 1e-21
Identities = 92/301 (30%), Positives = 132/301 (43%), Gaps = 26/301 (8%)
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGM 69
AG FG HP DT+K +LQSQ I R + ++ +G +G Y+G+
Sbjct: 42 AGAFGEGMM----HPVDTLKTRLQSQI--IMNATQRQKSILQMLRTVWVGDGLKGFYRGI 95
Query: 70 GAPLATVAALNAVLFT-VRGQMEALFRSHPGASLTVHQQVFCGAGAGLAV-SFLVCPTEL 127
+ A A F + + + SHP SL H F G + SF+ P E+
Sbjct: 96 APGVTGSLATGATYFGFIESTKKWIEESHP--SLAGHWAHFIAGAVGDTLGSFIYVPCEV 153
Query: 128 IKCRLQAQSTLAGSGTA----AVAVKYGGPMD-------VARHVLRSEGGARGLFKGLVP 176
IK R+Q Q T + + +V V+ G M A + E G +GL+ G
Sbjct: 154 IKQRMQIQGTSSSWSSYISRNSVPVQPRGDMYGYYTGMFQAGCSIWKEQGPKGLYAGYWS 213
Query: 177 TMAREVPGNAAMFGAYEASKQLLAGG----PDTSGLGRGSLIVAGGLAGASFWFFVYPTD 232
T+AR+VP M YE K L G P +V GGLAG + P D
Sbjct: 214 TLARDVPFAGLMVVFYEGLKDLTDQGKKKFPQYGVNSSIEGLVLGGLAGGLSAYLTTPLD 273
Query: 233 VVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMT 292
VVK+ +QV K+ G +DA +I EG +G ++G P + +PA+A F+A E
Sbjct: 274 VVKTRLQVQGSTI-KYKGWLDAVGQIWRKEGPQGFFRGSVPRVMWYLPASALTFMAVEFL 332
Query: 293 R 293
R
Sbjct: 333 R 333
Score = 39.3 bits (90), Expect = 0.003
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 5/61 (8%)
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
L G G + P D +K +LQ Q G +Y G +DAV Q EGP+G ++
Sbjct: 255 LVLGGLAGGLSAYLTTPLDVVKTRLQVQ-----GSTIKYKGWLDAVGQIWRKEGPQGFFR 309
Query: 68 G 68
G
Sbjct: 310 G 310
Score = 35.8 bits (81), Expect = 0.029
Identities = 21/79 (26%), Positives = 37/79 (46%), Gaps = 2/79 (2%)
Query: 217 GGLAGASFWFFVYPTDVVKSVI--QVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPA 274
GG+AGA ++P D +K+ + Q+ + + R + +G+KG Y+G P
Sbjct: 39 GGIAGAFGEGMMHPVDTLKTRLQSQIIMNATQRQKSILQMLRTVWVGDGLKGFYRGIAPG 98
Query: 275 MFRSVPANAACFLAYEMTR 293
+ S+ A F E T+
Sbjct: 99 VTGSLATGATYFGFIESTK 117
>At5g01340 unknown protein
Length = 309
Score = 98.6 bits (244), Expect = 4e-21
Identities = 84/281 (29%), Positives = 124/281 (43%), Gaps = 20/281 (7%)
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGM 69
+G+ GG + P D IK +LQ ++ Y G + + EG R L+KG+
Sbjct: 19 SGSLGGVVEACCLQPIDVIKTRLQLD------RVGAYKGIAHCGSKVVRTEGVRALWKGL 72
Query: 70 GAPLATVAALNAVLFTVRGQM-EALFRSHPGASLTVHQQVFCGAGAGLAVSF-LVCPTEL 127
P AT L L M + F+ ++ + G GAG+ + +V P E+
Sbjct: 73 -TPFATHLTLKYTLRMGSNAMFQTAFKDSETGKVSNRGRFLSGFGAGVLEALAIVTPFEV 131
Query: 128 IKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAA 187
+K RLQ Q G + KY GP+ AR ++R E GL+ G PT+ R A
Sbjct: 132 VKIRLQQQK-----GLSPELFKYKGPIHCARTIVREES-ILGLWSGAAPTVMRNGTNQAV 185
Query: 188 MFGAYEASKQLLAGGPDTSG--LGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQV---DD 242
MF A A LL + G L +++G LAG + F P DVVK+ + D
Sbjct: 186 MFTAKNAFDILLWNKHEGDGKILQPWQSMISGFLAGTAGPFCTGPFDVVKTRLMAQSRDS 245
Query: 243 YKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANA 283
++ G + A R I A EG+ L++G P + R P A
Sbjct: 246 EGGIRYKGMVHAIRTIYAEEGLVALWRGLLPRLMRIPPGQA 286
Score = 68.2 bits (165), Expect = 5e-12
Identities = 52/174 (29%), Positives = 81/174 (45%), Gaps = 10/174 (5%)
Query: 19 LIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAA 78
L + PF+ +K++LQ Q P +L +Y G I + + E GL+ G +
Sbjct: 123 LAIVTPFEVVKIRLQQQKGLSP-ELFKYKGPIHCARTIVREESILGLWSGAAPTVMRNGT 181
Query: 79 LNAVLFTVRGQMEAL-FRSHPGAS--LTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQ 135
AV+FT + + L + H G L Q + G AG A F P +++K RL AQ
Sbjct: 182 NQAVMFTAKNAFDILLWNKHEGDGKILQPWQSMISGFLAGTAGPFCTGPFDVVKTRLMAQ 241
Query: 136 STLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMF 189
S + G ++Y G + R + EG L++GL+P + R PG A M+
Sbjct: 242 SRDSEGG-----IRYKGMVHAIRTIYAEEGLV-ALWRGLLPRLMRIPPGQAIMW 289
Score = 33.5 bits (75), Expect = 0.14
Identities = 17/59 (28%), Positives = 32/59 (53%), Gaps = 2/59 (3%)
Query: 215 VAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGP 273
V+G L G + P DV+K+ +Q+D + + G +++ +EG++ L+KG P
Sbjct: 18 VSGSLGGVVEACCLQPIDVIKTRLQLD--RVGAYKGIAHCGSKVVRTEGVRALWKGLTP 74
>At3g21390 unknown protein
Length = 335
Score = 97.1 bits (240), Expect = 1e-20
Identities = 85/318 (26%), Positives = 134/318 (41%), Gaps = 43/318 (13%)
Query: 7 DLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIP------GQL-PRYSGAIDAVKQTLAA 59
D +AG GA +V P D IK++ Q Q P QL P+Y+G K
Sbjct: 18 DASAGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTKDIFRE 77
Query: 60 EGPRGLYKGMGAPLATVAALNAVLFTVRGQMEALF----RSHPGASLTVHQQVFCGAGAG 115
EG G ++G L V ++ F V ++++ ++ A L+ + GA AG
Sbjct: 78 EGLSGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAENHAQLSPYLSYISGALAG 137
Query: 116 LAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLV 175
A + P +L++ + LA G V Y ++++ G +GL+ GL
Sbjct: 138 CAATVGSYPFDLLR------TVLASQGEPKV---YPNMRSAFLSIVQTRG-IKGLYAGLS 187
Query: 176 PTMAREVPGNAAMFGAYEASKQLL------------AGGPDTSGLGRGSLIVAGGLAGAS 223
PT+ +P FG Y+ K+ + + L L + G +G
Sbjct: 188 PTLIEIIPYAGLQFGTYDTFKRWSMVYNKRYRSSSSSSTNPSDSLSSFQLFLCGLASGTV 247
Query: 224 FWFFVYPTDVVKSVIQVDDY-KYPKFSGSI---------DAFRRILASEGIKGLYKGFGP 273
+P DVVK QV+ ++PK+ + D +IL SEG GLYKG P
Sbjct: 248 SKLVCHPLDVVKKRFQVEGLQRHPKYGARVELNAYKNMFDGLGQILRSEGWHGLYKGIVP 307
Query: 274 AMFRSVPANAACFLAYEM 291
+ ++ PA A F+AYE+
Sbjct: 308 STIKAAPAGAVTFVAYEL 325
Score = 41.2 bits (95), Expect = 7e-04
Identities = 27/90 (30%), Positives = 38/90 (42%), Gaps = 11/90 (12%)
Query: 216 AGGLAGASFWFFVYPTDVVKSVIQV-----------DDYKYPKFSGSIDAFRRILASEGI 264
AGG+AGA P DV+K QV D PK++G + I EG+
Sbjct: 21 AGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTKDIFREEGL 80
Query: 265 KGLYKGFGPAMFRSVPANAACFLAYEMTRS 294
G ++G PA+ VP + F +S
Sbjct: 81 SGFWRGNVPALLMVVPYTSIQFAVLHKVKS 110
>At5g58970 uncoupling protein AtUCP2
Length = 305
Score = 95.5 bits (236), Expect = 3e-20
Identities = 84/278 (30%), Positives = 126/278 (45%), Gaps = 22/278 (7%)
Query: 24 PFDTIKVKLQSQ---PTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAALN 80
P DT KV+LQ Q PT LP+Y G+I + EG GL+KG+ A L
Sbjct: 32 PLDTAKVRLQLQRKIPTGDGENLPKYRGSIGTLATIAREEGISGLWKGVIAGLHRQCIYG 91
Query: 81 AVLFTVRGQMEALF-RSHPGASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLA 139
+ + ++ L S + ++Q++ G + PT+L+K RLQ++ L
Sbjct: 92 GLRIGLYEPVKTLLVGSDFIGDIPLYQKILAALLTGAIAIIVANPTDLVKVRLQSEGKLP 151
Query: 140 GSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLL 199
A V +Y G +D +++ EG L+ GL P +AR NAA +Y+ K+ +
Sbjct: 152 ----AGVPRRYAGAVDAYFTIVKLEG-VSALWTGLGPNIARNAIVNAAELASYDQIKETI 206
Query: 200 AGGPDTSGLGRGSLIVAGGLAGASFWFFVY----PTDVVKSVIQVDDYKYPKFSGSIDAF 255
P R S ++ LAG + FF P DVVKS + D + ++D F
Sbjct: 207 MKIP----FFRDS-VLTHLLAGLAAGFFAVCIGSPIDVVKSRMMGDS----TYRNTVDCF 257
Query: 256 RRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTR 293
+ + +EGI YKGF P R NA FL E +
Sbjct: 258 IKTMKTEGIMAFYKGFLPNFTRLGTWNAIMFLTLEQVK 295
Score = 77.4 bits (189), Expect = 9e-15
Identities = 57/204 (27%), Positives = 90/204 (43%), Gaps = 8/204 (3%)
Query: 98 PGASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVA 157
P ++ + C A A P + K RLQ Q + +G KY G +
Sbjct: 6 PRIEISFLETFICSAFAACFAELCTIPLDTAKVRLQLQRKIP-TGDGENLPKYRGSIGTL 64
Query: 158 RHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAG 217
+ R EG GL+KG++ + R+ G YE K LL G + I+A
Sbjct: 65 ATIAREEG-ISGLWKGVIAGLHRQCIYGGLRIGLYEPVKTLLVGSDFIGDIPLYQKILAA 123
Query: 218 GLAGASFWFFVYPTDVVKSVIQVDDYKYP-----KFSGSIDAFRRILASEGIKGLYKGFG 272
L GA PTD+VK +Q + K P +++G++DA+ I+ EG+ L+ G G
Sbjct: 124 LLTGAIAIIVANPTDLVKVRLQSEG-KLPAGVPRRYAGAVDAYFTIVKLEGVSALWTGLG 182
Query: 273 PAMFRSVPANAACFLAYEMTRSAL 296
P + R+ NAA +Y+ + +
Sbjct: 183 PNIARNAIVNAAELASYDQIKETI 206
Score = 72.4 bits (176), Expect = 3e-13
Identities = 55/200 (27%), Positives = 88/200 (43%), Gaps = 15/200 (7%)
Query: 1 MGDVA--KDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLA 58
+GD+ + + A GA +IV +P D +KV+LQS+ G RY+GA+DA +
Sbjct: 111 IGDIPLYQKILAALLTGAIAIIVANPTDLVKVRLQSEGKLPAGVPRRYAGAVDAYFTIVK 170
Query: 59 AEGPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAV 118
EG L+ G+G +A A +NA Q++ P +V + G AG
Sbjct: 171 LEGVSALWTGLGPNIARNAIVNAAELASYDQIKETIMKIPFFRDSVLTHLLAGLAAGFFA 230
Query: 119 SFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTM 178
+ P +++K R+ ST Y +D +++E G +KG +P
Sbjct: 231 VCIGSPIDVVKSRMMGDST------------YRNTVDCFIKTMKTE-GIMAFYKGFLPNF 277
Query: 179 AREVPGNAAMFGAYEASKQL 198
R NA MF E K++
Sbjct: 278 TRLGTWNAIMFLTLEQVKKV 297
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/91 (25%), Positives = 37/91 (40%), Gaps = 8/91 (8%)
Query: 4 VAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPR 63
V L AG G + +G P D +K ++ T Y +D +T+ EG
Sbjct: 216 VLTHLLAGLAAGFFAVCIGSPIDVVKSRMMGDST--------YRNTVDCFIKTMKTEGIM 267
Query: 64 GLYKGMGAPLATVAALNAVLFTVRGQMEALF 94
YKG + NA++F Q++ +F
Sbjct: 268 AFYKGFLPNFTRLGTWNAIMFLTLEQVKKVF 298
>At5g51050 calcium-binding transporter-like protein
Length = 487
Score = 92.0 bits (227), Expect = 3e-19
Identities = 88/298 (29%), Positives = 132/298 (43%), Gaps = 38/298 (12%)
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGM 69
AG GAA P D +KV LQ Q T + +A+K G RG ++G
Sbjct: 214 AGGIAGAASRTATAPLDRLKVLLQIQKTD--------ARIREAIKLIWKQGGVRGFFRGN 265
Query: 70 GAPLATVAALNAVLFTVRGQMEALFRSHPG---ASLTVHQQVFCGAGAGLAVSFLVCPTE 126
G + VA +A+ F + + G A + ++F G AG + P +
Sbjct: 266 GLNIVKVAPESAIKFYAYELFKNAIGENMGEDKADIGTTVRLFAGGMAGAVAQASIYPLD 325
Query: 127 LIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNA 186
L+K RLQ ++ AG VAV G + + +L EG R +KGL P++ +P
Sbjct: 326 LVKTRLQTYTSQAG-----VAVPRLGTL--TKDILVHEG-PRAFYKGLFPSLLGIIPYAG 377
Query: 187 AMFGAYEASKQL--------LAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVI 238
AYE K L GP LG G++ G GA+ VYP VV++ +
Sbjct: 378 IDLAAYETLKDLSRTYILQDAEPGPLVQ-LGCGTI---SGALGATC---VYPLQVVRTRM 430
Query: 239 QVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
Q + + SG FRR ++ EG + LYKG P + + VPA + ++ YE + +L
Sbjct: 431 QAERAR-TSMSG---VFRRTISEEGYRALYKGLLPNLLKVVPAASITYMVYEAMKKSL 484
Score = 63.2 bits (152), Expect = 2e-10
Identities = 53/198 (26%), Positives = 84/198 (41%), Gaps = 26/198 (13%)
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
L AG GA +P D +K +LQ+ + +PR K L EGPR YK
Sbjct: 307 LFAGGMAGAVAQASIYPLDLVKTRLQTYTSQAGVAVPRLG---TLTKDILVHEGPRAFYK 363
Query: 68 GMGAPLATVAALNAVLFTVRGQMEALFRSH------PGASLTVHQQVFCGAGAGLAVSFL 121
G+ L + + ++ L R++ PG + Q+ CG +G +
Sbjct: 364 GLFPSLLGIIPYAGIDLAAYETLKDLSRTYILQDAEPGPLV----QLGCGTISGALGATC 419
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
V P ++++ R+QA+ V R + SE G R L+KGL+P + +
Sbjct: 420 VYPLQVVRTRMQAERARTSMS------------GVFRRTI-SEEGYRALYKGLLPNLLKV 466
Query: 182 VPGNAAMFGAYEASKQLL 199
VP + + YEA K+ L
Sbjct: 467 VPAASITYMVYEAMKKSL 484
Score = 45.4 bits (106), Expect = 4e-05
Identities = 27/89 (30%), Positives = 48/89 (53%), Gaps = 6/89 (6%)
Query: 210 RGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSI-DAFRRILASEGIKGLY 268
R + +AGG+AGA+ P D +K ++Q+ K I +A + I G++G +
Sbjct: 208 RSNYFIAGGIAGAASRTATAPLDRLKVLLQIQ-----KTDARIREAIKLIWKQGGVRGFF 262
Query: 269 KGFGPAMFRSVPANAACFLAYEMTRSALG 297
+G G + + P +A F AYE+ ++A+G
Sbjct: 263 RGNGLNIVKVAPESAIKFYAYELFKNAIG 291
>At4g32400 adenylate translocator (brittle-1) - like protein
Length = 392
Score = 90.1 bits (222), Expect = 1e-18
Identities = 79/292 (27%), Positives = 122/292 (41%), Gaps = 23/292 (7%)
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
L +G GA V P +TI+ L + + + + EG GL++
Sbjct: 114 LLSGAVAGAVSRTVVAPLETIRTHLMVGSGG--------NSSTEVFSDIMKHEGWTGLFR 165
Query: 68 GMGAPLATVAALNAV-LFTVRGQMEALFRSHPGAS-LTVHQQVFCGAGAGLAVSFLVCPT 125
G + VA AV LF + L H S + + + GA AG++ + L P
Sbjct: 166 GNLVNVIRVAPARAVELFVFETVNKKLSPPHGQESKIPIPASLLAGACAGVSQTLLTYPL 225
Query: 126 ELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGN 185
EL+K RL Q + Y G D ++R EG L++GL P++ VP
Sbjct: 226 ELVKTRLTIQRGV-----------YKGIFDAFLKIIREEGPTE-LYRGLAPSLIGVVPYA 273
Query: 186 AAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYK- 244
A + AY++ ++ +G ++ G LAGA +P +V + +QV
Sbjct: 274 ATNYFAYDSLRKAYRSFSKQEKIGNIETLLIGSLAGALSSTATFPLEVARKHMQVGAVSG 333
Query: 245 YPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
+ + A IL EGI G YKG GP+ + VPA F+ YE + L
Sbjct: 334 RVVYKNMLHALVTILEHEGILGWYKGLGPSCLKLVPAAGISFMCYEACKKIL 385
>At5g01500 unknown protein
Length = 415
Score = 89.4 bits (220), Expect = 2e-18
Identities = 78/288 (27%), Positives = 131/288 (45%), Gaps = 21/288 (7%)
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYS-GAIDAVKQTLAAEGPRGLYKG 68
AG F GAA V P D IK+ +Q+ Q + + G I+A+ EG +G +KG
Sbjct: 121 AGAFAGAAAKSVTAPLDRIKLLMQTHGVRAGQQSAKKAIGFIEAITLIGKEEGIKGYWKG 180
Query: 69 MGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVCPTELI 128
+ + +AV + LFR G L+V ++ GA AG+ + + P +++
Sbjct: 181 NLPQVIRIVPYSAVQLFAYETYKKLFRGKDG-QLSVLGRLGAGACAGMTSTLITYPLDVL 239
Query: 129 KCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAM 188
+ RL AV Y VA ++LR EG A + GL P++ P A
Sbjct: 240 RLRL------------AVEPGYRTMSQVALNMLREEGVA-SFYNGLGPSLLSIAPYIAIN 286
Query: 189 FGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKF 248
F ++ K+ L P+ S ++ +A A YP D ++ +Q+ K +
Sbjct: 287 FCVFDLVKKSL---PEKYQQKTQSSLLTAVVAAAIATGTCYPLDTIRRQMQL---KGTPY 340
Query: 249 SGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
+DAF I+A EG+ GLY+GF P +S+P ++ +++ + +
Sbjct: 341 KSVLDAFSGIIAREGVVGLYRGFVPNALKSMPNSSIKLTTFDIVKKLI 388
Score = 49.7 bits (117), Expect = 2e-06
Identities = 43/194 (22%), Positives = 78/194 (40%), Gaps = 24/194 (12%)
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
L AG G ++ +P D ++++L +P Y L EG Y
Sbjct: 219 LGAGACAGMTSTLITYPLDVLRLRLAVEPG--------YRTMSQVALNMLREEGVASFYN 270
Query: 68 GMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVC-PTE 126
G+G L ++A A+ F V + + +S P Q A A++ C P +
Sbjct: 271 GLGPSLLSIAPYIAINFCV---FDLVKKSLPEKYQQKTQSSLLTAVVAAAIATGTCYPLD 327
Query: 127 LIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNA 186
I+ ++Q + T Y +D ++ EG GL++G VP + +P ++
Sbjct: 328 TIRRQMQLKGT-----------PYKSVLDAFSGIIAREGVV-GLYRGFVPNALKSMPNSS 375
Query: 187 AMFGAYEASKQLLA 200
++ K+L+A
Sbjct: 376 IKLTTFDIVKKLIA 389
Score = 43.1 bits (100), Expect = 2e-04
Identities = 30/84 (35%), Positives = 40/84 (46%), Gaps = 5/84 (5%)
Query: 212 SLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYK-----YPKFSGSIDAFRRILASEGIKG 266
+L AG AGA+ P D +K ++Q + K G I+A I EGIKG
Sbjct: 117 ALFFAGAFAGAAAKSVTAPLDRIKLLMQTHGVRAGQQSAKKAIGFIEAITLIGKEEGIKG 176
Query: 267 LYKGFGPAMFRSVPANAACFLAYE 290
+KG P + R VP +A AYE
Sbjct: 177 YWKGNLPQVIRIVPYSAVQLFAYE 200
>At5g66380 unknown protein
Length = 308
Score = 88.6 bits (218), Expect = 4e-18
Identities = 75/296 (25%), Positives = 120/296 (40%), Gaps = 20/296 (6%)
Query: 6 KDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGL 65
++ TAG G A + H D ++ + Q LP Y AV EG RGL
Sbjct: 8 ENATAGAVAGFATVAAMHSLDVVRTRFQVNDGR-GSSLPTYKNTAHAVFTIARLEGLRGL 66
Query: 66 YKGM-GAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVCP 124
Y G A + + + F + R L+ + A AG V P
Sbjct: 67 YAGFFPAVIGSTVSWGLYFFFYGRAKQRYARGRDDEKLSPALHLASAAEAGALVCLCTNP 126
Query: 125 TELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPG 184
L+K RLQ Q+ L + Y G +D R +++ EG R L+KG+VP + V
Sbjct: 127 IWLVKTRLQLQTPLHQTQP------YSGLLDAFRTIVKEEG-PRALYKGIVPGLVL-VSH 178
Query: 185 NAAMFGAYEASKQLLAGGPDTSG--------LGRGSLIVAGGLAGASFWFFVYPTDVVKS 236
A F AYE ++++ + L GG + + YP V+++
Sbjct: 179 GAIQFTAYEELRKIIVDLKERRRKSESTDNLLNSADYAALGGSSKVAAVLLTYPFQVIRA 238
Query: 237 VIQV--DDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYE 290
+Q P++ S+ R EG++G Y+G + ++VPA++ F+ YE
Sbjct: 239 RLQQRPSTNGIPRYIDSLHVIRETARYEGLRGFYRGLTANLLKNVPASSITFIVYE 294
Score = 68.9 bits (167), Expect = 3e-12
Identities = 60/210 (28%), Positives = 92/210 (43%), Gaps = 19/210 (9%)
Query: 5 AKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRG 64
A L + GA + +P +K +LQ Q TP+ P YSG +DA + + EGPR
Sbjct: 107 ALHLASAAEAGALVCLCTNPIWLVKTRLQLQ-TPLHQTQP-YSGLLDAFRTIVKEEGPRA 164
Query: 65 LYKGMGAPLATVAALNAVLFTVRGQMEALF-----RSHPGAS----LTVHQQVFCGAGAG 115
LYKG+ P + + A+ FT ++ + R S L G +
Sbjct: 165 LYKGI-VPGLVLVSHGAIQFTAYEELRKIIVDLKERRRKSESTDNLLNSADYAALGGSSK 223
Query: 116 LAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLV 175
+A L P ++I+ RLQ + + G +Y + V R R EG RG ++GL
Sbjct: 224 VAAVLLTYPFQVIRARLQQRPSTNG------IPRYIDSLHVIRETARYEG-LRGFYRGLT 276
Query: 176 PTMAREVPGNAAMFGAYEASKQLLAGGPDT 205
+ + VP ++ F YE +LL P T
Sbjct: 277 ANLLKNVPASSITFIVYENVLKLLKQHPTT 306
>At5g61810 peroxisomal Ca-dependent solute carrier - like protein
Length = 478
Score = 87.4 bits (215), Expect = 8e-18
Identities = 75/294 (25%), Positives = 122/294 (40%), Gaps = 25/294 (8%)
Query: 5 AKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRG 64
+K L AG GA P D +KV LQ Q T + G + +K+ + G
Sbjct: 205 SKLLLAGGIAGAVSRTATAPLDRLKVALQVQRTNL--------GVVPTIKKIWREDKLLG 256
Query: 65 LYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVCP 124
++G G +A VA +A+ F ++ + G + ++ G AG + P
Sbjct: 257 FFRGNGLNVAKVAPESAIKFAAYEMLKPIIGGADG-DIGTSGRLLAGGLAGAVAQTAIYP 315
Query: 125 TELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPG 184
+L+K RLQ + G+ K + + G R ++GL P++ +P
Sbjct: 316 MDLVKTRLQTFVSEVGTPKLWKLTKD----------IWIQEGPRAFYRGLCPSLIGIIPY 365
Query: 185 NAAMFGAYEASKQLLAGG--PDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDD 242
AYE K L DT+ G + G +GA VYP V+++ +Q D
Sbjct: 366 AGIDLAAYETLKDLSRAHFLHDTAEPGPLIQLGCGMTSGALGASCVYPLQVIRTRMQADS 425
Query: 243 YKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
K + F + L EG+KG Y+G P F+ +P+ + +L YE + L
Sbjct: 426 SK----TSMGQEFLKTLRGEGLKGFYRGIFPNFFKVIPSASISYLVYEAMKKNL 475
Score = 56.6 bits (135), Expect = 2e-08
Identities = 52/208 (25%), Positives = 86/208 (41%), Gaps = 31/208 (14%)
Query: 2 GDVAKD--LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAA 59
GD+ L AG GA +P D +K +LQ+ + + P+ K
Sbjct: 291 GDIGTSGRLLAGGLAGAVAQTAIYPMDLVKTRLQTFVSEVG--TPKLW---KLTKDIWIQ 345
Query: 60 EGPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSH-------PGASLTVHQQVFCGA 112
EGPR Y+G+ L + + ++ L R+H PG + Q+ CG
Sbjct: 346 EGPRAFYRGLCPSLIGIIPYAGIDLAAYETLKDLSRAHFLHDTAEPGPLI----QLGCGM 401
Query: 113 GAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFK 172
+G + V P ++I+ R+QA S+ G + LR EG +G ++
Sbjct: 402 TSGALGASCVYPLQVIRTRMQADSSKTSMGQEFLKT------------LRGEG-LKGFYR 448
Query: 173 GLVPTMAREVPGNAAMFGAYEASKQLLA 200
G+ P + +P + + YEA K+ LA
Sbjct: 449 GIFPNFFKVIPSASISYLVYEAMKKNLA 476
Score = 43.1 bits (100), Expect = 2e-04
Identities = 26/88 (29%), Positives = 43/88 (48%), Gaps = 4/88 (4%)
Query: 210 RGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYK 269
R L++AGG+AGA P D +K +QV G + ++I + + G ++
Sbjct: 204 RSKLLLAGGIAGAVSRTATAPLDRLKVALQVQRTNL----GVVPTIKKIWREDKLLGFFR 259
Query: 270 GFGPAMFRSVPANAACFLAYEMTRSALG 297
G G + + P +A F AYEM + +G
Sbjct: 260 GNGLNVAKVAPESAIKFAAYEMLKPIIG 287
>At1g07030 unknown protein
Length = 326
Score = 87.4 bits (215), Expect = 8e-18
Identities = 82/294 (27%), Positives = 126/294 (41%), Gaps = 26/294 (8%)
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQS-QPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLY 66
+ AG+ G+ + + P DTIK +Q+ +P P+ + G +A + + EGP LY
Sbjct: 40 MIAGSIAGSVEHMAMFPVDTIKTHMQALRPCPL-----KPVGIREAFRSIIQKEGPSALY 94
Query: 67 KGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVCPTE 126
+G+ A +AV F+ + + + H G A ++ + P +
Sbjct: 95 RGIWAMGLGAGPAHAVYFSFYEVSKKYLSAGDQNNSVAH--AMSGVFATISSDAVFTPMD 152
Query: 127 LIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNA 186
++K RLQ G GT Y G D + VLR EG + T+ P A
Sbjct: 153 MVKQRLQM-----GEGT------YKGVWDCVKRVLREEG-IGAFYASYRTTVLMNAPFTA 200
Query: 187 AMFGAYEASKQ-LLAGGPDTSGLGRGSLI--VAGGLAGASFWFFVYPTDVVKSVIQVDDY 243
F YEA+K+ L+ PD G L+ AG AG P DVVK+ +Q
Sbjct: 201 VHFATYEAAKKGLMEFSPDRISDEEGWLVHATAGAAAGGLAAAVTTPLDVVKTQLQCQGV 260
Query: 244 -KYPKFSGSI--DAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRS 294
+F+ S R I+ +G +GL +G+ P M PA A C+ YE +S
Sbjct: 261 CGCDRFTSSSISHVLRTIVKKDGYRGLLRGWLPRMLFHAPAAAICWSTYEGVKS 314
Score = 58.9 bits (141), Expect = 3e-09
Identities = 51/201 (25%), Positives = 83/201 (40%), Gaps = 17/201 (8%)
Query: 96 SHPGASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMD 155
+H G L Q + G+ AG + P + IK +QA +K G +
Sbjct: 30 AHDG--LKFWQFMIAGSIAGSVEHMAMFPVDTIKTHMQA--------LRPCPLKPVGIRE 79
Query: 156 VARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIV 215
R +++ EG + L++G+ P +A F YE SK+ L+ G + + +
Sbjct: 80 AFRSIIQKEGPS-ALYRGIWAMGLGAGPAHAVYFSFYEVSKKYLSAGDQNNSVAHA---M 135
Query: 216 AGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAM 275
+G A S P D+VK +Q+ + Y G D +R+L EGI Y + +
Sbjct: 136 SGVFATISSDAVFTPMDMVKQRLQMGEGTY---KGVWDCVKRVLREEGIGAFYASYRTTV 192
Query: 276 FRSVPANAACFLAYEMTRSAL 296
+ P A F YE + L
Sbjct: 193 LMNAPFTAVHFATYEAAKKGL 213
>At5g42130 mitochondrial carrier protein-like
Length = 412
Score = 86.7 bits (213), Expect = 1e-17
Identities = 81/291 (27%), Positives = 118/291 (39%), Gaps = 27/291 (9%)
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGM 69
AG GA + P D IK KLQ++ G YS DA+ +T A+G G Y G+
Sbjct: 120 AGGLAGAFTYVTLLPLDAIKTKLQTK-----GASQVYSNTFDAIVKTFQAKGILGFYSGV 174
Query: 70 GAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVCPTELIK 129
A + +AV F ++L P TV GA + S ++ P ELI
Sbjct: 175 SAVIVGSTFSSAVYFGTCEFGKSLLSKFPDFP-TVLIPPTAGAMGNIISSAIMVPKELIT 233
Query: 130 CRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMF 189
R+QA ++ G V +L +G GL+ G T+ R +P +
Sbjct: 234 QRMQAGAS-------------GRSYQVLLKILEKDG-ILGLYAGYSATLLRNLPAGVLSY 279
Query: 190 GAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKS-------VIQVDD 242
++E K + S L + G LAGA P DVVK+ V VD
Sbjct: 280 SSFEYLKAAVLEKTKQSHLEPLQSVCCGALAGAISASITTPLDVVKTRLMTQIHVEAVDK 339
Query: 243 YKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTR 293
++G ++IL EG G +G GP + S +A + A+E R
Sbjct: 340 LGGAMYTGVAGTVKQILTEEGWVGFTRGMGPRVVHSACFSAIGYFAFETAR 390
Score = 52.0 bits (123), Expect = 4e-07
Identities = 52/202 (25%), Positives = 87/202 (42%), Gaps = 27/202 (13%)
Query: 102 LTVHQQVFCGAGAG-LAVSFL---VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVA 157
L+V ++ GAGAG LA +F + P + IK +LQ T + Y D
Sbjct: 108 LSVWERAIIGAGAGGLAGAFTYVTLLPLDAIKTKLQ---------TKGASQVYSNTFDAI 158
Query: 158 RHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLI--V 215
+++G G + G+ + +A FG E K LL+ PD + LI
Sbjct: 159 VKTFQAKG-ILGFYSGVSAVIVGSTFSSAVYFGTCEFGKSLLSKFPDFPTV----LIPPT 213
Query: 216 AGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSG-SIDAFRRILASEGIKGLYKGFGPA 274
AG + + P +++ +Q SG S +IL +GI GLY G+
Sbjct: 214 AGAMGNIISSAIMVPKELITQRMQAGA------SGRSYQVLLKILEKDGILGLYAGYSAT 267
Query: 275 MFRSVPANAACFLAYEMTRSAL 296
+ R++PA + ++E ++A+
Sbjct: 268 LLRNLPAGVLSYSSFEYLKAAV 289
>At2g37890 mitochondrial carrier like protein
Length = 337
Score = 85.1 bits (209), Expect = 4e-17
Identities = 82/302 (27%), Positives = 120/302 (39%), Gaps = 23/302 (7%)
Query: 6 KDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGL 65
++L AG GA P + + Q Q G + + + EG R
Sbjct: 43 QNLLAGGIAGAISKTCTAPLARLTILFQLQGMQSEGAVLSRPNLRREASRIINEEGYRAF 102
Query: 66 YKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQ-------QVFCGAGAGLAV 118
+KG + AV F + F S+P + G AG+
Sbjct: 103 WKGNLVTVVHRIPYTAVNFYAYEKYNLFFNSNPVVQSFIGNTSGNPIVHFVSGGLAGITA 162
Query: 119 SFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTM 178
+ P +L++ RL AQ A+ Y G R + R EG GL+KGL T+
Sbjct: 163 ATATYPLDLVRTRLAAQRN---------AIYYQGIEHTFRTICREEG-ILGLYKGLGATL 212
Query: 179 AREVPGNAAMFGAYEASKQLL-AGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSV 237
P A F AYE+ K + P+ S L +V+GGLAGA YP D+V+
Sbjct: 213 LGVGPSLAINFAAYESMKLFWHSHRPNDSDLVVS--LVSGGLAGAVSSTATYPLDLVRRR 270
Query: 238 IQVDDY---KYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRS 294
+QV+ +G F+ I SEG KG+Y+G P ++ VP F+ Y+ R
Sbjct: 271 MQVEGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEYYKVVPGVGIVFMTYDALRR 330
Query: 295 AL 296
L
Sbjct: 331 LL 332
Score = 77.0 bits (188), Expect = 1e-14
Identities = 56/203 (27%), Positives = 90/203 (43%), Gaps = 11/203 (5%)
Query: 2 GDVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEG 61
G+ +G G +P D ++ +L +Q I Y G + EG
Sbjct: 146 GNPIVHFVSGGLAGITAATATYPLDLVRTRLAAQRNAI-----YYQGIEHTFRTICREEG 200
Query: 62 PRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFL 121
GLYKG+GA L V A+ F M+ + SH + + G AG S
Sbjct: 201 ILGLYKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPNDSDLVVSLVSGGLAGAVSSTA 260
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
P +L++ R+Q + G+G A V G +H+ +SE G +G+++G++P +
Sbjct: 261 TYPLDLVRRRMQVE----GAGGRA-RVYNTGLFGTFKHIFKSE-GFKGIYRGILPEYYKV 314
Query: 182 VPGNAAMFGAYEASKQLLAGGPD 204
VPG +F Y+A ++LL PD
Sbjct: 315 VPGVGIVFMTYDALRRLLTSLPD 337
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,222,929
Number of Sequences: 26719
Number of extensions: 261828
Number of successful extensions: 1091
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 534
Number of HSP's gapped (non-prelim): 185
length of query: 297
length of database: 11,318,596
effective HSP length: 99
effective length of query: 198
effective length of database: 8,673,415
effective search space: 1717336170
effective search space used: 1717336170
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0381.7


