

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
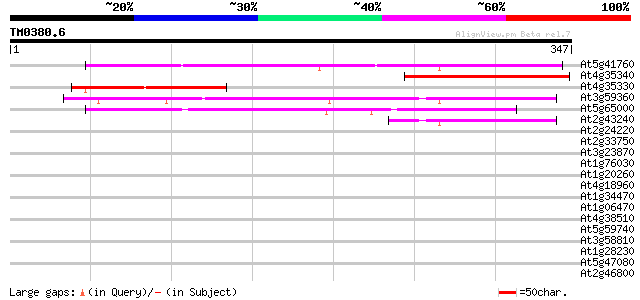
Query= TM0380.6
(347 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g41760 CMP-sialic acid transporter-like protein 219 2e-57
At4g35340 UDP-galactose transporter - like protein 178 3e-45
At4g35330 putative protein 149 2e-36
At3g59360 transporter-like protein 136 2e-32
At5g65000 unknown protein 82 4e-16
At2g43240 unknown protein 71 8e-13
At2g24220 hypothetical protein 41 0.001
At2g33750 putative purine permease (PUP2) 40 0.002
At3g23870 unknown protein 35 0.046
At1g76030 vacuolar-type H+-ATPase subunit B1 (VHA-B1) 33 0.30
At1g20260 vacuolar-type H+-ATPase subunit B3 (VHA-B3) 33 0.30
At4g18960 floral homeotic protein agamous 32 0.67
At1g34470 unknown protein 32 0.67
At1g06470 integral membrane protein, putative 31 0.87
At4g38510 vacuolar-type H+-ATPase subunit B2 (VHA-B2) 31 1.1
At5g59740 protein serine /threonine kinase - like protein 30 1.9
At3g58810 zinc transporter -like protein 30 1.9
At1g28230 purine permease like protein 30 1.9
At5g47080 casein kinase II beta chain 29 3.3
At2g46800 putative zinc transporter 29 3.3
>At5g41760 CMP-sialic acid transporter-like protein
Length = 340
Score = 219 bits (558), Expect = 2e-57
Identities = 125/304 (41%), Positives = 180/304 (59%), Gaps = 11/304 (3%)
Query: 48 VTFALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDD 107
V LTILTSSQ IL S+ G Y+Y T F+ E K IS + L R T
Sbjct: 9 VAVLLTILTSSQGILTTLSQSDGGYKYDYATVPFLAEVFKLIISGLFLWREMRTSSSTT- 67
Query: 108 NRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKK 167
+R+TT V ++ IP+ +YL+ N +Q+ YVD YQI+ N I++TG+L+R+ LK+
Sbjct: 68 SRITTDWKSVRLFVIPSLIYLIHNNVQFATLTYVDTSTYQIMGNLKIVTTGILFRLFLKR 127
Query: 168 RLSEIQWAAFILLTAGCTTAQLN----SNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEA 223
+LS++QW A LL G TT+Q+ ++ D + P QG+++ I A LS AG+YTE
Sbjct: 128 KLSKLQWMAIGLLAVGTTTSQVKGCGEASCDSLFTAPIQGYLLGILSAGLSALAGIYTEF 187
Query: 224 IIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAVMNKG-----FFHGYSFITVLMIF 278
++K R + + QN LY FG FN ++ DF KG F GYS T L++
Sbjct: 188 LMK-RNNDTLYWQNLQLYTFGSLFNVARLIADDFRHGFEKGPWWQRIFDGYSITTWLVVL 246
Query: 279 NHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIY 338
N +G+ VS +MKYADNIVKVYSTS+AMLLT V S++LF F +L FLG ++ ++++
Sbjct: 247 NLGSTGLLVSWLMKYADNIVKVYSTSMAMLLTMVASIYLFSFKPTLQLFLGIVICIMSLH 306
Query: 339 LHSA 342
++ A
Sbjct: 307 MYFA 310
>At4g35340 UDP-galactose transporter - like protein
Length = 102
Score = 178 bits (452), Expect = 3e-45
Identities = 89/102 (87%), Positives = 97/102 (94%)
Query: 245 MGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTS 304
M FNAVAI++QDFDAV NKGFFHGYSFIT+LMI NHALSGIAVSMVMKYADNIVKVYSTS
Sbjct: 1 MAFNAVAIVIQDFDAVANKGFFHGYSFITLLMILNHALSGIAVSMVMKYADNIVKVYSTS 60
Query: 305 VAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSAGKMQ 346
VAMLLTAVVSVFLF FHLSLAFFLG+ VVSV++YLHSAGK++
Sbjct: 61 VAMLLTAVVSVFLFNFHLSLAFFLGSTVVSVSVYLHSAGKLR 102
>At4g35330 putative protein
Length = 193
Score = 149 bits (376), Expect = 2e-36
Identities = 78/98 (79%), Positives = 86/98 (87%), Gaps = 3/98 (3%)
Query: 39 QVQWKRK--SMVTFALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALG 96
++ WKRK +VT ALTILTSSQAILIVWSKRAGKYEYSVTTANF+V TLKCA+SL+AL
Sbjct: 11 RINWKRKIRGVVTCALTILTSSQAILIVWSKRAGKYEYSVTTANFLV-TLKCALSLLALT 69
Query: 97 RIWNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQ 134
RIW EGVTDDNRL+TT DEV V+PIPAALYL KNLLQ
Sbjct: 70 RIWKNEGVTDDNRLSTTFDEVKVFPIPAALYLFKNLLQ 107
>At3g59360 transporter-like protein
Length = 405
Score = 136 bits (342), Expect = 2e-32
Identities = 90/323 (27%), Positives = 160/323 (48%), Gaps = 23/323 (7%)
Query: 34 DSHRTQVQWKRKSMVTFALT---ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAI 90
D H+ +V K++ + + +L Q +L+ SK GK+ +S + NF+ E K
Sbjct: 28 DDHKIRVSSKQRVLNVLLVVGDCMLVGLQPVLVYMSKVDGKFNFSPISVNFLTEIAKVIF 87
Query: 91 SLVAL------GRIWNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAP 144
++V L ++ K ++ + + V++ +PA LY + N L++ + Y +
Sbjct: 88 AIVMLLIQARHQKVGEKPLLSVSTFVQAARNNVLL-AVPALLYAINNYLKFTMQLYFNPA 146
Query: 145 GYQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNSDHV----LQTP 200
++L N ++ VL ++++K+R S IQW A LL G + QL S + +
Sbjct: 147 TVKMLSNLKVLVIAVLLKMVMKRRFSIIQWEALALLLIGISVNQLRSLPEGATAIGIPLA 206
Query: 201 FQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQDFDAV 260
+V + + A V+ E +K + +I +QN +LY +G FN + IL V
Sbjct: 207 TGAYVCTVIFVTVPSMASVFNEYALKSQYDTSIYLQNLFLYGYGAIFNFLGIL----GTV 262
Query: 261 MNKG-----FFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSV 315
+ KG G+S T+ +I N+A GI S KYAD I+K YS++VA + T + S
Sbjct: 263 IYKGPGSFDILQGHSRATMFLILNNAAQGILSSFFFKYADTILKKYSSTVATIFTGIASA 322
Query: 316 FLFGFHLSLAFFLGTIVVSVAIY 338
LFG +++ F LG +V ++++
Sbjct: 323 ALFGHVITMNFLLGISIVFISMH 345
>At5g65000 unknown protein
Length = 325
Score = 82.0 bits (201), Expect = 4e-16
Identities = 70/275 (25%), Positives = 131/275 (47%), Gaps = 15/275 (5%)
Query: 48 VTFALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDD 107
V F +LT + SKR + + VT++ E +K +L+ + R + +G+ +
Sbjct: 17 VLFYSILLTLQYGAQPLISKRCIRKDVIVTSSVLTCEIVKVICALILMARNGSLKGLAKE 76
Query: 108 NRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKK 167
T + + +PAA+Y ++N L + +D+ + IL I T IIL++
Sbjct: 77 ---WTLMGSLTASGLPAAIYALQNSLLQISYRSLDSLTFSILNQTKIFFTAFFTFIILRQ 133
Query: 168 RLSEIQWAAFILLTAGCTTAQLNSNSD------HVLQTPFQGWVMAIAMALLSGFAGVYT 221
+ S +Q A LL + S+ + Q F G + +A ++LSG A
Sbjct: 134 KQSILQIGALCLLIMAAVLLSVGEGSNKDSSGINADQKLFYGIIPVLAASVLSGLASSLC 193
Query: 222 E--AIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQ-DFDAVMNKGFFHGYSFITVLMIF 278
+ + +KK S + V+ + + G V+ L D +A+ GFFHG++ +T++ +
Sbjct: 194 QWASQVKKHSSYLMTVE---MSIVGSLCLLVSTLKSPDGEAIKKYGFFHGWTALTLVPVI 250
Query: 279 NHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVV 313
++AL GI V +V +A + K + A+L+TA++
Sbjct: 251 SNALGGILVGLVTSHAGGVRKGFVIVSALLVTALL 285
>At2g43240 unknown protein
Length = 806
Score = 71.2 bits (173), Expect = 8e-13
Identities = 42/109 (38%), Positives = 62/109 (56%), Gaps = 9/109 (8%)
Query: 235 VQNFWLYVFGMGFNAVAILVQDFDAVMNKG-----FFHGYSFITVLMIFNHALSGIAVSM 289
VQN +LY +G FN + IL V+ KG G+S T+ +I N+A GI S
Sbjct: 301 VQNLFLYGYGAIFNFLGIL----GTVIYKGPGSFDILQGHSRATMFLILNNAAQGILSSF 356
Query: 290 VMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIY 338
KYAD I+K YS++VA + T + S LFG L++ F LG +V ++++
Sbjct: 357 FFKYADTILKKYSSTVATIFTGIASAALFGHILTMNFLLGISIVFISMH 405
Score = 37.0 bits (84), Expect = 0.016
Identities = 27/109 (24%), Positives = 53/109 (47%), Gaps = 8/109 (7%)
Query: 35 SHRTQVQWKRKSMVTFALT---ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAIS 91
+++ +V K++++ F + +L Q +L+ SK GK+ +S + NF+ E K +
Sbjct: 27 NYKIRVSSKQRALNVFLVVGDCMLVGLQPVLVYMSKVDGKFNFSPISVNFLTEIAKVIFA 86
Query: 92 LVAL--GRIWNKEGVTDDNRLTTTLDEV---IVYPIPAALYLVKNLLQY 135
+V L K G L+T + ++ +PA LY + N L++
Sbjct: 87 MVMLLFQARHQKVGEKPLLSLSTFVQAARNNMLLAVPAGLYAINNYLKF 135
>At2g24220 hypothetical protein
Length = 315
Score = 40.8 bits (94), Expect = 0.001
Identities = 45/215 (20%), Positives = 94/215 (42%), Gaps = 14/215 (6%)
Query: 117 VIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQWAA 176
V+ Y + L NL+ Y +AY+ A +L + ++ + + +I+K L+ +
Sbjct: 60 VLSYVVLGFLSAADNLMYAYAYAYLPASTSSLLASSSLAFSALFGYLIVKNPLNASVINS 119
Query: 177 FILLTAGCTTAQLNSNSDHVLQTP----FQGWVMAIAMALLSGFAGVYTEAI-IKKRPSR 231
+++T L+S+SD F G+ I + L G +E + +K R
Sbjct: 120 IVVITGAMAIIALDSSSDRYSYISNSQYFAGFFWDIMGSALHGLIFALSELLFVKLLGRR 179
Query: 232 NINV---QNFWLYVFGMGFNAVAILV-QDFDAVMN--KGFFHGYSFITVLMIFN---HAL 282
+ +V Q + + F + ++V DF + + K F G S T +++++ L
Sbjct: 180 SFHVALEQQVMVSLTAFAFTTIGMVVSNDFQGMSHEAKSFKGGESLYTQVLVWSAVTFQL 239
Query: 283 SGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFL 317
+ + V+ A ++ +V + +T+V +V L
Sbjct: 240 GVLGATAVLFLASTVMAGVLNAVRVPITSVAAVIL 274
>At2g33750 putative purine permease (PUP2)
Length = 356
Score = 40.0 bits (92), Expect = 0.002
Identities = 28/105 (26%), Positives = 46/105 (43%), Gaps = 4/105 (3%)
Query: 131 NLLQYYIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLN 190
N L Y AY+ ++ + + T + ++K++ + A +LLT G LN
Sbjct: 94 NYLYSYGLAYIPVSTASLIISAQLGFTALFAFFMVKQKFTPFTINAIVLLTGGAVVLALN 153
Query: 191 SNSDHVLQTPFQ----GWVMAIAMALLSGFAGVYTEAIIKKRPSR 231
S+SD + + G++M + ALL GF E KK R
Sbjct: 154 SDSDKLANETHKEYVVGFIMTLGAALLYGFILPLVELSYKKSGQR 198
>At3g23870 unknown protein
Length = 335
Score = 35.4 bits (80), Expect = 0.046
Identities = 49/206 (23%), Positives = 79/206 (37%), Gaps = 32/206 (15%)
Query: 139 AYVDAPGYQI--LKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNSDHV 196
AY AP + L +II + VL ILK++L IL G TT L++ +
Sbjct: 73 AYAFAPAILVTPLGALSIIFSAVLAHFILKEKLHMFGILGCILCVVGSTTIVLHAPHEQK 132
Query: 197 LQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILVQD 256
+++ Q W +AI L VY+ I+ VAIL+
Sbjct: 133 IESVKQIWQLAIEPGFL-----VYSAVIV----------------------IVVAILIFY 165
Query: 257 FDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMKY---ADNIVKVYSTSVAMLLTAVV 313
++ K Y I LM +S AV++ +K N K ++T + +L+ A
Sbjct: 166 YEPRYGKTHMIVYVGICSLMGSLTVMSVKAVAIAIKLTFSGTNQFKYFNTWIFILVVATC 225
Query: 314 SVFLFGFHLSLAFFLGTIVVSVAIYL 339
+ + T V+S Y+
Sbjct: 226 CILQINYLNKALDTFNTAVISPVYYV 251
>At1g76030 vacuolar-type H+-ATPase subunit B1 (VHA-B1)
Length = 486
Score = 32.7 bits (73), Expect = 0.30
Identities = 34/115 (29%), Positives = 49/115 (42%), Gaps = 25/115 (21%)
Query: 1 MEYRKIKDE-------DKVRDSDVESVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT 53
MEYR + DKV+ + + VN + DG T+ R QV
Sbjct: 17 MEYRTVSGVAGPLVILDKVKGPKYQEI-VNIRLGDGSTR----RGQV------------L 59
Query: 54 ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+ +A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 60 EVDGEKAVVQVFEGTSG-IDNKFTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 113
>At1g20260 vacuolar-type H+-ATPase subunit B3 (VHA-B3)
Length = 330
Score = 32.7 bits (73), Expect = 0.30
Identities = 34/115 (29%), Positives = 49/115 (42%), Gaps = 25/115 (21%)
Query: 1 MEYRKIKDE-------DKVRDSDVESVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT 53
MEYR + DKV+ + + VN + DG T+ R QV
Sbjct: 17 MEYRTVSGVAGPLVILDKVKGPKYQEI-VNIRLGDGSTR----RGQV------------L 59
Query: 54 ILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+ +A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 60 EVDGEKAVVQVFEGTSG-IDNKFTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 113
>At4g18960 floral homeotic protein agamous
Length = 284
Score = 31.6 bits (70), Expect = 0.67
Identities = 16/42 (38%), Positives = 24/42 (57%)
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAIS 91
+ L++L ++ LIV+S R YEYS + +E K AIS
Sbjct: 81 YELSVLCDAEVALIVFSSRGRLYEYSNNSVKGTIERYKKAIS 122
>At1g34470 unknown protein
Length = 368
Score = 31.6 bits (70), Expect = 0.67
Identities = 25/89 (28%), Positives = 38/89 (42%), Gaps = 2/89 (2%)
Query: 139 AYVDAPGYQI--LKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNSNSDHV 196
AY AP + L +II + L +IL ++L +L G T L++ +
Sbjct: 84 AYAFAPAILVTPLGALSIIISAALAHVILHEKLHTFGLLGCVLCVVGSITIVLHAPQEQE 143
Query: 197 LQTPFQGWVMAIAMALLSGFAGVYTEAII 225
+ + Q W +A A L A V AII
Sbjct: 144 IDSVLQVWNLATEPAFLLYAAAVVGAAII 172
>At1g06470 integral membrane protein, putative
Length = 414
Score = 31.2 bits (69), Expect = 0.87
Identities = 33/151 (21%), Positives = 67/151 (43%), Gaps = 15/151 (9%)
Query: 201 FQGWVMAIAMALLSGFAGVYTEAIIKKRPSRNINVQNFWLY------VFGMGFNAVAILV 254
F G+V + A++SGF T+ +++K ++N +++ V + +++L+
Sbjct: 224 FWGFVFVMLAAVMSGFRWCMTQVLLQK---ETFGLKNPFIFMSCVAPVMAIATGLLSLLL 280
Query: 255 QDFDAVM-NKGFFHGYSFITV--LMIFNHALSGIAV---SMVMKYADNIVKVYSTSVAML 308
+ NK F G F LM+F AL+ V +++ + + V
Sbjct: 281 DPWSEFRDNKYFDSGAHFARTCFLMLFGGALAFCMVLTEYVLVSVTSAVTVTIAGVVKEA 340
Query: 309 LTAVVSVFLFGFHLSLAFFLGTIVVSVAIYL 339
+T VV+VF F + +G +++ V + L
Sbjct: 341 VTIVVAVFYFHDEFTWLKGVGLMIIMVGVSL 371
>At4g38510 vacuolar-type H+-ATPase subunit B2 (VHA-B2)
Length = 487
Score = 30.8 bits (68), Expect = 1.1
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Query: 59 QAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRIWNKEGVTDDN 108
+A++ V+ +G + TT F E LK +SL LGRI+N G DN
Sbjct: 66 KAVVQVFEGTSG-IDNKYTTVQFTGEVLKTPVSLDMLGRIFNGSGKPIDN 114
>At5g59740 protein serine /threonine kinase - like protein
Length = 295
Score = 30.0 bits (66), Expect = 1.9
Identities = 32/148 (21%), Positives = 59/148 (39%), Gaps = 11/148 (7%)
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTT-----ANFMVETLKCAISLVALGRIWNKEGV 104
FA++ + S+ I V ++ + Y V + F+V + S V+ G + + V
Sbjct: 21 FAVSGIMSTLVIYGVLQEKIMRVPYGVNKEFFKHSLFLVFCNRLTTSAVSAGALLASKKV 80
Query: 105 TDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTGVLYRII 164
D + V Y + + ++ QY YV P + K +I V +I
Sbjct: 81 LDP------VAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMVWGTLI 134
Query: 165 LKKRLSEIQWAAFILLTAGCTTAQLNSN 192
++K+ + L+T GC+ L N
Sbjct: 135 MQKKYKGFDYLVAFLVTLGCSVFILFPN 162
>At3g58810 zinc transporter -like protein
Length = 432
Score = 30.0 bits (66), Expect = 1.9
Identities = 9/34 (26%), Positives = 23/34 (67%)
Query: 221 TEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILV 254
++ +I+++ RN+N+Q +L+V G +V +++
Sbjct: 278 SDVLIEQKKQRNVNIQGAYLHVLGDSIQSVGVMI 311
>At1g28230 purine permease like protein
Length = 356
Score = 30.0 bits (66), Expect = 1.9
Identities = 33/160 (20%), Positives = 65/160 (40%), Gaps = 8/160 (5%)
Query: 76 VTTANFMVETLKCAISLVALGRIWNKEGVTDDNRLTTT-LDEVIVYPIPAALYLVKNLLQ 134
++TA F + + +S ++ R ++ R T L E ++ + L+ L
Sbjct: 41 LSTAGFPIILIPLLVSFLSRRRSNRNPNNAENKRKTKLFLMETPLFIASIVIGLLTGLDN 100
Query: 135 Y---YIFAYVDAPGYQILKNFNIISTGVLYRIILKKRLSEIQWAAFILLTAGCTTAQLNS 191
Y Y AY+ ++ + + +++K++ + A +LLT G L+S
Sbjct: 101 YLYSYGLAYLPVSTSSLIIGTQLAFNALFAFLLVKQKFTPFSINAVVLLTVGIGILALHS 160
Query: 192 NSDHVLQTPFQ----GWVMAIAMALLSGFAGVYTEAIIKK 227
+ D + + G++M + ALL F E KK
Sbjct: 161 DGDKPAKESKKEYVVGFLMTVVAALLYAFILPLVELTYKK 200
>At5g47080 casein kinase II beta chain
Length = 287
Score = 29.3 bits (64), Expect = 3.3
Identities = 14/28 (50%), Positives = 18/28 (64%)
Query: 13 RDSDVESVAVNNTVSDGETKVDSHRTQV 40
RDS S++ NNTVSD E+ DS + V
Sbjct: 64 RDSRSASLSKNNTVSDDESDTDSEESDV 91
>At2g46800 putative zinc transporter
Length = 398
Score = 29.3 bits (64), Expect = 3.3
Identities = 11/34 (32%), Positives = 21/34 (61%)
Query: 221 TEAIIKKRPSRNINVQNFWLYVFGMGFNAVAILV 254
T+ K++ RNIN+Q +L+V G +V +++
Sbjct: 245 TQVAAKEKRKRNINLQGAYLHVLGDSIQSVGVMI 278
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,858,234
Number of Sequences: 26719
Number of extensions: 258862
Number of successful extensions: 899
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 864
Number of HSP's gapped (non-prelim): 32
length of query: 347
length of database: 11,318,596
effective HSP length: 100
effective length of query: 247
effective length of database: 8,646,696
effective search space: 2135733912
effective search space used: 2135733912
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0380.6


