

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0378b.5
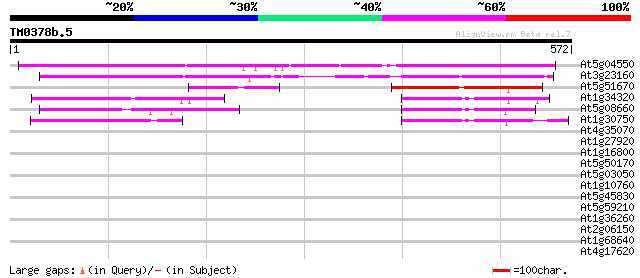
(572 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g04550 unknown protein 375 e-104
At3g23160 hypothetical protein 283 2e-76
At5g51670 unknown protein 149 3e-36
At1g34320 hypothetical protein 82 1e-15
At5g08660 putative protein 71 2e-12
At1g30750 unknown protein 66 6e-11
At4g35070 unknown protein 32 0.95
At1g27920 hypothetical protein 32 1.2
At1g16800 hypothetical protein 32 1.2
At5g50170 putative protein 30 2.8
At5g03050 unknown protein 30 2.8
At1g10760 unknown protein 30 2.8
At5g45830 tumor-related protein-like 30 3.6
At5g59210 unknown protein 30 4.7
At1g36260 unknown protein 30 4.7
At2g06150 putative Athila retroelement ORF1 protein 29 6.2
At1g68640 bZip transcription factor PERIANTHIA (PAN) / AtbZip46 29 6.2
At4g17620 hypothetical protein 29 8.1
>At5g04550 unknown protein
Length = 599
Score = 375 bits (962), Expect = e-104
Identities = 231/598 (38%), Positives = 336/598 (55%), Gaps = 63/598 (10%)
Query: 10 SWLSALWPVSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAG 69
+W LW +K V+GVL EVA L+ K+V+LWQSLSD+ V LR+ I +S G
Sbjct: 5 TWFRNLWRFPKKHDGHKDKAVLGVLAFEVASLLSKLVHLWQSLSDKNVARLRDEITHSTG 64
Query: 70 VRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQW 129
++KLVSEDDD+++ L+ +E+++N ++VA+ VARL R+C DP FE+ +
Sbjct: 65 IKKLVSEDDDFIVRLIRDEMMENVENVAKAVARLARKCNDPKLKCFENCFSDMMKTGADP 124
Query: 130 SGWDYRGKKMERKVKRMEKFVAAMTQLCQELEVLAEVEQTLRRMQANPVLHRGKLPEFQK 189
GW + KKM++K K+ME+F+++ L QE E+LA++EQT +RM++N L E+QK
Sbjct: 125 YGWQFGWKKMDKKAKKMERFISSNASLYQETEILADLEQTFKRMKSNESA-TDNLLEYQK 183
Query: 190 KVTLQRQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILERIILVFGNNH------------ 237
KVT +R EV+NLRD+S WNR+YDY V LLVRS+FTIL R VFG ++
Sbjct: 184 KVTWKRHEVKNLRDVSLWNRTYDYTVILLVRSVFTILSRTKHVFGISYRVEASDVSSADS 243
Query: 238 ------------LATKQNENESPN---------------TTANNLLRSQSFSVFMHSSV- 269
L +++ES + + RS S F+ S+
Sbjct: 244 DFIGRSHSVSTILTPVSHKSESSGLPRFASGPLGRFTGPASGSAATRSTKMSDFLSGSLS 303
Query: 270 --HPSENDL-------YGFHSGHLGRRLASNSGILVDKKQRKKKQQQALHPPALCRDRLN 320
P L + F+SG LG ++ S SG L+ + KK Q P++ +
Sbjct: 304 AESPKSGPLVAEKHKRFKFYSGQLG-KITSKSGPLIGMGKNNKKMGQTPERPSISSVKKQ 362
Query: 321 SESKQFGPIGPFKSCMSVANNSPVIDSFVQTNGGSMRLTDSH-MENVDKMKRVEKSSLSN 379
++ + +GPFK CM V+++ S NG + H +E+ VE +L +
Sbjct: 363 LKANRLTQVGPFKGCM-VSHDGITPLSTRARNGARNSSAEHHILEDNSNSVHVENLTLPS 421
Query: 380 RIRIYSKLSMSNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLP 439
R KLS A TLG A L LHYAN+I++IE+ V+SPHL + RDDLY MLP
Sbjct: 422 R----PKLS-----DAAPNTLGTACLALHYANVIIVIERFVASPHLIGDDARDDLYNMLP 472
Query: 440 ATIRAVLRHKLKGRKKS-KSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFE 498
A++R LR +LK K+ SS VY+ +A EW+ +A IL+WL PLAH+ I+W S RS+E
Sbjct: 473 ASVRTSLRERLKPYSKNLSSSTVYDPGLAREWTDAMAGILEWLGPLAHNMIKWQSERSYE 532
Query: 499 KEHATLKANILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASARS 556
+ + +I+L QTL+FANQ KTEA I +LLVGLNYV R ++ + EC S+++
Sbjct: 533 HQSLVSRTHIVLAQTLFFANQQKTEAIITELLVGLNYVWRFGRELNAKALQECTSSKT 590
>At3g23160 hypothetical protein
Length = 522
Score = 283 bits (723), Expect = 2e-76
Identities = 180/528 (34%), Positives = 287/528 (54%), Gaps = 62/528 (11%)
Query: 31 VGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVL 90
+G+L EVA +M K ++L +SLSD E+ L+ + +S GVRKLVS D+++L++L ++E L
Sbjct: 34 IGILSFEVANVMSKTIHLHRSLSDTEISKLKAEVFHSEGVRKLVSSDENHLLDLSVSEKL 93
Query: 91 DNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKMERKVKRMEKFV 150
D+ VA V+RLG++C +P FEH + + K ME VK+ME+FV
Sbjct: 94 DDLSRVASVVSRLGKKCNEPALQGFEHVYEDIVNGAIDFRKLGFLVKDMESMVKKMERFV 153
Query: 151 AAMTQLCQELEVLAEVEQTLRRMQANPVLHRGKLPEFQKKVTLQRQEVRNLRDMSPWNRS 210
A L E+EV+ E+EQ + ++Q + H+ + F++K+ QRQ+V++LRD S WN++
Sbjct: 154 NATCSLYCEMEVMNELEQAIVKLQRSQ-QHQESVKAFEQKLMWQRQDVKSLRDGSLWNQT 212
Query: 211 YDYVVRLLVRSLFTILERIILVFGNNHLATKQN---ENESPNTTANNLLRSQSFSVFMHS 267
YD VV +L R++ TI RI VFG L K++ + + A+ + S+S + F S
Sbjct: 213 YDKVVEMLARTVCTIYGRIETVFGGLGLRGKKDVTLKRDRSKNEASKAVNSRSVAGFKDS 272
Query: 268 SVHPSENDLYGFHSGHLGRRLASNSGILVDKKQRKKKQQQALHPPALCRDRLNSESKQFG 327
SE D + +G +N G +
Sbjct: 273 --RRSEADEF-TRAGDFNFPCGTNPGRM-------------------------------- 297
Query: 328 PIGPFKSCMSVANNSPVIDSFVQTNGGSMRLTDSHMENVDKMKRVEKSSLSNRIRIYSKL 387
F C+++ D +GG + S +M R K +R+ ++
Sbjct: 298 ----FMECLAMNRTIGDDDDDDDEDGGRITFPLS----TARMIRSNKFGFKSRLTQHASA 349
Query: 388 SMSNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLR 447
S T+G +AL+LHYAN+++++E+++ PHL E RDDLY+MLP +++ L+
Sbjct: 350 S----------TIGGSALSLHYANVVIVVEKLLKYPHLIGEEARDDLYQMLPTSLKTTLK 399
Query: 448 HKLKGRKKSKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLK-A 506
L+ K+ S +Y+A +A +W + IL WLAPLAH+ IRW S R+FE+++ +K
Sbjct: 400 ASLRSYLKNIS--IYDAPLAHDWKETIDGILSWLAPLAHNMIRWQSERNFEQQNQIVKRT 457
Query: 507 NILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASA 554
N+LL+QTLYFA++ KTEAAI LLVGLNY+C + + ++CAS+
Sbjct: 458 NVLLLQTLYFADREKTEAAICKLLVGLNYICHYEQQQNA--LLDCASS 503
>At5g51670 unknown protein
Length = 369
Score = 149 bits (377), Expect = 3e-36
Identities = 78/158 (49%), Positives = 106/158 (66%), Gaps = 8/158 (5%)
Query: 390 SNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHK 449
S LKP TLG A + LHYAN+IV++E+M+ P L + RDDLY MLPA++R+ LR +
Sbjct: 200 SRLLKPPETTLGGAGVALHYANLIVVMEKMIKQPQLVGLDARDDLYSMLPASVRSSLRSR 259
Query: 450 LKGRKKSKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLKAN-- 507
LKG + + + +A EW A L +IL WL PLA + IRW S RSFE++H N
Sbjct: 260 LKGVGFTAT----DGGLATEWKAALGRILRWLLPLAQNMIRWQSERSFEQQHMATATNSQ 315
Query: 508 --ILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKV 543
++LVQTL FA++VKTEAAI +LLVGLNY+ R + ++
Sbjct: 316 NRVMLVQTLVFADKVKTEAAITELLVGLNYIWRFEREM 353
Score = 58.9 bits (141), Expect = 7e-09
Identities = 36/93 (38%), Positives = 52/93 (55%), Gaps = 5/93 (5%)
Query: 183 KLPEFQKKVTLQRQEVRNLRDMSPWNRSYDYVVRLLVRSLFTILERIILVFGNNHLATKQ 242
K+ + Q K+ Q+Q V+ L+D S WN+S+D VV +L RS+FT L R+ VF ++
Sbjct: 96 KVIDLQNKIERQKQHVKYLKDRSLWNKSFDTVVLILARSVFTALARLKSVF-----SSAA 150
Query: 243 NENESPNTTANNLLRSQSFSVFMHSSVHPSEND 275
T ++L RS S S + VHPS ND
Sbjct: 151 ATGYMVPTVVSSLPRSLSSSSSSMNLVHPSPND 183
Score = 33.9 bits (76), Expect = 0.25
Identities = 21/70 (30%), Positives = 40/70 (57%), Gaps = 2/70 (2%)
Query: 31 VGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLV--SEDDDYLMELVLNE 88
VGVL EVA +M K+++L SL+D +L+ R+ + ++ + E++DY + + +
Sbjct: 37 VGVLSFEVARVMTKLLHLTHSLTDSNLLTPRDNSLRKQSLQIGIEFEEEEDYENKKDVMK 96
Query: 89 VLDNFQSVAR 98
V+D + R
Sbjct: 97 VIDLQNKIER 106
>At1g34320 hypothetical protein
Length = 657
Score = 81.6 bits (200), Expect = 1e-15
Identities = 59/204 (28%), Positives = 98/204 (47%), Gaps = 10/204 (4%)
Query: 23 ASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLM 82
A+ K + +L EVA ++K NL SLS + + L+E ++ S GV+ L+S+D D L+
Sbjct: 147 ATTVKGNKISILSFEVANTIVKGANLMHSLSKDSITHLKEVVLPSEGVQNLISKDMDELL 206
Query: 83 ELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKMERK 142
+ + + + + V R G RC DP YH + F +++ + ++ E
Sbjct: 207 RIAAADKREELRIFSGEVVRFGNRCKDPQYHNLDRFFDRLGS---EFTPQKHLKQEAETI 263
Query: 143 VKRMEKFVAAMTQLCQELEVLAEVEQTLRRM---QANP-VLHRG---KLPEFQKKVTLQR 195
+ +M FV L EL L EQ +R + NP RG L + ++ Q+
Sbjct: 264 MHQMMSFVHFTADLYHELHALDRFEQDYQRKIQEEENPSTAQRGVGDTLAILRTELKSQK 323
Query: 196 QEVRNLRDMSPWNRSYDYVVRLLV 219
+ VRNL+ S W+R + V+ LV
Sbjct: 324 KHVRNLKKKSLWSRILEEVMEKLV 347
Score = 72.8 bits (177), Expect = 5e-13
Identities = 56/163 (34%), Positives = 88/163 (53%), Gaps = 18/163 (11%)
Query: 400 LGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSS 459
LG A L LHYAN+I I+ +VS RD LY+ LP +I++ LR +++ + +
Sbjct: 380 LGSAGLALHYANIITQIDTLVSRSSTMPASTRDALYQGLPPSIKSALRSRIQSFQVKEEL 439
Query: 460 WVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHAT--LKAN-------ILL 510
V +I AE + + L WL P+A +T + H + E A+ +AN IL
Sbjct: 440 TV--PQIKAE----MEKTLQWLVPVATNTTKAHHGFGWVGEWASSGSEANQRPAGQTILR 493
Query: 511 VQTLYFANQVKTEAAIVDLLVGLNYV---CRIDTKVGVRDRVE 550
+ TL+ A++ KTEA I+DL+V L+++ R T G+R V+
Sbjct: 494 IDTLHHADKEKTEAYILDLVVWLHHLVTQVRATTGYGLRSPVK 536
>At5g08660 putative protein
Length = 649
Score = 70.9 bits (172), Expect = 2e-12
Identities = 61/214 (28%), Positives = 101/214 (46%), Gaps = 18/214 (8%)
Query: 31 VGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVL 90
+G+L EVA ++K NL +SLS + L+ I+ S GV+ LVS D D L+ LV +
Sbjct: 149 LGILAFEVANTIVKSSNLIESLSKRNIEHLKGTILYSEGVQNLVSNDFDELLRLVAADKR 208
Query: 91 DNFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKMERK-----VKR 145
Q + V R G R D +H Q YF + ++ ++ V +
Sbjct: 209 QELQVFSGEVVRFGNRSKDFQWHNL--------QRYFDRISKELTPQRQLKEDAVLVVDQ 260
Query: 146 MEKFVAAMTQLCQELEVL----AEVEQTLRRMQANPVLHRGK-LPEFQKKVTLQRQEVRN 200
+ V +L QEL+VL + EQ R + + +G L + ++ QR+ V++
Sbjct: 261 LMVLVQYTAELYQELQVLYRLEKDYEQKRREEENSANSSKGDGLAILKTELKAQRKVVKS 320
Query: 201 LRDMSPWNRSYDYVVRLLVRSLFTILERIILVFG 234
L+ S W+R ++ V+ LV + +L I +FG
Sbjct: 321 LKKKSLWSRGFEEVMEKLVDIVHFLLLEIHNIFG 354
Score = 66.2 bits (160), Expect = 5e-11
Identities = 48/146 (32%), Positives = 72/146 (48%), Gaps = 15/146 (10%)
Query: 400 LGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSS 459
LG A L LHYAN+IV I+ +V+ RD LY+ LP I+ LR K+K K
Sbjct: 372 LGPAGLALHYANIIVQIDTLVARASSITSNARDSLYQSLPPGIKLALRSKIKSFNVDKEL 431
Query: 460 WVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATL---------KANILL 510
V +I E + + L WL P+A +T + H + E A +IL
Sbjct: 432 SV--TQIKDE----MERTLHWLVPVAGNTTKAHHGFGWVGEWANTGTDFTSKPSGGDILR 485
Query: 511 VQTLYFANQVKTEAAIVDLLVGLNYV 536
++TLY A++ KTE I+ ++ L ++
Sbjct: 486 IETLYHASKEKTEIYILGQIIWLQHL 511
>At1g30750 unknown protein
Length = 744
Score = 65.9 bits (159), Expect = 6e-11
Identities = 53/183 (28%), Positives = 86/183 (46%), Gaps = 32/183 (17%)
Query: 400 LGDAALTLHYANMIVLIEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSS 459
LG+A L+LHYAN+I I+ + S P RD LY LPAT++ LR +L+ + +
Sbjct: 282 LGEAGLSLHYANLIQQIDNIASRPSSLPSNVRDTLYNALPATVKTALRPRLQTLDQEEEL 341
Query: 460 WVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLK-------------A 506
V EI AE + + L WL P A +T + H + E A +
Sbjct: 342 SV--PEIKAE----MEKSLQWLVPFAENTTKAHQGFGWVGEWANSRIEFGKGKGKGENNG 395
Query: 507 NILLVQTLYFANQVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASARSFHGVGLRKNG 566
N +QTL+ A++ ++ +++L+V L+ R+ +S + HGV L++
Sbjct: 396 NPTRLQTLHHADKPIVDSYVLELVVWLH-------------RLMKSSKKRAHGVKLQETN 442
Query: 567 MYS 569
S
Sbjct: 443 HVS 445
Score = 50.8 bits (120), Expect = 2e-06
Identities = 40/157 (25%), Positives = 77/157 (48%), Gaps = 7/157 (4%)
Query: 22 GASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYL 81
G + ++ V +L EVA + K L QSLS+E + +++ +++S V+KLVS D L
Sbjct: 116 GVTSSRGGKVTILAFEVANTIAKGAALLQSLSEENLKFMKKDMLHSEEVKKLVSTDTTEL 175
Query: 82 MELVLNEVLDNFQSVARTVARLGRRCVDPVYHRFE-HFVRSPAQNYFQWSGWDYRGKKME 140
L ++ + + V R G C D +H + +F++ +N D +M+
Sbjct: 176 QILAASDKREELDLFSGEVIRFGNMCKDLQWHNLDRYFMKLDTENSQHKLLKDDAEARMQ 235
Query: 141 RKVKRMEKFVAAMTQLCQEL-EVLAEVEQTLRRMQAN 176
V +A +T + ++L +V++ + QT+ + N
Sbjct: 236 ELVT-----LARITSIIEKLVDVVSYIRQTIVEVFGN 267
>At4g35070 unknown protein
Length = 265
Score = 32.0 bits (71), Expect = 0.95
Identities = 26/125 (20%), Positives = 62/125 (48%), Gaps = 15/125 (12%)
Query: 112 YHRFEHFVRSPAQNYFQWSGWDYRGKKMERKVKRMEKFVAAMTQLCQEL---EVLAEVEQ 168
+H+ +SP+Q++ +ME++ + +++F+ + + + + E+E
Sbjct: 59 HHQHHQQQQSPSQSFL--------AAQMEKQKQEIDQFIKIQNERLRYVLQEQRKREMEM 110
Query: 169 TLRRMQANPVLHRGKLPEFQKKVTLQRQEVRN-LRDMSPWNRSYDYVVR---LLVRSLFT 224
LR+M++ +L + E K + E+ + LR M N+++ + R +V++L T
Sbjct: 111 ILRKMESKALLLMSQKEEEMSKALNKNMELEDLLRKMEMENQTWQRMARENEAIVQTLNT 170
Query: 225 ILERI 229
LE++
Sbjct: 171 TLEQV 175
>At1g27920 hypothetical protein
Length = 592
Score = 31.6 bits (70), Expect = 1.2
Identities = 15/44 (34%), Positives = 23/44 (52%)
Query: 267 SSVHPSENDLYGFHSGHLGRRLASNSGILVDKKQRKKKQQQALH 310
+ +HPS NDLYG L +G +V ++ K K+ + LH
Sbjct: 242 TKIHPSLNDLYGISKNISDDILKKLNGTVVSLEEEKHKRLEKLH 285
>At1g16800 hypothetical protein
Length = 1939
Score = 31.6 bits (70), Expect = 1.2
Identities = 18/65 (27%), Positives = 32/65 (48%)
Query: 245 NESPNTTANNLLRSQSFSVFMHSSVHPSENDLYGFHSGHLGRRLASNSGILVDKKQRKKK 304
N S N +N +S+ S + SS + D G + + ASN G + D ++K+
Sbjct: 1830 NSSKNENSNEWKKSKKASSKLDSSKRANPTDKIGQQDRQINKGNASNQGGVEDMISKRKQ 1889
Query: 305 QQQAL 309
Q++A+
Sbjct: 1890 QREAV 1894
>At5g50170 putative protein
Length = 1027
Score = 30.4 bits (67), Expect = 2.8
Identities = 14/41 (34%), Positives = 24/41 (58%)
Query: 41 LMLKVVNLWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYL 81
L L + W+S S E+VL+ ++ I+N GV++L D +
Sbjct: 123 LSLSLQGKWESTSGEKVLNDKQDIINLEGVKELEGSPKDLI 163
>At5g03050 unknown protein
Length = 140
Score = 30.4 bits (67), Expect = 2.8
Identities = 20/62 (32%), Positives = 33/62 (52%), Gaps = 6/62 (9%)
Query: 58 LSLREGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARL---GRRCVDPVYHR 114
+SL I N KLV+ D+ MEL + ++LD +S +TV+ L G+ + + +
Sbjct: 33 ISLDHTIENEEAETKLVASDE---MELSIAQILDKIESFTQTVSNLLETGKTMLKELSNE 89
Query: 115 FE 116
FE
Sbjct: 90 FE 91
>At1g10760 unknown protein
Length = 1399
Score = 30.4 bits (67), Expect = 2.8
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 12/79 (15%)
Query: 132 WDYRGKKMERKVKRMEKFVAAMTQLCQELEVLAEVEQTLRRM----------QANPVLHR 181
W+ +GK+M K E++ AA T+L +E+ A VE ++ ++N
Sbjct: 224 WERKGKQMYNPEKEKEEYEAARTELREEMMRGASVEDLRAKLLKKDNSNESPKSNGTSSS 283
Query: 182 GKLPEFQKKVTLQRQEVRN 200
G+ E +KKV+ Q + +N
Sbjct: 284 GR--EEKKKVSKQPERKKN 300
>At5g45830 tumor-related protein-like
Length = 283
Score = 30.0 bits (66), Expect = 3.6
Identities = 20/83 (24%), Positives = 35/83 (42%), Gaps = 6/83 (7%)
Query: 49 WQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCV 108
W SL + + L++ + R ED+D + + +++ +F++ A A L RC
Sbjct: 18 WMSLQSQRIPELKQLLAQR---RSHGDEDNDNKLRKLTGKIIGDFKNYAAKRADLAHRCS 74
Query: 109 DPVYHRFEHFVRSPAQNYFQWSG 131
Y SP +N W G
Sbjct: 75 SNYY---APTWNSPLENALIWMG 94
>At5g59210 unknown protein
Length = 434
Score = 29.6 bits (65), Expect = 4.7
Identities = 28/115 (24%), Positives = 48/115 (41%), Gaps = 14/115 (12%)
Query: 290 SNSGILVDKKQRKKKQQQALHPPALCRDRLNSESKQFGPIGPFKSCMSVANNSPVIDSFV 349
S SG V + K+K ++ LH A+ +RL S + K M + N S I+
Sbjct: 251 SESGQKVFSVEDKEKLEKQLHDMAVALERLESSRQ--------KLLMEIDNQSSEIEKLF 302
Query: 350 QTNGGSMRLTDSHMENVDKMKRVE---KSSLSNRIRIYSKLSMSNRLKPASFTLG 401
+ N L+ S+ E+++ + E K L + + L + SF+ G
Sbjct: 303 EENS---NLSASYQESINISNQWENQVKECLKQNVELREVLDKLRTEQAGSFSRG 354
>At1g36260 unknown protein
Length = 409
Score = 29.6 bits (65), Expect = 4.7
Identities = 17/66 (25%), Positives = 31/66 (46%), Gaps = 4/66 (6%)
Query: 416 IEQMVSSPHLTDPEPRDDLYKMLPATIRAVLRHKLKGRKKSKSSWVYNAE-IAAEWSAVL 474
IE++++SP D + AT+R++ K K+ +SW +E + W
Sbjct: 51 IEELLASPGRAGLPRLDPNQPPVAATVRSIFEKKF---KEPHASWSQTSEAVVDRWFETF 107
Query: 475 AQILDW 480
AQ+ +W
Sbjct: 108 AQVYNW 113
>At2g06150 putative Athila retroelement ORF1 protein
Length = 485
Score = 29.3 bits (64), Expect = 6.2
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 8/81 (9%)
Query: 48 LWQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRC 107
L++ LS E LREG G + DY+M++ E + F S T R+ R
Sbjct: 163 LFEGLSAAE---LREG---GLGYLSFTIDGRDYIMKIKTLETMFGFPSGKGTKPRIIREE 216
Query: 108 VDPVYHRFEHFVRSPAQNYFQ 128
+ + +F+R+PA YFQ
Sbjct: 217 LKALSK--SNFIRNPAIRYFQ 235
>At1g68640 bZip transcription factor PERIANTHIA (PAN) / AtbZip46
Length = 452
Score = 29.3 bits (64), Expect = 6.2
Identities = 30/134 (22%), Positives = 62/134 (45%), Gaps = 20/134 (14%)
Query: 49 WQSLSDEEVLSLREGIVNSAGVRKLVSEDDDYLMELVLNEVLDNFQSVARTVARLGRRCV 108
W+ + LR G+ + G D+D + ++++ V+ ++ + R + +G + V
Sbjct: 243 WKEEHQRMINDLRSGVNSQLG-------DND--LRVLVDAVMSHYDEIFR-LKGIGTK-V 291
Query: 109 DPVYHRFEHFVRSPAQNYFQWSGWDYRGKKMERKVKRMEKFVAAMTQLCQELEVLAEVEQ 168
D V+H ++PA+ +F W G G + +K + V +T Q+L + ++Q
Sbjct: 292 D-VFHMLSGMWKTPAERFFMWLG----GFRSSELLKILGNHVDPLTD--QQLIGICNLQQ 344
Query: 169 TLRRMQANPVLHRG 182
+ QA L +G
Sbjct: 345 S--SQQAEDALSQG 356
>At4g17620 hypothetical protein
Length = 1225
Score = 28.9 bits (63), Expect = 8.1
Identities = 40/167 (23%), Positives = 66/167 (38%), Gaps = 13/167 (7%)
Query: 372 VEKSSLSNRIRIYSKLSMSNRLKPASFTLGDAALTLHYANMIVLIEQMVSSPHLTDPEPR 431
+ +L+ + Y + L F+LG + L + V I + ++ + D +
Sbjct: 379 ITNKTLAKLVYGYKRHDNLPELSKLLFSLGGSRLCADVIDACVAIGWLEAAHDILD-DMN 437
Query: 432 DDLYKMLPATIRAVLRHKLKGRKKSKSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRW 491
Y M AT R VL G KSK + NAE+ + I D P +
Sbjct: 438 SAGYPMELATYRMVL----SGYYKSKM--LRNAEVLLKQMTKAGLITD---PSNEIVV-- 486
Query: 492 HSVRSFEKEHATLKANILLVQTLYFANQVKTEAAIVDLLVGLNYVCR 538
S + EK+ + LLVQ + Q+K + + +L L Y C+
Sbjct: 487 -SPETEEKDSENTELRDLLVQEINAGKQMKAPSMLYELNSSLYYFCK 532
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,446,036
Number of Sequences: 26719
Number of extensions: 510345
Number of successful extensions: 1419
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1383
Number of HSP's gapped (non-prelim): 25
length of query: 572
length of database: 11,318,596
effective HSP length: 105
effective length of query: 467
effective length of database: 8,513,101
effective search space: 3975618167
effective search space used: 3975618167
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0378b.5


