

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0377b.7
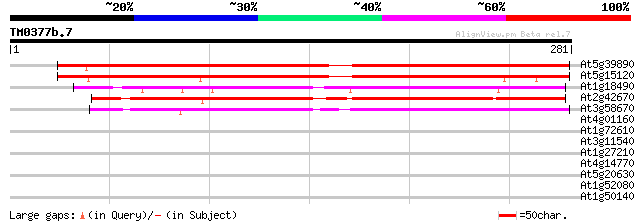
(281 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g39890 unknown protein 337 6e-93
At5g15120 unknown protein 305 1e-83
At1g18490 unknown protein 204 6e-53
At2g42670 unknown protein 199 1e-51
At3g58670 unknown protein 194 3e-50
At4g01160 predicted protein 33 0.22
At1g72610 germin-like protein 30 1.9
At3g11540 spindly (gibberellin signal transduction protein) 29 3.2
At1g27210 unknown protein 29 3.2
At4g14770 hypothetical protein 28 5.5
At5g20630 germin-like protein (GLP3b) 28 7.2
At1g52080 unknown protein 27 9.4
At1g50140 hypothetical protein, 5' partial 27 9.4
>At5g39890 unknown protein
Length = 276
Score = 337 bits (863), Expect = 6e-93
Identities = 166/259 (64%), Positives = 196/259 (75%), Gaps = 14/259 (5%)
Query: 25 SSGSKRNRRRQKK--MPPVQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVG 82
S+ K+ +RR KK + PVQKLFDTCK+VFA G +G +P ++I+ L+AVLD I+PEDVG
Sbjct: 29 SNSRKKIQRRSKKTLICPVQKLFDTCKKVFADGKSGTVPSQENIEMLRAVLDEIKPEDVG 88
Query: 83 LKPDMPHFRSSSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFG 142
+ P M +FRS+ R P +TYLHIY C +FS+ IFCLPPSGVIPLHNHP MTVFSKLLFG
Sbjct: 89 VNPKMSYFRSTVTGRSPLVTYLHIYACHRFSICIFCLPPSGVIPLHNHPEMTVFSKLLFG 148
Query: 143 TMHIKSYDWVVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMH 202
TMHIKSYDWV D P S + RLAKVKVD+DFTAPC+ SILYP DGGNMH
Sbjct: 149 TMHIKSYDWVPDSPQPSS-----------DTRLAKVKVDSDFTAPCDTSILYPADGGNMH 197
Query: 203 CFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWLQEKEQ 262
CFTA TACAVLDV+GPPYSD GRHCTYY +YPFS SV+G+ + EEE+ GY WL+E+E+
Sbjct: 198 CFTAKTACAVLDVIGPPYSDPAGRHCTYYFDYPFSSFSVDGVVVAEEEKEGYAWLKEREE 257
Query: 263 L-EDLEVDGKMYSGPKIHE 280
EDL V MYSGP I E
Sbjct: 258 KPEDLTVTALMYSGPTIKE 276
>At5g15120 unknown protein
Length = 293
Score = 305 bits (782), Expect = 1e-83
Identities = 154/279 (55%), Positives = 193/279 (68%), Gaps = 34/279 (12%)
Query: 25 SSGSKRNRRRQKKM----------------PPVQKLFDTCKEVFASGGTGFIPPPQDIQR 68
+S K+N+ + KKM V++LF+TCKEVF++GG G IP IQ+
Sbjct: 26 NSVKKKNKNKNKKMMMTWRRKKIDSPADGITAVRRLFNTCKEVFSNGGPGVIPSEDKIQQ 85
Query: 69 LQAVLDAIRPEDVGLKPDMPHFRSSS---AQRIPKITYLHIYECEKFSMGIFCLPPSGVI 125
L+ +LD ++PEDVGL P MP+FR +S A+ P ITYLH+++C++FS+GIFCLPPSGVI
Sbjct: 86 LREILDDMKPEDVGLTPTMPYFRPNSGVEARSSPPITYLHLHQCDQFSIGIFCLPPSGVI 145
Query: 126 PLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTENQALEMRLAKVKVDADFT 185
PLHNHPGMTVFSKLLFGTMHIKSYDWVVD P + RLAK+KVD+ FT
Sbjct: 146 PLHNHPGMTVFSKLLFGTMHIKSYDWVVDAPMRDS-----------KTRLAKLKVDSTFT 194
Query: 186 APCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGIS 245
APCN SILYPEDGGNMH FTA+TACAVLDVLGPPY + EGRHCTY+ +P +S E
Sbjct: 195 APCNASILYPEDGGNMHRFTAITACAVLDVLGPPYCNPEGRHCTYFLEFPLDKLSSEDDD 254
Query: 246 I--PEEERNGYEWLQEKEQ--LEDLEVDGKMYSGPKIHE 280
+ EEE+ GY WLQE++ + V G +Y GPK+ +
Sbjct: 255 VLSSEEEKEGYAWLQERDDNPEDHTNVVGALYRGPKVED 293
>At1g18490 unknown protein
Length = 282
Score = 204 bits (518), Expect = 6e-53
Identities = 114/263 (43%), Positives = 156/263 (58%), Gaps = 26/263 (9%)
Query: 33 RRQKKMPPVQKLFDTCKEVFASGGTGFIPPPQD--IQRLQAVLDAIRPEDVGLKP----D 86
R Q+K P VQ+L+D CKE F TG P P IQ+L +VLD++ P DVGL+ D
Sbjct: 27 RNQEKSPKVQELYDLCKETF----TGKAPSPASMAIQKLCSVLDSVSPADVGLEEVSQDD 82
Query: 87 MPHFRSSSAQRIPK-------ITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKL 139
+ S R + IT+L I+EC+ F+M IFC P S VIPLH+HP M VFSK+
Sbjct: 83 DRGYGVSGVSRFNRVGRWAQPITFLDIHECDTFTMCIFCFPTSSVIPLHDHPEMAVFSKI 142
Query: 140 LFGTMHIKSYDWVVDLPPESPTIVKPTENQ--ALEMRLAKVKVDADFTAPCNPSILYPED 197
L+G++H+K+YDWV E P I+ + +L RLAK+ D T LYP+
Sbjct: 143 LYGSLHVKAYDWV-----EPPCIITQDKGVPGSLPARLAKLVSDKVITPQSEIPALYPKT 197
Query: 198 GGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNNYPFSDISVEG--ISIPEEERNGYE 255
GGN+HCFTA+T CAVLD+L PPY + GR C+YY +YPFS ++E + E + + Y
Sbjct: 198 GGNLHCFTALTPCAVLDILSPPYKESVGRSCSYYMDYPFSTFALENGMKKVDEGKEDEYA 257
Query: 256 WLQEKEQLEDLEVDGKMYSGPKI 278
WL + + +DL + Y+GP I
Sbjct: 258 WLVQIDTPDDLHMRPGSYTGPTI 280
>At2g42670 unknown protein
Length = 241
Score = 199 bits (506), Expect = 1e-51
Identities = 104/247 (42%), Positives = 149/247 (60%), Gaps = 23/247 (9%)
Query: 42 QKLFDTCKEVFASGGTGFIPPPQD-IQRLQAVLDAIRPEDVGLKPDMPHFRSSSA----- 95
Q+L++TCK F+S G P +D +++++ VL+ I+P DVG++ D RS S
Sbjct: 6 QRLYNTCKASFSSDG----PITEDALEKVRNVLEKIKPSDVGIEQDAQLARSRSGPLNER 61
Query: 96 ----QRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDW 151
Q P I YLH++EC+ FS+GIFC+PPS +IPLHNHPGMTV SKL++G+MH+KSYDW
Sbjct: 62 NGSNQSPPAIKYLHLHECDSFSIGIFCMPPSSMIPLHNHPGMTVLSKLVYGSMHVKSYDW 121
Query: 152 VVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACA 211
+ P + +P + + R AK+ D + TA + LYP+ GGN+HCF A+T CA
Sbjct: 122 L------EPQLTEPEDPS--QARPAKLVKDTEMTAQSPVTTLYPKSGGNIHCFKAITHCA 173
Query: 212 VLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWLQEKEQLEDLEVDGK 271
+LD+L PPYS RHCTY+ D+ E + + E WL+E + +D +
Sbjct: 174 ILDILAPPYSSEHDRHCTYFRKSRREDLPGE-LEVDGEVVTDVTWLEEFQPPDDFVIRRI 232
Query: 272 MYSGPKI 278
Y GP I
Sbjct: 233 PYRGPVI 239
>At3g58670 unknown protein
Length = 242
Score = 194 bits (494), Expect = 3e-50
Identities = 98/249 (39%), Positives = 144/249 (57%), Gaps = 20/249 (8%)
Query: 41 VQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIRPEDVGLK---------PDMPHFR 91
+Q+LF+TCK + G + + +++ VL+ I+P DVGL+ P + R
Sbjct: 5 IQRLFNTCKSSLSPNGP---VSEEALDKVRNVLEKIKPSDVGLEQEAQLVRNWPGPGNER 61
Query: 92 SSSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDW 151
+ + +P I YL ++EC+ FS+GIFC+PP +IPLHNHPGMTV SKL++G+MH+KSYDW
Sbjct: 62 NGNHHSLPAIKYLQLHECDSFSIGIFCMPPGSIIPLHNHPGMTVLSKLVYGSMHVKSYDW 121
Query: 152 VVDLPPESPTIVKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACA 211
P+ + P L+ R AK+ D D T+P + LYP GGN+HCF A+T CA
Sbjct: 122 A---EPDQSELDDP-----LQARPAKLVKDIDMTSPSPATTLYPTTGGNIHCFKAITHCA 173
Query: 212 VLDVLGPPYSDYEGRHCTYYNNYPFSDISVEGISIPEEERNGYEWLQEKEQLEDLEVDGK 271
+ D+L PPYS GRHC Y+ P D+ E + E + WL+E + ++ +
Sbjct: 174 IFDILSPPYSSTHGRHCNYFRKSPMLDLPGEIEVMNGEVISNVTWLEEYQPPDNFVIWRV 233
Query: 272 MYSGPKIHE 280
Y GP I +
Sbjct: 234 PYRGPVIRK 242
>At4g01160 predicted protein
Length = 527
Score = 32.7 bits (73), Expect = 0.22
Identities = 16/58 (27%), Positives = 29/58 (49%)
Query: 176 AKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDVLGPPYSDYEGRHCTYYNN 233
A V VD DF+ P++ + ++ FT A ++LG P+ + ++C Y+ N
Sbjct: 457 ASVTVDYDFSVRSKPTMEFVGKFKGIYTFTRGKAVGCRNLLGIPWDIFTAKNCPYFIN 514
>At1g72610 germin-like protein
Length = 208
Score = 29.6 bits (65), Expect = 1.9
Identities = 12/26 (46%), Positives = 17/26 (65%)
Query: 119 LPPSGVIPLHNHPGMTVFSKLLFGTM 144
L P GVIP+H HPG + +L G++
Sbjct: 91 LAPKGVIPMHTHPGASEVLFVLTGSI 116
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 28.9 bits (63), Expect = 3.2
Identities = 17/66 (25%), Positives = 32/66 (47%), Gaps = 1/66 (1%)
Query: 109 CEKFSMGIFCLPPSGVIPLHNHPGMTVFSKLLFGTMHIKSYDWVVDLPPESPTIVKPTEN 168
CE MG+ C+ +G + HN G+++ +K+ G + K+ D V L + + V
Sbjct: 751 CESLYMGVPCVTMAGSVHAHN-VGVSLLTKVGLGHLVAKNEDEYVQLSVDLASDVTALSK 809
Query: 169 QALEMR 174
+ +R
Sbjct: 810 LRMSLR 815
>At1g27210 unknown protein
Length = 625
Score = 28.9 bits (63), Expect = 3.2
Identities = 19/53 (35%), Positives = 30/53 (55%), Gaps = 4/53 (7%)
Query: 163 VKPTENQALEMRLAKVKVDADFTAPCNPSILYPEDGGNMHCFTAVTACAVLDV 215
V+ T N+AL + +V D A +PS +DG N+ CF++VT + +DV
Sbjct: 291 VRETMNRALNLWK---EVSTDDEASLSPSRSSTDDG-NIGCFSSVTRSSTIDV 339
>At4g14770 hypothetical protein
Length = 658
Score = 28.1 bits (61), Expect = 5.5
Identities = 16/53 (30%), Positives = 26/53 (48%), Gaps = 8/53 (15%)
Query: 79 EDVGLKPDMPHFRSSSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHP 131
+DV ++P + SS + K +Y CE F+ G++C+ P I N P
Sbjct: 351 QDVPVEPALQELNLSSPK---KKSY-----CECFAAGVYCIEPCSCIDCFNKP 395
>At5g20630 germin-like protein (GLP3b)
Length = 211
Score = 27.7 bits (60), Expect = 7.2
Identities = 12/26 (46%), Positives = 16/26 (61%)
Query: 119 LPPSGVIPLHNHPGMTVFSKLLFGTM 144
L GVIPLH HPG + ++ GT+
Sbjct: 94 LAGGGVIPLHTHPGASEVLVVIQGTI 119
>At1g52080 unknown protein
Length = 573
Score = 27.3 bits (59), Expect = 9.4
Identities = 15/62 (24%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Query: 63 PQDIQRLQAVLDAIRPEDVGLKPDMPHFRSSSAQRIPKITYLH-IYECEKFSMGIFCLPP 121
P++I+ L+ + +R E+ LK D+ + + ++ YL I C ++ + + PP
Sbjct: 318 PEEIETLREDCNRLRSENEELKKDVEQLQGDRCTDLEQLVYLRWINACLRYELRTY-QPP 376
Query: 122 SG 123
+G
Sbjct: 377 AG 378
>At1g50140 hypothetical protein, 5' partial
Length = 601
Score = 27.3 bits (59), Expect = 9.4
Identities = 27/125 (21%), Positives = 52/125 (41%), Gaps = 18/125 (14%)
Query: 18 LPRETITSSGSKRNRRRQKKMPPVQKLFDTCKEVFASGGTGFIPPPQDIQRLQAVLDAIR 77
LPRE++ S ++ + + P Q L + K+ + + P +I ++
Sbjct: 259 LPRESLEISIARLRKLEDNSLKPSQNLKNIAKDEYERNFVSAVVAPGEI--------GVK 310
Query: 78 PEDVGLKPDMPHFRSSSAQRIPKITYLHIYECEKFSMGIFCLPPSGVIPLHNHP--GMTV 135
ED+G D+ + + ++ L + E F+ G P G++ L P G T+
Sbjct: 311 FEDIGALEDV-------KKALNELVILPMRRPELFARGNLLRPCKGIL-LFGPPGTGKTL 362
Query: 136 FSKLL 140
+K L
Sbjct: 363 LAKAL 367
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,360,659
Number of Sequences: 26719
Number of extensions: 338586
Number of successful extensions: 755
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 732
Number of HSP's gapped (non-prelim): 13
length of query: 281
length of database: 11,318,596
effective HSP length: 98
effective length of query: 183
effective length of database: 8,700,134
effective search space: 1592124522
effective search space used: 1592124522
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0377b.7


