

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0375a.8
(606 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
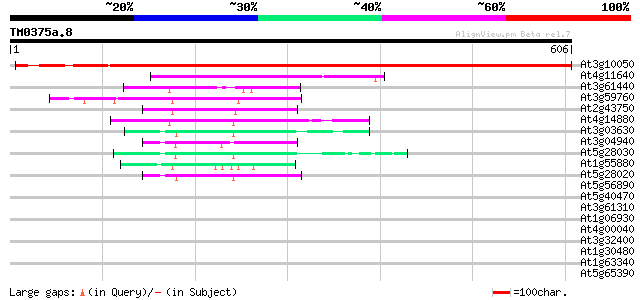
Score E
Sequences producing significant alignments: (bits) Value
At3g10050 threonine dehydratase/deaminase (OMR1) 827 0.0
At4g11640 unknown protein 135 5e-32
At3g61440 cysteine synthase (AtcysC1) 62 7e-10
At3g59760 cysteine synthase 61 2e-09
At2g43750 cysteine synthase (cpACS1) 57 4e-08
At4g14880 cytosolic O-acetylserine(thiol)lyase (EC 4.2.99.8) 52 1e-06
At3g03630 O-acetylserine (thiol) lyase 49 1e-05
At3g04940 cysteine synthase (AtcysD1) 47 3e-05
At5g28030 cysteine synthase - like 47 4e-05
At1g55880 unknown protein 45 9e-05
At5g28020 cysteine synthase 45 1e-04
At5g56890 unknown protein 33 0.60
At5g40470 unknown protein 33 0.60
At3g61310 DNA-binding protein like 32 0.78
At1g06930 hypothetical protein 31 1.7
At4g00040 chalcone synthase like protein 31 2.3
At3g32400 putative protein 31 2.3
At1g30480 DNA damage repair protein, putative 31 2.3
At1g63340 unknown protein 30 3.0
At5g65390 arabinogalactan protein AGP7 30 3.9
>At3g10050 threonine dehydratase/deaminase (OMR1)
Length = 592
Score = 827 bits (2135), Expect = 0.0
Identities = 424/601 (70%), Positives = 489/601 (80%), Gaps = 21/601 (3%)
Query: 7 STTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSAAVG 66
S +H H PS P + H + R + +A +TP P
Sbjct: 12 SLRSHIHRPSKPVV-----------GFTHFSSRSRIAVAVLSRDETSMTPPPPK------ 54
Query: 67 DHLSSPRLRVSPDSLQYEPGYLGAVPDRSR-SDGGSDSDAMLTAMANILSAKVYDVAIES 125
L PRL+VSP+SLQY GYLGAVP+R+ ++ GS ++AM + NILS KVYD+AIES
Sbjct: 55 --LPLPRLKVSPNSLQYPAGYLGAVPERTNEAENGSIAEAM-EYLTNILSTKVYDIAIES 111
Query: 126 PLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLEKGVICSSAGNHAQ 185
PLQLA KLS+R+GV ++LKREDLQPVFSFKLRGAYNMMVKLP + L KGVICSSAGNHAQ
Sbjct: 112 PLQLAKKLSKRLGVRMYLKREDLQPVFSFKLRGAYNMMVKLPADQLAKGVICSSAGNHAQ 171
Query: 186 GVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGIEEGRT 245
GVAL+A +L C AV+ MPVTTP+IKW++VE LGATVVL GDSYD+AQA+AK R EEG T
Sbjct: 172 GVALSASKLGCTAVIVMPVTTPEIKWQAVENLGATVVLFGDSYDQAQAHAKIRAEEEGLT 231
Query: 246 FVPAFDHPDVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIG 305
F+P FDHPDVI GQGTVGMEI RQ P+HAIFVPVGGGGLIAGIAAYVK VSP+VKIIG
Sbjct: 232 FIPPFDHPDVIAGQGTVGMEITRQAKGPLHAIFVPVGGGGLIAGIAAYVKRVSPEVKIIG 291
Query: 306 VEPTDANTMALSLHHDQRVILDQVGGFADGVAVKEVGEETFRLCKDLIDGVVLVNRDSIC 365
VEP DAN MALSLHH +RVILDQVGGFADGVAVKEVGEETFR+ ++L+DGVVLV RD+IC
Sbjct: 292 VEPADANAMALSLHHGERVILDQVGGFADGVAVKEVGEETFRISRNLMDGVVLVTRDAIC 351
Query: 366 ASIKDMFEENRSILEPSGALALAGAEAYCKYYGIKGGNVVAITSGANMNFDKLRVVTELA 425
ASIKDMFEE R+ILEP+GALALAGAEAYCKYYG+K NVVAITSGANMNFDKLR+VTELA
Sbjct: 352 ASIKDMFEEKRNILEPAGALALAGAEAYCKYYGLKDVNVVAITSGANMNFDKLRIVTELA 411
Query: 426 NVGRKQEAVLATVMPEEPGSFKQFCELVGQANITEFTYRFTSSDKAVVLYSVGVHTVSDL 485
NVGR+QEAVLAT+MPE+PGSFKQFCELVG NI+EF YR +S +AVVLYSVGVHT +L
Sbjct: 412 NVGRQQEAVLATLMPEKPGSFKQFCELVGPMNISEFKYRCSSEKEAVVLYSVGVHTAGEL 471
Query: 486 SAMKERMKSSHLKTHDLTENDLVKDHLRYMMGGRSDVENEVLCRFIFPERPGALMKFLDS 545
A+++RM+SS LKT +LT +DLVKDHLRY+MGGRS V +EVLCRF FPERPGALM FLDS
Sbjct: 472 KALQKRMESSQLKTVNLTTSDLVKDHLRYLMGGRSTVGDEVLCRFTFPERPGALMNFLDS 531
Query: 546 FSPRWNISLFHYRGEGETRANILVGIQVPRTEMDEFHERANRLGYEYKVVNDDDDFQLLM 605
FSPRWNI+LFHYRG+GET AN+LVGIQVP EM+EF RA LGY+Y +V+DDD F+LLM
Sbjct: 532 FSPRWNITLFHYRGQGETGANVLVGIQVPEQEMEEFKNRAKALGYDYFLVSDDDYFKLLM 591
Query: 606 H 606
H
Sbjct: 592 H 592
>At4g11640 unknown protein
Length = 346
Score = 135 bits (341), Expect = 5e-32
Identities = 87/264 (32%), Positives = 136/264 (50%), Gaps = 14/264 (5%)
Query: 153 SFKLRGAYNMMVKLPKELLEKGVICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWK 212
+FK RGA N ++ L E KGV+ S+GNHA ++LAAK A + +P P K
Sbjct: 72 AFKFRGACNAVLSLDAEQAAKGVVTHSSGNHAAALSLAAKIQGIPAYIVVPKGAPKCKVD 131
Query: 213 SVEKLGATVVLIGDSYDEAQAYAKKRGIEEGRTFVPAFDHPDVIMGQGTVGMEIMRQMVD 272
+V + G V+ + + A K E G + ++ +I GQGT+ +E++ Q +
Sbjct: 132 NVIRYGGKVIWSEATMSSREEIASKVLQETGSVLIHPYNDGRIISGQGTIALELLEQ-IQ 190
Query: 273 PVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGF 332
+ AI VP+ GGGLI+G+A K++ P ++II EP A+ A S + + L
Sbjct: 191 EIDAIVVPISGGGLISGVALAAKSIKPSIRIIAAEPKGADDAAQSKVAGKIITLPVTNTI 250
Query: 333 ADGVAVKEVGEETFRLCKDLIDGVVLVNRDSICASIKDMFEENRSILEPSGALALAGAEA 392
ADG+ +G+ T+ + +DL+D VV + I ++K +E + +EPSGA+ LA +
Sbjct: 251 ADGLRA-SLGDLTWPVVRDLVDDVVTLEECEIIEAMKMCYEILKVSVEPSGAIGLAAVLS 309
Query: 393 Y----------CKYYGI--KGGNV 404
CK GI GGNV
Sbjct: 310 NSFRNNPSCRDCKNIGIVLSGGNV 333
>At3g61440 cysteine synthase (AtcysC1)
Length = 368
Score = 62.4 bits (150), Expect = 7e-10
Identities = 57/211 (27%), Positives = 90/211 (42%), Gaps = 32/211 (15%)
Query: 124 ESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELL----EKGVICSS 179
++PL K++E + K+E QP S K R A M+ K+ L + +I +
Sbjct: 59 KTPLVFLNKVTEGCEAYVAAKQEHFQPTCSIKDRPAIAMIADAEKKKLIIPGKTTLIEPT 118
Query: 180 AGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRG 239
+GN +A A ++ MP T + ++ GA +VL D A+
Sbjct: 119 SGNMGISLAFMAAMKGYRIIMTMPSYTSLERRVTMRSFGAELVLT----DPAKGM----- 169
Query: 240 IEEGRTFVPAFD----HPDVIMGQ------------GTVGMEIMRQMVDPVHAIFVPVGG 283
G T A+D PD M Q T G EI + V + +G
Sbjct: 170 ---GGTVKKAYDLLDSTPDAFMCQQFANPANTQIHFDTTGPEIWEDTLGNVDIFVMGIGS 226
Query: 284 GGLIAGIAAYVKTVSPQVKIIGVEPTDANTM 314
GG ++G+ Y+K+ +P VKI GVEP ++N +
Sbjct: 227 GGTVSGVGRYLKSKNPNVKIYGVEPAESNIL 257
>At3g59760 cysteine synthase
Length = 430
Score = 61.2 bits (147), Expect = 2e-09
Identities = 72/295 (24%), Positives = 127/295 (42%), Gaps = 29/295 (9%)
Query: 44 IAATISKPAEITPSPSSSSAAVGDHLSSPRLRVSPD----SLQYEPGYLGAVPDRSRSDG 99
+AAT S + P SSSS++ + R R SP+ S + A+ +SRS
Sbjct: 34 MAATSSSALLLNPLTSSSSSS-----TLRRFRCSPEISSLSFSSASDFSLAMKRQSRSFA 88
Query: 100 -GSDSDAMLTAMA----------NILSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDL 148
GS+ D + A NI V + ++P+ +++ N+ K E +
Sbjct: 89 DGSERDPSVVCEAVKRETGPDGLNIAD-NVSQLIGKTPMVYLNSIAKGCVANIAAKLEIM 147
Query: 149 QPVFSFKLRGAYNMMVKLP-KELLEKG---VICSSAGNHAQGVALAAKRLNCNAVVAMPV 204
+P S K R Y+M+ K + G ++ ++GN G+A A ++ MP
Sbjct: 148 EPCCSVKDRIGYSMVTDAEQKGFISPGKSVLVEPTSGNTGIGLAFIAASRGYRLILTMPA 207
Query: 205 TTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGIEEGRT---FVPAFDHP-DVIMGQG 260
+ + ++ GA +VL + A K I + + FD+P + +
Sbjct: 208 SMSMERRVLLKAFGAELVLTDPAKGMTGAVQKAEEILKNTPDAYMLQQFDNPANPKIHYE 267
Query: 261 TVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMA 315
T G EI V +G GG I G+ ++K +P+ ++IGVEPT+++ ++
Sbjct: 268 TTGPEIWDDTKGKVDIFVAGIGTGGTITGVGRFIKEKNPKTQVIGVEPTESDILS 322
>At2g43750 cysteine synthase (cpACS1)
Length = 392
Score = 56.6 bits (135), Expect = 4e-08
Identities = 47/176 (26%), Positives = 80/176 (44%), Gaps = 8/176 (4%)
Query: 144 KREDLQPVFSFKLRGAYNMMVKLP-KELLEKG---VICSSAGNHAQGVALAAKRLNCNAV 199
K E ++P S K R Y+M+ K L+ G ++ S++GN G+A A +
Sbjct: 105 KLEIMEPCCSVKDRIGYSMITDAEEKGLITPGKSVLVESTSGNTGIGLAFIAASKGYKLI 164
Query: 200 VAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGIEE---GRTFVPAFDHP-DV 255
+ MP + + + GA +VL + A K I + + FD+P +
Sbjct: 165 LTMPASMSLERRVLLRAFGAELVLTEPAKGMTGAIQKAEEILKKTPNSYMLQQFDNPANP 224
Query: 256 IMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDA 311
+ T G EI + + +G GG I G+ ++K P++K+IGVEPT++
Sbjct: 225 KIHYETTGPEIWEDTRGKIDILVAGIGTGGTITGVGRFIKERKPELKVIGVEPTES 280
>At4g14880 cytosolic O-acetylserine(thiol)lyase (EC 4.2.99.8)
Length = 322
Score = 51.6 bits (122), Expect = 1e-06
Identities = 71/289 (24%), Positives = 120/289 (40%), Gaps = 23/289 (7%)
Query: 110 MANILSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKE 169
MA+ ++ V ++ +PL ++E + K E ++P S K R ++M+ K+
Sbjct: 1 MASRIAKDVTELIGNTPLVYLNNVAEGCVGRVAAKLEMMEPCSSVKDRIGFSMISDAEKK 60
Query: 170 LL----EKGVICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIG 225
L E +I ++GN G+A A ++ MP + + + G +VL
Sbjct: 61 GLIKPGESVLIEPTSGNTGVGLAFTAAAKGYKLIITMPASMSTERRIILLAFGVELVLTD 120
Query: 226 DSYDEAQAYAKKRGI---EEGRTFVPAFDHP-DVIMGQGTVGMEIMRQMVDPVHAIFVPV 281
+ A AK I + F++P + + T G EI + + +
Sbjct: 121 PAKGMKGAIAKAEEILAKTPNGYMLQQFENPANPKIHYETTGPEIWKGTGGKIDGFVSGI 180
Query: 282 GGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVG-GFADGVAVKE 340
G GG I G Y+K + VK+ GVEP ++ ++ ++ +G GF V
Sbjct: 181 GTGGTITGAGKYLKEQNANVKLYGVEPVESAILSGGKPGPHKI--QGIGAGFIPSV---- 234
Query: 341 VGEETFRLCKDLIDGVVLVNRD-SICASIKDMFEENRSILEPSGALALA 388
L DLID VV V+ D SI + + +E + SGA A A
Sbjct: 235 -------LNVDLIDEVVQVSSDESIDMARQLALKEGLLVGISSGAAAAA 276
>At3g03630 O-acetylserine (thiol) lyase
Length = 404
Score = 48.5 bits (114), Expect = 1e-05
Identities = 59/276 (21%), Positives = 109/276 (39%), Gaps = 37/276 (13%)
Query: 125 SPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLEKGVICS------ 178
+P+ ++++ ++ K E ++P S K R +M+ E G I
Sbjct: 110 TPMVYLNRVTDGCLADIAAKLESMEPCRSVKDRIGLSMI----NEAENSGAITPRKTVLV 165
Query: 179 --SAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAK 236
+ GN G+A A +V MP + + + LGA +VL A K
Sbjct: 166 EPTTGNTGLGIAFVAAAKGYKLIVTMPASINIERRMLLRALGAEIVLTNPEKGLKGAVDK 225
Query: 237 KRGI---EEGRTFVPAFDHP-DVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAA 292
+ I + FD+ + + T G EI + V +G GG + G
Sbjct: 226 AKEIVLKTKNAYMFQQFDNTANTKIHFETTGPEIWEDTMGNVDIFVAGIGTGGTVTGTGG 285
Query: 293 YVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVKEVGEETFRLCKDL 352
++K ++ +K++GVEP+ ++ VI G+ G+ ++ +E F++
Sbjct: 286 FLKMMNKDIKVVGVEPS-----------ERSVISGDNPGYLPGILDVKLLDEVFKVS--- 331
Query: 353 IDGVVLVNRDSICASIKDMFEENRSILEPSGALALA 388
N ++I + + EE + SGA A+A
Sbjct: 332 -------NGEAIEMARRLALEEGLLVGISSGAAAVA 360
>At3g04940 cysteine synthase (AtcysD1)
Length = 324
Score = 47.0 bits (110), Expect = 3e-05
Identities = 49/183 (26%), Positives = 76/183 (40%), Gaps = 22/183 (12%)
Query: 144 KREDLQPVFSFKLRGAYNMMVKLPKELLEKGVIC--------SSAGNHAQGVALAAKRLN 195
K E ++P S K R AY M+ K+ +KG+I +++GN G+A
Sbjct: 38 KLEMMEPCSSVKERIAYGMI----KDAEDKGLITPGKSTLIEATSGNTGIGLAFIGAAKG 93
Query: 196 CNAVVAMPVTTPDIKWKSVEKLGATVVLIGDS------YDEAQAYAKKRGIEEGRTFVPA 249
V+ MP + + + LGA V L S D+A+ K +
Sbjct: 94 YKVVLTMPSSMSLERKIILLALGAEVHLTDPSKGVQGIIDKAEEICSKN---PDSIMLEQ 150
Query: 250 FDHPDVIMGQ-GTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEP 308
F +P T G EI R V + VG GG ++G ++K + K+ GVEP
Sbjct: 151 FKNPSNPQTHYRTTGPEIWRDSAGEVDILVAGVGTGGTLSGSGRFLKEKNKDFKVYGVEP 210
Query: 309 TDA 311
T++
Sbjct: 211 TES 213
>At5g28030 cysteine synthase - like
Length = 323
Score = 46.6 bits (109), Expect = 4e-05
Identities = 77/329 (23%), Positives = 127/329 (38%), Gaps = 55/329 (16%)
Query: 113 ILSAKVYDVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLE 172
++ V ++ +P+ K+ + + K E ++P S K R AY+M+ K+ +
Sbjct: 6 LIKNDVTELIGNTPMVYLNKIVDGCVARIAAKLEMMEPCSSIKDRIAYSMI----KDAED 61
Query: 173 KGVIC--------SSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATVVLI 224
KG+I ++ GN G+A ++ MP T + + LGA V L
Sbjct: 62 KGLITPGKSTLIEATGGNTGIGLASIGASRGYKVILLMPSTMSLERRIILRALGAEVHLT 121
Query: 225 GDSYDEAQAYAKKRGI---EEGRTFVPAFDHPD-VIMGQGTVGMEIMRQMVDPVHAIFVP 280
S K + I G F +P+ + T G EI R V +
Sbjct: 122 DISIGIKGQLEKAKEILSKTPGGYIPHQFINPENPEIHYRTTGPEIWRDSAGKVDILVAG 181
Query: 281 VGGGGLIAGIAAYVKTVSPQVKIIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVKE 340
VG GG + G ++K + +K+ VEP+++ AV
Sbjct: 182 VGTGGTVTGTGKFLKEKNKDIKVCVVEPSES-------------------------AVLS 216
Query: 341 VGEETFRLCKDLIDGVVLVNRDSICASIKDMFEENRSILEPSGALALAGAEAYCKYYGIK 400
G+ L + + G + N D SI D I++ +G A+ K IK
Sbjct: 217 GGKPGPHLIQGIGSGEIPANLD---LSIVD------EIIQVTGEEAIETT----KLLAIK 263
Query: 401 GGNVVAITSGANMNFDKLRVVTELANVGR 429
G +V I+SGA+ L+V NVG+
Sbjct: 264 EGLLVGISSGASA-AAALKVAKRPENVGK 291
>At1g55880 unknown protein
Length = 421
Score = 45.4 bits (106), Expect = 9e-05
Identities = 64/241 (26%), Positives = 89/241 (36%), Gaps = 56/241 (23%)
Query: 120 DVAIESPLQLAPKLSERIGVNLWLKREDLQPVFSFKLRGAYNMMVKLPKELLEKG----- 174
D +PL LSE G + K E L P S K R A VK+ +E LE G
Sbjct: 46 DAIGNTPLIRINSLSEATGCEILGKCEFLNPGGSVKDRVA----VKIIQEALESGKLFPG 101
Query: 175 --VICSSAGNHAQGVALAAKRLNCNAVVAMPVTTPDIKWKSVEKLGATV-------VLIG 225
V SAG+ A +A A C V +P K + +E LGA+V +
Sbjct: 102 GIVTEGSAGSTAISLATVAPAYGCKCHVVIPDDAAIEKSQIIEALGASVERVRPVSITHK 161
Query: 226 DSY--------DEAQAYAKKR-------GIEEGRT---FVPAFDHPDVIMGQGT------ 261
D Y DEA A KR GI + +T V P + T
Sbjct: 162 DHYVNIARRRADEANELASKRRLGSETNGIHQEKTNGCTVEEVKEPSLFSDSVTGGFFAD 221
Query: 262 --------------VGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVE 307
G EI Q + A G GG +AG++ +++ + +VK ++
Sbjct: 222 QFENLANYRAHYEGTGPEIWHQTQGNIDAFVAAAGTGGTLAGVSRFLQDKNERVKCFLID 281
Query: 308 P 308
P
Sbjct: 282 P 282
>At5g28020 cysteine synthase
Length = 323
Score = 45.1 bits (105), Expect = 1e-04
Identities = 46/184 (25%), Positives = 75/184 (40%), Gaps = 16/184 (8%)
Query: 144 KREDLQPVFSFKLRGAYNMMVKLPKELLEKGVICS--------SAGNHAQGVALAAKRLN 195
K E ++P S K R AY+M+ K+ +KG+I +AGN G+A
Sbjct: 37 KLEMMEPCSSVKDRIAYSMI----KDAEDKGLITPGKSTLIEPTAGNTGIGLACMGAARG 92
Query: 196 CNAVVAMPVTTPDIKWKSVEKLGATVVLIGDSYDEAQAYAKKRGI---EEGRTFVPAFDH 252
++ MP T + + LGA + L K I G F++
Sbjct: 93 YKVILVMPSTMSLERRIILRALGAELHLSDQRIGLKGMLEKTEAILSKTPGGYIPQQFEN 152
Query: 253 P-DVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVEPTDA 311
P + + T G EI R V + VG GG G+ ++K + +K+ VEP ++
Sbjct: 153 PANPEIHYRTTGPEIWRDSAGKVDILVAGVGTGGTATGVGKFLKEQNKDIKVCVVEPVES 212
Query: 312 NTMA 315
++
Sbjct: 213 PVLS 216
>At5g56890 unknown protein
Length = 1113
Score = 32.7 bits (73), Expect = 0.60
Identities = 23/69 (33%), Positives = 30/69 (43%), Gaps = 7/69 (10%)
Query: 11 HPHHPSPPPLLRRHAFHKMPTNVLH---PNPRRKPFIAATIS-KPAEITPSPSS---SSA 63
HP PSPPPL H H+ + P+P I+ S + +TP P S S
Sbjct: 334 HPSSPSPPPLSSHHQHHQERKKIADSPAPSPLPPHLISPKKSNRKGSMTPPPQSHHAPSP 393
Query: 64 AVGDHLSSP 72
+ D L SP
Sbjct: 394 PIPDSLISP 402
>At5g40470 unknown protein
Length = 496
Score = 32.7 bits (73), Expect = 0.60
Identities = 21/75 (28%), Positives = 38/75 (50%), Gaps = 5/75 (6%)
Query: 301 VKIIGVEPTDANTMAL--SLHHDQRVILDQVGGFAD--GVAVKEVGEETFRLCKDLIDGV 356
V + G++P D + L S +++ L G + G+ +K + E R C+ ++D V
Sbjct: 207 VSLFGIQPDDNDVTWLWKSCRKLKKLSLRSCGSIGEEIGLCLKNLEEIELRTCRSIVD-V 265
Query: 357 VLVNRDSICASIKDM 371
VL+ IC S+K +
Sbjct: 266 VLLKVSEICESLKSL 280
>At3g61310 DNA-binding protein like
Length = 354
Score = 32.3 bits (72), Expect = 0.78
Identities = 32/109 (29%), Positives = 46/109 (41%), Gaps = 18/109 (16%)
Query: 17 PPPLLRRHAFHKMP-----TNVL-----HPNPRRKPFIAAT--ISKPAEITPSPSSSSAA 64
PPP + FH TN + +PNP P +T +S P + SP+ SSAA
Sbjct: 26 PPPPQTQPTFHGSQGFHHFTNSISPFGSNPNPNPNPGGVSTGFVSPPLPVDSSPADSSAA 85
Query: 65 VGDHLSSPRLRVSPDSLQYEPGYLGAVPDRSRSDGGSDSDAMLTAMANI 113
L V+P S P + DGGS S A+ +++N+
Sbjct: 86 AAGAL------VAPPSGDTSVKRKRGRPRKYGQDGGSVSLALSPSISNV 128
>At1g06930 hypothetical protein
Length = 169
Score = 31.2 bits (69), Expect = 1.7
Identities = 18/63 (28%), Positives = 33/63 (51%), Gaps = 5/63 (7%)
Query: 16 SPPPLLRRHAFHKMPTNVLHPNPRR-KPFIAATISKPAEITPSPSSSSAAVGDHLSSPRL 74
+PPP F+ P++ H +P++ ++ +SK + SP+SSS++ + S L
Sbjct: 70 TPPPSY----FYASPSSTKHVSPKKTNTLFSSLLSKNRSVPSSPASSSSSSSSSVPSSPL 125
Query: 75 RVS 77
R S
Sbjct: 126 RTS 128
>At4g00040 chalcone synthase like protein
Length = 385
Score = 30.8 bits (68), Expect = 2.3
Identities = 28/105 (26%), Positives = 44/105 (41%), Gaps = 20/105 (19%)
Query: 303 IIGVEPTDANTMALSLHHDQRVILDQVGGFADGVAVKEVGEE--TFRLCKDLIDGV---- 356
IIG +PT++ + + LH + L Q G DG + EE TF+L +DL +
Sbjct: 224 IIGADPTESESPFMELHCAMQQFLPQTQGVIDG----RLSEEGITFKLGRDLPQKIEDNV 279
Query: 357 ------VLVNRDSICASIKDMFEENRSILEPSGALALAGAEAYCK 395
++ S + D+F + P G L+G E K
Sbjct: 280 EEFCKKLVAKAGSGALELNDLF----WAVHPGGPAILSGLETKLK 320
>At3g32400 putative protein
Length = 397
Score = 30.8 bits (68), Expect = 2.3
Identities = 27/82 (32%), Positives = 39/82 (46%), Gaps = 4/82 (4%)
Query: 1 MAALRFSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSS 60
M L T P P PPPLL+ H + ++ L P K +A T + P P P
Sbjct: 1 MELLSSIATEGPPPPPPPPLLQPH-HSALSSSPLPPPLPPKKLLATTNTPPP--PPPPLH 57
Query: 61 SSAAVGDHLSSPRLRVSPDSLQ 82
S++ +G SS L+ SP +L+
Sbjct: 58 SNSQMGAPTSSLVLK-SPHNLK 78
>At1g30480 DNA damage repair protein, putative
Length = 387
Score = 30.8 bits (68), Expect = 2.3
Identities = 18/55 (32%), Positives = 26/55 (46%), Gaps = 3/55 (5%)
Query: 18 PPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSAAVGDHLSSP 72
PP LR+ P +L P + KP ++A P PS SS S + + S+P
Sbjct: 34 PPTLRKPPAFAPPQTILRPLNKPKPIVSAPYKPP---PPSNSSQSVLIPANESAP 85
>At1g63340 unknown protein
Length = 398
Score = 30.4 bits (67), Expect = 3.0
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 5/88 (5%)
Query: 248 PAFDHPDVIMGQGTVGMEIMRQMVDPVHAIFVPVGGGGLIAGIAAYVKTVSPQVKIIGVE 307
P H ++G G G+ R++ H++ V G I G+ AY V P + ++
Sbjct: 8 PTTSHHVAVIGAGAAGLVAARELRREGHSVVV-FERGNQIGGVWAYTPNVEPDP--LSID 64
Query: 308 PTDANTMALSLHHDQRVILD-QVGGFAD 334
PT + SL+ R I+ + GF D
Sbjct: 65 PT-RPVIHSSLYSSLRTIIPRECMGFTD 91
>At5g65390 arabinogalactan protein AGP7
Length = 130
Score = 30.0 bits (66), Expect = 3.9
Identities = 30/122 (24%), Positives = 49/122 (39%), Gaps = 11/122 (9%)
Query: 6 FSTTTHPHHPSPPPLLRRHAFHKMPTNVLHPNPRRKPFIAATISKPAEITPSPSSSSAAV 65
F+T+ P+P P P V P P P A T + P ++P+P+SS +
Sbjct: 15 FTTSCLAQAPAPSPTTT-----VTPPPVATPPPAATP--APTTTPPPAVSPAPTSSPPSS 67
Query: 66 GDHLSSPRLRVSPDSLQ---YEPGYLGAVP-DRSRSDGGSDSDAMLTAMANILSAKVYDV 121
SS SP + + PG L P D S + + ++++A +Y V
Sbjct: 68 APSPSSDAPTASPPAPEGPGVSPGELAPTPSDASAPPPNAALTNKAFVVGSLVAAIIYAV 127
Query: 122 AI 123
+
Sbjct: 128 VL 129
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,676,631
Number of Sequences: 26719
Number of extensions: 607852
Number of successful extensions: 1796
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1756
Number of HSP's gapped (non-prelim): 44
length of query: 606
length of database: 11,318,596
effective HSP length: 105
effective length of query: 501
effective length of database: 8,513,101
effective search space: 4265063601
effective search space used: 4265063601
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0375a.8


