

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371b.8
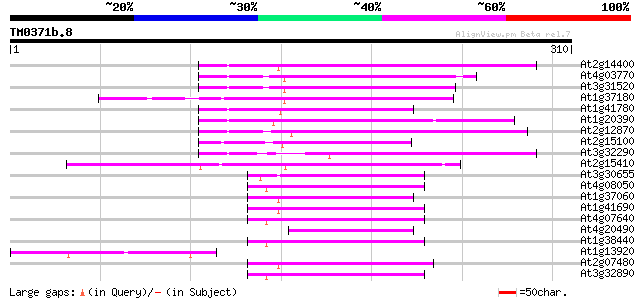
(310 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g14400 putative retroelement pol polyprotein 79 4e-15
At4g03770 hypothetical protein 73 2e-13
At3g31520 hypothetical protein 69 2e-12
At1g37180 hypothetical protein 67 1e-11
At1g41780 hypothetical protein 65 4e-11
At1g20390 hypothetical protein 65 5e-11
At2g12870 putative retroelement pol polyprotein 64 1e-10
At2g15100 putative retroelement pol polyprotein 62 4e-10
At3g32290 hypothetical protein 55 4e-08
At2g15410 putative retroelement pol polyprotein 54 1e-07
At3g30655 hypothetical protein, 3' partial 50 1e-06
At4g08050 50 2e-06
At1g37060 Athila retroelment ORF 1, putative 50 2e-06
At1g41690 hypothetical protein 49 5e-06
At4g07640 putative athila transposon protein 47 1e-05
At4g20490 putative protein 47 2e-05
At1g38440 hypothetical protein 45 4e-05
At1g13920 hypothetical protein 45 4e-05
At2g07480 F9A16.15 45 5e-05
At3g32890 Athila ORF 1, putative 45 7e-05
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 78.6 bits (192), Expect = 4e-15
Identities = 54/192 (28%), Positives = 92/192 (47%), Gaps = 6/192 (3%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVII----AASDAVKCRMFPS 160
PF+ + S+ I D+ K L L+SY+G DPK +L F + A D C++F
Sbjct: 144 PFTPQITSLRIRDSRK-LNLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSE 202
Query: 161 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 220
A+ WFT L GSISNF + S FL Q+S ++ ++ DL+N+ Q E+L+ ++
Sbjct: 203 NLCGQALMWFTQLEPGSISNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFI 262
Query: 221 ARYSAASDKVEDEEPRACALAFKNGL-LPGGLNSKLARKPARSMEEMRARASTYILDEED 279
++ K+ ++ A + GL L ++++ RA+ ++ E++
Sbjct: 263 TKFKYVLSKLSRISQQSALSALRKGLWYDSRFKEDLILHKLDTIQDALFRANNWMEVEDE 322
Query: 280 DAFKGKRAKLEK 291
KR K K
Sbjct: 323 KESFAKRDKQAK 334
>At4g03770 hypothetical protein
Length = 464
Score = 72.8 bits (177), Expect = 2e-13
Identities = 48/161 (29%), Positives = 82/161 (50%), Gaps = 14/161 (8%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAAS-------DAVKCRM 157
PF+ + + I D K L ++ ++G SDPK HL F ++ +A + DA C +
Sbjct: 218 PFTHMISNAIISDPGK-LRIEYFNGSSDPKGHLKLF---IISVARAKFRPEERDAGLCHL 273
Query: 158 FPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLK 217
F K A+ WFT L S+ +F++ S+ FL Q+S + + DL+++ QQ E L+
Sbjct: 274 FVEHLKGPALNWFTRLKGNSVDSFQELSTLFLKQYSVLIDPSTSDADLWSLSQQPNEPLR 333
Query: 218 EYMARYSAASDKVEDEEPRACALAFKNGLLPGGLNSKLARK 258
+++A++ + KVE A K L L ++L+ K
Sbjct: 334 DFLAKFRSTLAKVEGINDLAALSTLKKALC---LRARLSAK 371
>At3g31520 hypothetical protein
Length = 528
Score = 69.3 bits (168), Expect = 2e-12
Identities = 45/149 (30%), Positives = 76/149 (50%), Gaps = 11/149 (7%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAAS-------DAVKCRM 157
PF+ + + I D K L ++ ++G SDPK HL F ++ +A + DA C +
Sbjct: 226 PFTHRISNAIISDPGK-LRIEYFNGSSDPKRHLKSF---IISVARTKFRPEERDAGLCHL 281
Query: 158 FPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLK 217
F K A+ WF+ L S+ +F++ S+ FL Q+S + + DL+++ QQ E L+
Sbjct: 282 FVEHLKGPALCWFSRLEGNSMDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLR 341
Query: 218 EYMARYSAASDKVEDEEPRACALAFKNGL 246
+++A+ + KVE A A K L
Sbjct: 342 DFLAKLRSTLAKVEGINDVAALSALKKAL 370
>At1g37180 hypothetical protein
Length = 661
Score = 67.0 bits (162), Expect = 1e-11
Identities = 47/200 (23%), Positives = 83/200 (41%), Gaps = 14/200 (7%)
Query: 50 PEQQPAVTQEQWRNLMRSIGNIQQRNEHLQAQLDFYRREQRDDGSRKADSVAEFRPFSED 109
P Q+P+ + ++L I + + QA E+ + +R+ PF+
Sbjct: 252 PRQEPSAVNSELQSLQDQIWAMNAKVH--QATTSAPEVEKVIEATRRT-------PFTPR 302
Query: 110 VESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAAS----DAVKCRMFPSTFKST 165
+ + I + + L Y+G DP +HL F + + DA C++F
Sbjct: 303 ISKLRIRE-FRDFKLPVYNGKGDPNEHLTSFQVIVRRVPLEPYEEDAGLCKLFSENLSGP 361
Query: 166 AMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSA 225
A+ WFT L GSI NF+ S+ F+ Q+ +T L+N+ Q + L+ Y+ +
Sbjct: 362 ALTWFTQLEEGSIDNFKQLSTAFIKQYGYFIKSDITEAHLWNLSQSADDPLRTYIIVFKE 421
Query: 226 ASDKVEDEEPRACALAFKNG 245
++ A KNG
Sbjct: 422 IMVQIPSLSDSTAQFALKNG 441
>At1g41780 hypothetical protein
Length = 246
Score = 65.5 bits (158), Expect = 4e-11
Identities = 37/123 (30%), Positives = 63/123 (51%), Gaps = 5/123 (4%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIA----ASDAVKCRMFPS 160
PF+ + + I D K L ++ +SG SDPK HL F + SDA C +F
Sbjct: 125 PFTRKISNAIISDPEK-LRIEYFSGSSDPKGHLKSFKISVARAKFKPEESDAGLCHLFVE 183
Query: 161 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 220
K + WF+ L S+ +F++ S+ FL Q+S + + DL+++ QQ E L +++
Sbjct: 184 HLKGPVLDWFSRLEGNSVDSFQELSTLFLKQYSVLIDPGTSDVDLWSLSQQPNEPLSDFL 243
Query: 221 ARY 223
++
Sbjct: 244 TKF 246
>At1g20390 hypothetical protein
Length = 1791
Score = 65.1 bits (157), Expect = 5e-11
Identities = 48/180 (26%), Positives = 85/180 (46%), Gaps = 7/180 (3%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKM----VIIAASDAVKCRMFPS 160
PF+ + +V+I K + L+SY+G +DPK+ L FN + + I DA +C++F
Sbjct: 156 PFTRRITNVSIRGAQK-IKLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIE 214
Query: 161 TFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYM 220
A WF+ L SI +F +S FL ++ + DL++I Q ESL+ ++
Sbjct: 215 HLTGPAHNWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFV 274
Query: 221 ARYSAASDKVEDEEPRACALAFKNGL-LPGGLNSKLARKPARSMEEMRARASTYILDEED 279
R+ + + A +A +N + + ++E+ RAS +I EE+
Sbjct: 275 DRFKLVVTNITVPD-EAAIVALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEE 333
>At2g12870 putative retroelement pol polyprotein
Length = 411
Score = 63.9 bits (154), Expect = 1e-10
Identities = 53/191 (27%), Positives = 82/191 (42%), Gaps = 14/191 (7%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVK--------CR 156
PF+ + S I D K L +D ++G S PK HL F +I AA D K C
Sbjct: 61 PFTRKISSAIISDPGK-LRIDYFNGSSYPKGHLKSF----IIYAARDKFKPEERDAGLCH 115
Query: 157 MFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESL 216
+F K + F L + +F + S+ FL QFS + +DL+++ QQ E L
Sbjct: 116 LFVEHLKGPVLVLFLRLEGNFVDSFEELSTLFLKQFSVLIDPGTLDSDLWSLSQQPNEPL 175
Query: 217 KEYMARYSAASDKVEDEEPRACALAFKNGL-LPGGLNSKLARKPARSMEEMRARASTYIL 275
+ + ++ + VE A A K L +L +++ + RAS Y+
Sbjct: 176 MDLLTKFRSTLASVEGIIDVAALSALKEALWYKSEFRKELNLSKPQTICDALHRASDYVS 235
Query: 276 DEEDDAFKGKR 286
EE+ KR
Sbjct: 236 HEEEMELLSKR 246
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 62.0 bits (149), Expect = 4e-10
Identities = 39/126 (30%), Positives = 58/126 (45%), Gaps = 13/126 (10%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAA--------SDAVKCR 156
PF+ + + I + + L Y+G D K+HL F +IA DA C+
Sbjct: 157 PFTPRISKLRIRE-FRDFKLPVYNGKGDLKEHLTSFQ----VIAGRVPLEPHEEDAGLCK 211
Query: 157 MFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESL 216
+F A+ WFT L GSI NF+ S+ F+ Q+ N +T L+N Q E L
Sbjct: 212 LFSENLFGLALTWFTQLEEGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPL 271
Query: 217 KEYMAR 222
+ Y+ R
Sbjct: 272 RTYIYR 277
>At3g32290 hypothetical protein
Length = 612
Score = 55.5 bits (132), Expect = 4e-08
Identities = 52/191 (27%), Positives = 84/191 (43%), Gaps = 26/191 (13%)
Query: 105 PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMVIIAASDAVKCRMFPSTFKS 164
PF+ + + I D K L ++ ++G SDPK HL K II S
Sbjct: 164 PFTHMISNAIISDPGK-LRIEYFNGSSDPKGHL-----KSFII----------------S 201
Query: 165 TAMAWFTTLPR---GSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMA 221
A A F R S+ +F++ S+ FL Q+S + + DL+++ QQ E L++++A
Sbjct: 202 VARAKFRQEERDAGNSVDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLRDFLA 261
Query: 222 RYSAASDKVEDEEPRACALAFKNGL-LPGGLNSKLARKPARSMEEMRARASTYILDEEDD 280
++ + KVE A A K L +L ++ + RAS Y+ EE+
Sbjct: 262 KFRSTLAKVEGINDVAALSALKKALWYKSEFRKELNLSKPLTIRDALHRASDYVSHEEEM 321
Query: 281 AFKGKRAKLEK 291
KR + K
Sbjct: 322 ELVAKRHEPSK 332
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 53.5 bits (127), Expect = 1e-07
Identities = 57/228 (25%), Positives = 95/228 (41%), Gaps = 12/228 (5%)
Query: 32 PSPSQVGSL-ERSPENSPAPEQQPAVTQEQWRNLMRSIGNIQQRNEHLQAQL-DFYRREQ 89
P+ VGS+ E + EN+P + +P V R G + + L+ L D +
Sbjct: 158 PASIVVGSIDEHTRENAPNRDNRPNVDTRARRTEDDDTGARDRELQQLRTTLKDTSSKMH 217
Query: 90 RDDGSR-KADSVAEFR---PFSEDVESVAIPDNMKTLVLDSYSGDSDPKDHLLYFNTKMV 145
R S + D V E PF+ + V I ++ + L +Y G DP+ L + +
Sbjct: 218 RAVSSTPEVDKVLEETQRSPFTSCISDVRIR-HISKIKLANYEGLVDPRPFLTSVSIAIG 276
Query: 146 IIAASD----AVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVT 201
SD A C++F A+ WF+ L SI + ++ FL + + +
Sbjct: 277 RAHFSDEDRDAGSCQLFVKHLSGAALTWFSRLEANSIDSVHALTTSFLKNYGVFMEKGAS 336
Query: 202 INDLYNIRQQEGESLKEYMARYSAASDKVEDEEPRACALAFKNGLLPG 249
DL+ + Q ESL+ ++ R+ V + A A A +N L G
Sbjct: 337 NVDLWTMAQTAKESLRSFIGRFKEIVTSVATPDDAAIA-ALRNALWHG 383
>At3g30655 hypothetical protein, 3' partial
Length = 660
Score = 50.4 bits (119), Expect = 1e-06
Identities = 33/103 (32%), Positives = 48/103 (46%), Gaps = 6/103 (5%)
Query: 132 DPKDHL-----LYFNTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSS 186
DP DHL LY TK+ ++ D K R+FP + A W TLP+ SI+ + D
Sbjct: 61 DPLDHLDEFERLYGLTKINGVS-EDGFKLRLFPFSLGDKAHLWEKTLPQNSITAWDDCKK 119
Query: 187 KFLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDK 229
FL +F +N N++ Q++ ES E R+ K
Sbjct: 120 AFLAKFFSNSRTARLRNEISGFTQKQNESFGEAWERFKGYQTK 162
>At4g08050
Length = 1428
Score = 50.1 bits (118), Expect = 2e-06
Identities = 30/102 (29%), Positives = 46/102 (44%), Gaps = 4/102 (3%)
Query: 132 DPKDHLLYF----NTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F N + + D K R+FP + A W L SI+ + D+
Sbjct: 61 DPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLSHDSITTWDDYKKA 120
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDK 229
FL +F +N N++Y Q+ GES E R+ +++
Sbjct: 121 FLSKFFSNARTARLRNEIYGFSQKTGESFCEAWERFKGYTNQ 162
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 50.1 bits (118), Expect = 2e-06
Identities = 29/96 (30%), Positives = 44/96 (45%), Gaps = 4/96 (4%)
Query: 132 DPKDHLLYFNTKMVII----AASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F+ + + D K R+FP + A W +LP+GSI+++ D
Sbjct: 61 DPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKA 120
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARY 223
FL +F +N ND+ Q E+ E R+
Sbjct: 121 FLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERF 156
>At1g41690 hypothetical protein
Length = 371
Score = 48.5 bits (114), Expect = 5e-06
Identities = 30/102 (29%), Positives = 44/102 (42%), Gaps = 4/102 (3%)
Query: 132 DPKDHLLYFNTKMVII----AASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F + + D K R+FP + A W TLP+GSI+ + D
Sbjct: 61 DPLDHLDEFERLCSLTKINGVSEDGFKLRLFPFSLGDKAHLWEKTLPQGSITTWDDCKKA 120
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDK 229
FL +F + N++ Q++ ES E R+ K
Sbjct: 121 FLAKFFSYSRTARLRNEISGFTQKQSESFCEAWERFKGYQTK 162
>At4g07640 putative athila transposon protein
Length = 866
Score = 47.0 bits (110), Expect = 1e-05
Identities = 30/102 (29%), Positives = 45/102 (43%), Gaps = 4/102 (3%)
Query: 132 DPKDHLLYF----NTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F N + ++D K R+FP + A W LP SI + D
Sbjct: 61 DPLDHLDEFDRLCNLTKINGVSADGFKLRLFPFSLGDKAHIWEKNLPHDSIITWDDCKKA 120
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDK 229
FL +F +N N++ Q+ GES E R+ +++
Sbjct: 121 FLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQ 162
>At4g20490 putative protein
Length = 853
Score = 46.6 bits (109), Expect = 2e-05
Identities = 21/69 (30%), Positives = 37/69 (53%)
Query: 155 CRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSKFLVQFSANKNQPVTINDLYNIRQQEGE 214
C++F A+ F+ L SI NF S+ FL Q+ + +DL+++ Q+ GE
Sbjct: 222 CQLFVEGLTGNALTCFSRLEANSIDNFTQLSTAFLKQYRVFIQPGASSSDLWSMTQENGE 281
Query: 215 SLKEYMARY 223
+L +Y+ R+
Sbjct: 282 TLNDYLGRF 290
>At1g38440 hypothetical protein
Length = 263
Score = 45.4 bits (106), Expect = 4e-05
Identities = 29/102 (28%), Positives = 44/102 (42%), Gaps = 4/102 (3%)
Query: 132 DPKDHLLYF----NTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F N + + D K R+FP + A W LP SI+ + D
Sbjct: 14 DPLDHLDEFDRLYNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSITTWDDCKKA 73
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDK 229
FL +F +N N++ Q+ ES E R+ +++
Sbjct: 74 FLSKFFSNARTARLRNEISGFSQKTSESFCEAWERFKGYTNQ 115
>At1g13920 hypothetical protein
Length = 345
Score = 45.4 bits (106), Expect = 4e-05
Identities = 32/130 (24%), Positives = 57/130 (43%), Gaps = 18/130 (13%)
Query: 1 RPHRVVLDSPVRTTGVQSPSPPPSPPVPPPP---PSPSQVGSLERSPENSPAPEQQPAVT 57
R + V + P +++ S P +PP PPPP PSP ++ E ++S ++ T
Sbjct: 180 RQRKPVSEPPRIQPPLRTRSEPRAPPPPPPPLLSPSPLRLPPRETKRQSSEHTSRKDDST 239
Query: 58 QEQWRNLMRSIGNIQQRNEHLQAQLDFYRREQRDDGSRKAD-------------SVAEFR 104
+ W + I+ R E L ++D + ++R+ RK D + FR
Sbjct: 240 ADAWEK--AELSKIKARYEKLNRKIDLWEAKKREKARRKLDISEQSELEQRRKRGLQRFR 297
Query: 105 PFSEDVESVA 114
+E +E +A
Sbjct: 298 EDTEYIEQIA 307
>At2g07480 F9A16.15
Length = 1012
Score = 45.1 bits (105), Expect = 5e-05
Identities = 28/107 (26%), Positives = 46/107 (42%), Gaps = 4/107 (3%)
Query: 132 DPKDHLLYFNTKMVII----AASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F+ + + ++ K R+FP + A W TLP SI + D
Sbjct: 56 DPLDHLDNFDRLCSLTKINGVSEESFKLRLFPFSLGDKAHLWEKTLPVKSIDTWDDCKKA 115
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDKVEDEE 234
FL +F +N + +++ Q+ ES E R+ + + E
Sbjct: 116 FLAKFFSNSRKARLRSEISGFNQKNSESFSEAWERFKGYTTQCPHHE 162
>At3g32890 Athila ORF 1, putative
Length = 755
Score = 44.7 bits (104), Expect = 7e-05
Identities = 29/102 (28%), Positives = 44/102 (42%), Gaps = 4/102 (3%)
Query: 132 DPKDHLLYF----NTKMVIIAASDAVKCRMFPSTFKSTAMAWFTTLPRGSISNFRDFSSK 187
DP DHL F N + + D K R+FP + A W LP SI+ + D
Sbjct: 14 DPLDHLDEFDRLYNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSITTWDDCKKA 73
Query: 188 FLVQFSANKNQPVTINDLYNIRQQEGESLKEYMARYSAASDK 229
FL +F +N N++ Q+ ES E R+ +++
Sbjct: 74 FLSKFFSNARTARLRNEISGFSQKTRESFCEAWERFKGYTNQ 115
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,617,493
Number of Sequences: 26719
Number of extensions: 368392
Number of successful extensions: 10266
Number of sequences better than 10.0: 408
Number of HSP's better than 10.0 without gapping: 286
Number of HSP's successfully gapped in prelim test: 128
Number of HSP's that attempted gapping in prelim test: 2910
Number of HSP's gapped (non-prelim): 2996
length of query: 310
length of database: 11,318,596
effective HSP length: 99
effective length of query: 211
effective length of database: 8,673,415
effective search space: 1830090565
effective search space used: 1830090565
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0371b.8


