

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371b.6
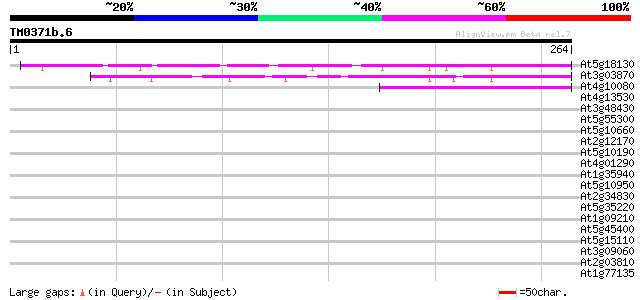
(264 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g18130 unknown protein 120 1e-27
At3g03870 unknown protein 109 2e-24
At4g10080 unknown protein 47 1e-05
At4g13530 unknown protein 39 0.004
At3g48430 putative zinc finger protein 33 0.12
At5g55300 DNA topoisomerase I 32 0.27
At5g10660 vacuolar calcium binding protein - like 32 0.46
At2g12170 hypothetical protein 29 2.3
At5g10190 unknown protein 29 3.0
At4g01290 hypothetical protein 28 3.9
At1g35940 hypothetical protein 28 3.9
At5g10950 putative protein 28 5.0
At2g34830 WRKY-type DNA binding protein-like 28 5.0
At5g35220 putative protein 28 6.6
At1g09210 putative calcium-binding protein, calreticulin 28 6.6
At5g45400 replication protein A1-like 27 8.6
At5g15110 pectate lyase -like protein 27 8.6
At3g09060 hypothetical protein 27 8.6
At2g03810 unknown protein 27 8.6
At1g77135 hypothetical protein 27 8.6
>At5g18130 unknown protein
Length = 280
Score = 120 bits (300), Expect = 1e-27
Identities = 96/291 (32%), Positives = 145/291 (48%), Gaps = 47/291 (16%)
Query: 6 DWVLLSDDG----SLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHD- 60
DW +L LD G E + K + FD +YF C P T+ LH
Sbjct: 5 DWEILPKINYKGLELDLGHEEDHEVTKMMRNTAKSFDSDYFIC--PIQDSVGKTEFLHQR 62
Query: 61 ---------------DPIQFVENTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFL 105
+P+ V+ + DH SE ++ E+ ++ F
Sbjct: 63 SSVVPTQLLQIPITWEPLSPVD--DKDHNKYLDPDFSEPDPELLTESFPSPRIT---FKK 117
Query: 106 QLENKFADMKMDSPKSSPRVLFPPPLDAGALKFEDK-----GEAMEVMVSPRRKVENEMA 160
E +FADMK+DSP + F PL + D GE+ + ++ + ++++
Sbjct: 118 SKETEFADMKIDSPAAR----FTSPLPQNDERHSDSEGGLGGESYDEIMGSEVEESSDLS 173
Query: 161 TMDCDREEVDPTDG--FNFWKWSLTGVGAICSFGVAAA--TICVLVFG--SQQKNKFHQN 214
+ ++EVD +G N WK L G+GAICSFGVAAA TICV G S + ++N
Sbjct: 174 S----KKEVDWDEGERTNLWKKGLNGIGAICSFGVAAAAATICVFFLGHNSSIQGGRNKN 229
Query: 215 HKIQFQIYTDD-KRIKQVVQQATKLNEAIAATRGIPLSRAHITYGGYYEAL 264
++FQIY+DD KR+ +VV+ ATKLNEAI+ +G+P++RA I++GGYY+AL
Sbjct: 230 QILRFQIYSDDNKRMNEVVKHATKLNEAISVMKGLPVARAQISFGGYYDAL 280
>At3g03870 unknown protein
Length = 266
Score = 109 bits (272), Expect = 2e-24
Identities = 87/252 (34%), Positives = 128/252 (50%), Gaps = 43/252 (17%)
Query: 39 MNYFYCTS-----PKSRDSSTTQLLHDDPIQF------VENTEDDHVGDNSDTTSEKIKA 87
M+YF C S PK+ ++++ +Q V + ++ + +N D +
Sbjct: 32 MDYFLCPSTKDPLPKTESPPRSKVVPKKILQVPIAWEAVLDQDNTKIPNNPDLEPDPHP- 90
Query: 88 VVVEADEQETLSVSQ-------FFLQLENKFADMKMDSPKSSPRVLFP-PPLDAGALKFE 139
+ D + TLS F + + +FADMK+D P R+ P P +D A K
Sbjct: 91 ---DPDSKPTLSTDSVSSPRVSFKIMKDTEFADMKIDPPA---RITSPLPQIDDAASKPS 144
Query: 140 DKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKWSLTGVGAICSFGVAAA--T 197
D +E R K E ++ + E+ + N WK L G+GAICSFGVAAA T
Sbjct: 145 D----LEGGDDLRDKKE---VVLEEEDEDGSGGERLNLWKAGLNGIGAICSFGVAAAAAT 197
Query: 198 ICVLVFGSQQ----KNKFHQNHKIQFQIYTDD-KRIKQVVQQATKLNEAIAATRGIPLSR 252
+CV G KNK N ++FQIY+DD KR+K+VV ATKLNEAI +G+P+ R
Sbjct: 198 VCVFFLGHNNIKICKNK---NQMLRFQIYSDDNKRMKEVVNHATKLNEAIFGMKGVPVVR 254
Query: 253 AHITYGGYYEAL 264
A I++GG+Y+ L
Sbjct: 255 AQISFGGHYDGL 266
>At4g10080 unknown protein
Length = 325
Score = 46.6 bits (109), Expect = 1e-05
Identities = 24/90 (26%), Positives = 46/90 (50%)
Query: 175 FNFWKWSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQ 234
F K+S+ +G + S +AAA + +++ G + N + + ++ DDK+ +V+ Q
Sbjct: 233 FVLLKYSVFRIGPVWSVSMAAAVMGLVLLGRRLYNMKKKAQRFHLKVTIDDKKASRVMSQ 292
Query: 235 ATKLNEAIAATRGIPLSRAHITYGGYYEAL 264
A +LNE R +P+ R + G + L
Sbjct: 293 AARLNEVFTEVRRVPVIRPALPSPGAWPVL 322
>At4g13530 unknown protein
Length = 269
Score = 38.5 bits (88), Expect = 0.004
Identities = 32/133 (24%), Positives = 59/133 (44%), Gaps = 23/133 (17%)
Query: 139 EDKGEAMEVMVSPRRKVENEMATMDCDREEV--DP----------TDGFNFWK------- 179
E + M+++ S RK E+ + ++ + V DP GF +WK
Sbjct: 123 ESIAQDMDLISSDERKEESLLHPVEGEGNSVSIDPGVKSGGGGGEEKGFVWWKIPIEVLK 182
Query: 180 WSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLN 239
+ + + I S +AAA + ++ G + N + +Q ++ DDK V A + N
Sbjct: 183 YCVLKINPIWSLSMAAAFVGFVMLGRRLYNMKKKTRSLQLKVLLDDK----VANHAARWN 238
Query: 240 EAIAATRGIPLSR 252
EAI+ + +P+ R
Sbjct: 239 EAISVVKRVPIIR 251
>At3g48430 putative zinc finger protein
Length = 1360
Score = 33.5 bits (75), Expect = 0.12
Identities = 30/109 (27%), Positives = 45/109 (40%), Gaps = 19/109 (17%)
Query: 5 NDWVLLSDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRD---------SSTT 55
NDW S +L + +P+SK D+NYFY ++ D +STT
Sbjct: 580 NDWTAASGSANLGQAARSL------HPQSKEKHDVNYFYNVPVQTMDHSVKTGDQKTSTT 633
Query: 56 Q--LLHDDPIQFVENTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQ 102
+ H D V GD+SD+ E K +V + + ET + Q
Sbjct: 634 SPTIAHKD--NDVLGMLASAYGDSSDSEEEDQKGLVTPSSKGETKTYDQ 680
>At5g55300 DNA topoisomerase I
Length = 916
Score = 32.3 bits (72), Expect = 0.27
Identities = 28/117 (23%), Positives = 49/117 (40%), Gaps = 20/117 (17%)
Query: 77 NSDTTSEKIKAVVVEADEQETLSVSQFFLQLENKFADMKMDSPKSSPR-----------V 125
NS +++ K KA V + ++ + + ++K++ SP S V
Sbjct: 320 NSTSSAAKPKARNVVSPRSRAMTKNTKKVTKDSKYSTSSKSSPSSGDGQKKWTTLVHNGV 379
Query: 126 LFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVD----PTDGFNFW 178
+FPPP +K KG+ +++ + + E+ATM E D P NFW
Sbjct: 380 IFPPPYKPHGIKILYKGKPVDLTIE-----QEEVATMFAVMRETDYYTKPQFRENFW 431
>At5g10660 vacuolar calcium binding protein - like
Length = 407
Score = 31.6 bits (70), Expect = 0.46
Identities = 28/105 (26%), Positives = 47/105 (44%), Gaps = 4/105 (3%)
Query: 27 GKSNPESKSDFDMNYFYCTSPK--SRDSSTTQLLHDDPIQFVENTEDDHVGDNSDTTSEK 84
G S+ +SK + N +S K S DSS H+D + V+ D H+ D+ + E+
Sbjct: 169 GNSSSKSKKEGSGNVPKKSSGKEISPDSSPLASAHEDEEEIVKVETDVHISDHGEEPKEE 228
Query: 85 IKAVVVEADEQ-ETLSVSQFFLQLENKFADMKMDSPKSSPRVLFP 128
K + DE E S E + ++ +D KS+ ++ P
Sbjct: 229 DKDQFAQPDESGEEKETSPVAASTEEQKGEL-IDEDKSTEQIEEP 272
>At2g12170 hypothetical protein
Length = 115
Score = 29.3 bits (64), Expect = 2.3
Identities = 14/37 (37%), Positives = 21/37 (55%)
Query: 143 EAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWK 179
E + VSPRRK+ + AT++ D V PT+ + K
Sbjct: 5 EKRVLRVSPRRKLSSTKATVELDNTSVAPTEDLIYAK 41
>At5g10190 unknown protein
Length = 488
Score = 28.9 bits (63), Expect = 3.0
Identities = 11/33 (33%), Positives = 19/33 (57%)
Query: 40 NYFYCTSPKSRDSSTTQLLHDDPIQFVENTEDD 72
++ YCT P+ RD + Q L + +Q + ED+
Sbjct: 432 SFLYCTYPRDRDRAKMQALIESEMQQLNEEEDE 464
>At4g01290 hypothetical protein
Length = 736
Score = 28.5 bits (62), Expect = 3.9
Identities = 33/129 (25%), Positives = 50/129 (38%), Gaps = 14/129 (10%)
Query: 16 LDGGDENQIFLGKSNPESKSDFDMNYFYC--TSPKSRDSSTTQLLHDDPIQFVENTEDDH 73
L G D+ Q F K+NP+ +DF T S S+T + P+ E+ E
Sbjct: 190 LQGADKLQTFDTKANPDLSTDFPFQGHATKRTDQLSSTSTTKSVTAVPPVLTCEDLEQSI 249
Query: 74 VGDNSDT-------TSEKIKAVVVEADEQETLSV----SQFFLQLENKFADMK-MDSPKS 121
+ + D+ + V+ +Q SV SQ L L + +D K D+
Sbjct: 250 LSEVGDSYHPPPPPVDQDTSVPSVKMTKQRKTSVDDQASQHLLSLLQRSSDPKSQDTQLL 309
Query: 122 SPRVLFPPP 130
S PPP
Sbjct: 310 SATERRPPP 318
>At1g35940 hypothetical protein
Length = 1678
Score = 28.5 bits (62), Expect = 3.9
Identities = 27/99 (27%), Positives = 41/99 (41%), Gaps = 8/99 (8%)
Query: 11 SDDGSLDGGDENQIFLGKSNPESKSDFDMNY-FYCTSPKSRDSSTTQLLHDDPIQFVENT 69
SD S D N +F G ES +D D NY T +S D +T Q + +
Sbjct: 198 SDTSSDDENMANMMFDGAVY-ESDTDVDQNYSLSSTDDESEDPTTMQTKSESMANVSTHE 256
Query: 70 EDDHVGDNS------DTTSEKIKAVVVEADEQETLSVSQ 102
+D V S DT S +I ++ + +T +V +
Sbjct: 257 VEDIVSQLSENDIELDTFSSQIDRFMIPTESTQTANVPE 295
>At5g10950 putative protein
Length = 395
Score = 28.1 bits (61), Expect = 5.0
Identities = 20/61 (32%), Positives = 31/61 (50%), Gaps = 6/61 (9%)
Query: 33 SKSDFDMNYFYCTSPKSRDSST-TQLLHDDPIQFVENTEDDHVGDNSDTTSEKIKAVVVE 91
SK + D S K + SS T+ H D + ED+ GD+ DT+SE+ K V++
Sbjct: 188 SKGNVDQKETSKASKKPKMSSKLTKRKHTD-----DQDEDEEAGDDIDTSSEEAKPKVLK 242
Query: 92 A 92
+
Sbjct: 243 S 243
>At2g34830 WRKY-type DNA binding protein-like
Length = 427
Score = 28.1 bits (61), Expect = 5.0
Identities = 33/141 (23%), Positives = 47/141 (32%), Gaps = 19/141 (13%)
Query: 31 PESKSDFDMNYFYCTSPKSRDS-STTQLLHDDPIQFVENTEDDH-----------VGDNS 78
P S + Y+ C+S K + + DP V +H G
Sbjct: 227 PIKGSPYPRGYYRCSSSKGCSARKQVERSRTDPNMLVITYTSEHNHPWPTQRNALAGSTR 286
Query: 79 DTTSEKIKAVVVEADEQETLSVSQFFLQLENKFADMKMDSPKSS--PRVLFPPPLDAGAL 136
++S + + T S S Q + D P +S P PP DA A+
Sbjct: 287 SSSSSSLNPSSKSSTAAATTSPSSRVFQNNSS-----KDEPNNSNLPSSSTHPPFDAAAI 341
Query: 137 KFEDKGEAMEVMVSPRRKVEN 157
K E+ E E M VEN
Sbjct: 342 KEENVEERQEKMEFDYNDVEN 362
>At5g35220 putative protein
Length = 548
Score = 27.7 bits (60), Expect = 6.6
Identities = 13/54 (24%), Positives = 21/54 (38%)
Query: 68 NTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFLQLENKFADMKMDSPKS 121
+ DD +G+N +T + +E E S S N+F K P +
Sbjct: 62 SNRDDSIGENGETHKSSVVKTATFEEEDEETSKSSSTTSSSNEFGSDKTSMPST 115
>At1g09210 putative calcium-binding protein, calreticulin
Length = 424
Score = 27.7 bits (60), Expect = 6.6
Identities = 19/68 (27%), Positives = 31/68 (44%), Gaps = 7/68 (10%)
Query: 27 GKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHDDPIQFVENTEDDHVGDNSDTTSEKIK 86
GK K+ FD +S+D+ D+P EDD GD+SD+ S+ +
Sbjct: 352 GKLKDAEKAAFDEAEKKNEEEESKDAPAESDAEDEP-------EDDEGGDDSDSESKAEE 404
Query: 87 AVVVEADE 94
V+++E
Sbjct: 405 TKSVDSEE 412
>At5g45400 replication protein A1-like
Length = 853
Score = 27.3 bits (59), Expect = 8.6
Identities = 16/60 (26%), Positives = 30/60 (49%), Gaps = 12/60 (20%)
Query: 192 GVAAATICVLVFGSQQKNKFHQ------------NHKIQFQIYTDDKRIKQVVQQATKLN 239
G++A + + + +Q + KF KI+ + Y+D++R+K V +A KLN
Sbjct: 666 GMSAKDLYYVKYENQDEEKFEDIIRSVAFTKYIFKLKIKEETYSDEQRVKATVVKAEKLN 725
>At5g15110 pectate lyase -like protein
Length = 472
Score = 27.3 bits (59), Expect = 8.6
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query: 54 TTQLLHDDPIQFVENTEDDHVGDNSD-TTSEKIKAVVVEADEQETLSV 100
T Q H DP + V++ + H +++D TT E+ E +E+E + +
Sbjct: 44 TLQAYHSDPYEIVDHFHERHYDNSTDVTTPEEDGDAKPEEEEKEFIEM 91
>At3g09060 hypothetical protein
Length = 687
Score = 27.3 bits (59), Expect = 8.6
Identities = 10/32 (31%), Positives = 16/32 (49%)
Query: 149 VSPRRKVENEMATMDCDREEVDPTDGFNFWKW 180
V+P + N + M C ++E + GF W W
Sbjct: 145 VAPNLQTYNVLIKMSCKKKEFEKARGFLDWMW 176
>At2g03810 unknown protein
Length = 439
Score = 27.3 bits (59), Expect = 8.6
Identities = 30/133 (22%), Positives = 53/133 (39%), Gaps = 19/133 (14%)
Query: 2 DTTNDWVLLSDD---------GSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDS 52
+ + D V +S D G + ++ Q L + N S S + + + R
Sbjct: 230 ENSKDEVAISQDNDSKECLTLGDILSREDEQKSLNQDNISSDSHEEQSPSQLQDKEKRSL 289
Query: 53 STTQLLHDDPIQFVENTEDDHVGDN-----SDTTSEKIKAVVVEADEQETLSVSQFFLQL 107
TT + + +E TE+ G+ S TTS++ E ++ ET + Q +
Sbjct: 290 ETTAIETE-----LEKTEEPKQGEEKLSSVSTTTSQEPNKTCNEPEKPETENHHQQNCLV 344
Query: 108 ENKFADMKMDSPK 120
EN + D K S +
Sbjct: 345 ENSYEDDKFSSSR 357
>At1g77135 hypothetical protein
Length = 585
Score = 27.3 bits (59), Expect = 8.6
Identities = 18/60 (30%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Query: 10 LSDDGSLDGGDE---NQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHDDPIQFV 66
++D S +GGD+ N+IF +F +++ +PK R S T D PI +V
Sbjct: 404 INDINSYNGGDQGYLNEIFTWWHRIPKHMNFLKHFWEGDTPKHRKSKTRLFGADPPILYV 463
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,325,720
Number of Sequences: 26719
Number of extensions: 268723
Number of successful extensions: 933
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 916
Number of HSP's gapped (non-prelim): 20
length of query: 264
length of database: 11,318,596
effective HSP length: 97
effective length of query: 167
effective length of database: 8,726,853
effective search space: 1457384451
effective search space used: 1457384451
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0371b.6


