

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371b.4
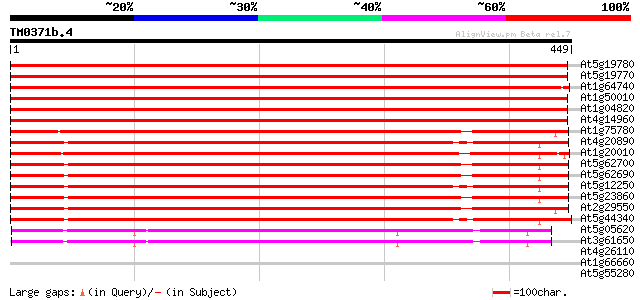
(449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g19780 tubulin alpha-5 chain 866 0.0
At5g19770 tubulin alpha-5 chain-like protein 866 0.0
At1g64740 unknown protein 845 0.0
At1g50010 tubulin alpha-2/alpha-4 chain, putative 834 0.0
At1g04820 tubulin alpha-2/alpha-4 chain 834 0.0
At4g14960 tubulin alpha-6 chain (TUA6) 833 0.0
At1g75780 tubulin beta-1 chain 398 e-111
At4g20890 tubulin beta-9 chain 394 e-110
At1g20010 beta tubulin 1, putative 393 e-110
At5g62700 tubulin beta-2/beta-3 chain (sp|P29512) 392 e-109
At5g62690 tubulin beta-2/beta-3 chain (sp|P29512) 392 e-109
At5g12250 tubulin beta-6 chain (sp|P29514) 392 e-109
At5g23860 beta tubulin 391 e-109
At2g29550 tubulin beta-7 chain 391 e-109
At5g44340 tubulin beta-4 chain (sp|P24636) 388 e-108
At5g05620 tubulin gamma-2 chain (sp|P38558) 248 5e-66
At3g61650 tubulin gamma-1 chain 245 4e-65
At4g26110 nucleosome assembly protein I-like protein 33 0.24
At1g66660 hypothetical protein 33 0.32
At5g55280 cell division protein FtsZ chloroplast homolog precurs... 33 0.42
>At5g19780 tubulin alpha-5 chain
Length = 450
Score = 866 bits (2237), Expect = 0.0
Identities = 418/446 (93%), Positives = 438/446 (97%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE+ISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDT+VG+++DAFNTFFSETG+GK
Sbjct: 1 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTTVGVAHDAFNTFFSETGAGK 60
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
HVPRA+FVDLEP+VIDEVRTG YRQLFHPEQLISGKEDAANNFARGHYTVG+EIV+LCLD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTVGKEIVDLCLD 120
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
R RKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA
Sbjct: 121 RVRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYNSVLSTHSLLEHTDV VLLDNEAIYDICRRSLDIERPTYTNLNRLISQ+ISSLTT
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLISQIISSLTT 240
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRFDGAINVD+TEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQ+S+PEIT+AVFEP++
Sbjct: 241 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQLSVPEITNAVFEPAS 300
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAV IKTKRTVQFVDWCPTGFKCGINYQPP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTVQFVDWCPTGFKCGINYQPP 360
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
TVVP GDLAKVQRAVCMISNNTAVAEVFSRID+KFDLMY+KRAFVHWYVGEGMEEGEFSE
Sbjct: 361 TVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 421 AREDLAALEKDYEEVGAEGEEGEEDE 446
AREDLAALEKDYEEVGAEG + EEDE
Sbjct: 421 AREDLAALEKDYEEVGAEGGDDEEDE 446
>At5g19770 tubulin alpha-5 chain-like protein
Length = 450
Score = 866 bits (2237), Expect = 0.0
Identities = 418/446 (93%), Positives = 438/446 (97%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE+ISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDT+VG+++DAFNTFFSETG+GK
Sbjct: 1 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTTVGVAHDAFNTFFSETGAGK 60
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
HVPRA+FVDLEP+VIDEVRTG YRQLFHPEQLISGKEDAANNFARGHYTVG+EIV+LCLD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTVGKEIVDLCLD 120
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
R RKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA
Sbjct: 121 RVRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYNSVLSTHSLLEHTDV VLLDNEAIYDICRRSLDIERPTYTNLNRLISQ+ISSLTT
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVAVLLDNEAIYDICRRSLDIERPTYTNLNRLISQIISSLTT 240
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRFDGAINVD+TEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQ+S+PEIT+AVFEP++
Sbjct: 241 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQLSVPEITNAVFEPAS 300
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAV IKTKRTVQFVDWCPTGFKCGINYQPP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTVQFVDWCPTGFKCGINYQPP 360
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
TVVP GDLAKVQRAVCMISNNTAVAEVFSRID+KFDLMY+KRAFVHWYVGEGMEEGEFSE
Sbjct: 361 TVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 421 AREDLAALEKDYEEVGAEGEEGEEDE 446
AREDLAALEKDYEEVGAEG + EEDE
Sbjct: 421 AREDLAALEKDYEEVGAEGGDDEEDE 446
>At1g64740 unknown protein
Length = 450
Score = 845 bits (2182), Expect = 0.0
Identities = 410/448 (91%), Positives = 432/448 (95%), Gaps = 1/448 (0%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE+ISIHIGQAGIQVGNSCWELYCLEHGIQPDG MPSD++VG +DAFNTFFSET SG+
Sbjct: 1 MREIISIHIGQAGIQVGNSCWELYCLEHGIQPDGTMPSDSTVGACHDAFNTFFSETSSGQ 60
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
HVPRA+F+DLEP+VIDEVRTG YRQLFHPEQLISGKEDAANNFARGHYTVGREIV+ CL+
Sbjct: 61 HVPRAVFLDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTVGREIVDTCLE 120
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
R RKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVD+GKKSKLGFTIYPSPQVSTA
Sbjct: 121 RLRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDFGKKSKLGFTIYPSPQVSTA 180
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTY+NLNRLISQ ISSLTT
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYSNLNRLISQTISSLTT 240
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRFDGAINVD+TEFQTNLVPYPRIHFMLSSYAPVIS+AKAYHEQ S+PEIT++VFEPSN
Sbjct: 241 SLRFDGAINVDITEFQTNLVPYPRIHFMLSSYAPVISSAKAYHEQFSVPEITTSVFEPSN 300
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MMAKCDPRHGKYMACCLMYRGDVVPKDVN AVA IK KRT+QFVDWCPTGFKCGINYQPP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNTAVAAIKAKRTIQFVDWCPTGFKCGINYQPP 360
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
+VVP GDLAKVQRAVCMISNNTAVAEVFSRID+KFDLMYSKRAFVHWYVGEGMEEGEFSE
Sbjct: 361 SVVPGGDLAKVQRAVCMISNNTAVAEVFSRIDHKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
Query: 421 AREDLAALEKDYEEVGAEGEEGEEDEEG 448
AREDLAALEKDYEEVG EG E ++DEEG
Sbjct: 421 AREDLAALEKDYEEVGGEGAE-DDDEEG 447
>At1g50010 tubulin alpha-2/alpha-4 chain, putative
Length = 450
Score = 834 bits (2154), Expect = 0.0
Identities = 400/446 (89%), Positives = 428/446 (95%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE ISIHIGQAGIQVGN+CWELYCLEHGIQPDG MPSD +VG +DAFNTFFSETG+GK
Sbjct: 1 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPSDKTVGGGDDAFNTFFSETGAGK 60
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
HVPRA+FVDLEP+VIDEVRTG YRQLFHPEQLISGKEDAANNFARGHYT+G+EIV+LCLD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 120
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
R RKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFT+YPSPQVST+
Sbjct: 121 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 180
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYNSVLSTHSLLEHTDV +LLDNEAIYDICRRSL IERPTYTNLNRL+SQVISSLT
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLSIERPTYTNLNRLVSQVISSLTA 240
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRFDGA+NVDVTEFQTNLVPYPRIHFMLSSYAPVISA KA+HEQ+S+ EIT++ FEP++
Sbjct: 241 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAFHEQLSVAEITNSAFEPAS 300
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAV IKTKRT+QFVDWCPTGFKCGINYQPP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTIQFVDWCPTGFKCGINYQPP 360
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
TVVP GDLAKVQRAVCMISN+T+VAEVFSRID+KFDLMY+KRAFVHWYVGEGMEEGEFSE
Sbjct: 361 TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 421 AREDLAALEKDYEEVGAEGEEGEEDE 446
AREDLAALEKDYEEVGAEG + E+DE
Sbjct: 421 AREDLAALEKDYEEVGAEGGDDEDDE 446
>At1g04820 tubulin alpha-2/alpha-4 chain
Length = 450
Score = 834 bits (2154), Expect = 0.0
Identities = 400/446 (89%), Positives = 428/446 (95%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE ISIHIGQAGIQVGN+CWELYCLEHGIQPDG MPSD +VG +DAFNTFFSETG+GK
Sbjct: 1 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPSDKTVGGGDDAFNTFFSETGAGK 60
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
HVPRA+FVDLEP+VIDEVRTG YRQLFHPEQLISGKEDAANNFARGHYT+G+EIV+LCLD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 120
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
R RKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFT+YPSPQVST+
Sbjct: 121 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 180
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYNSVLSTHSLLEHTDV +LLDNEAIYDICRRSL IERPTYTNLNRL+SQVISSLT
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLSIERPTYTNLNRLVSQVISSLTA 240
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRFDGA+NVDVTEFQTNLVPYPRIHFMLSSYAPVISA KA+HEQ+S+ EIT++ FEP++
Sbjct: 241 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAFHEQLSVAEITNSAFEPAS 300
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAV IKTKRT+QFVDWCPTGFKCGINYQPP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTIQFVDWCPTGFKCGINYQPP 360
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
TVVP GDLAKVQRAVCMISN+T+VAEVFSRID+KFDLMY+KRAFVHWYVGEGMEEGEFSE
Sbjct: 361 TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 421 AREDLAALEKDYEEVGAEGEEGEEDE 446
AREDLAALEKDYEEVGAEG + E+DE
Sbjct: 421 AREDLAALEKDYEEVGAEGGDDEDDE 446
>At4g14960 tubulin alpha-6 chain (TUA6)
Length = 450
Score = 833 bits (2151), Expect = 0.0
Identities = 399/446 (89%), Positives = 428/446 (95%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE ISIHIGQAGIQVGN+CWELYCLEHGIQPDG MP D +VG +DAFNTFFSETG+GK
Sbjct: 1 MRECISIHIGQAGIQVGNACWELYCLEHGIQPDGQMPGDKTVGGGDDAFNTFFSETGAGK 60
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
HVPRA+FVDLEP+VIDEVRTG YRQLFHPEQLISGKEDAANNFARGHYT+G+EIV+LCLD
Sbjct: 61 HVPRAVFVDLEPTVIDEVRTGTYRQLFHPEQLISGKEDAANNFARGHYTIGKEIVDLCLD 120
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
R RKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFT+YPSPQVST+
Sbjct: 121 RIRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTVYPSPQVSTS 180
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYNSVLSTHSLLEHTDV +LLDNEAIYDICRRSL+IERPTYTNLNRL+SQVISSLT
Sbjct: 181 VVEPYNSVLSTHSLLEHTDVSILLDNEAIYDICRRSLNIERPTYTNLNRLVSQVISSLTA 240
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRFDGA+NVDVTEFQTNLVPYPRIHFMLSSYAPVISA KA+HEQ+S+ EIT++ FEP++
Sbjct: 241 SLRFDGALNVDVTEFQTNLVPYPRIHFMLSSYAPVISAEKAFHEQLSVAEITNSAFEPAS 300
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAV IKTKRT+QFVDWCPTGFKCGINYQPP
Sbjct: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVGTIKTKRTIQFVDWCPTGFKCGINYQPP 360
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
TVVP GDLAKVQRAVCMISN+T+VAEVFSRID+KFDLMY+KRAFVHWYVGEGMEEGEFSE
Sbjct: 361 TVVPGGDLAKVQRAVCMISNSTSVAEVFSRIDHKFDLMYAKRAFVHWYVGEGMEEGEFSE 420
Query: 421 AREDLAALEKDYEEVGAEGEEGEEDE 446
AREDLAALEKDYEEVGAEG + E+DE
Sbjct: 421 AREDLAALEKDYEEVGAEGGDDEDDE 446
>At1g75780 tubulin beta-1 chain
Length = 447
Score = 398 bits (1023), Expect = e-111
Identities = 182/449 (40%), Positives = 287/449 (63%), Gaps = 11/449 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ + GQ G Q+G+ WE+ C EHG+ P G D S + + N +++E G+
Sbjct: 1 MREILHVQGGQCGNQIGSKFWEVICDEHGVDPTGRYNGD-SADLQLERINVYYNEASGGR 59
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
+VPRA+ +DLEP +D +R+G Y Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 60 YVPRAVLMDLEPGTMDSIRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDAVLD 119
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 120 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 179
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 180 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTC 239
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 240 SLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYISLTVPELTQQMWDAKN 299
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ M+RG + K+V+ + N++ K + FV+W P K + PP
Sbjct: 300 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQILNVQNKNSSYFVEWIPNNVKSSVCDIPP 359
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
T ++ A + N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 360 T--------GIKMASTFVGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 411
Query: 421 AREDLAALEKDYEEV--GAEGEEGEEDEE 447
A ++ L +Y++ EE E DEE
Sbjct: 412 AESNMNDLVSEYQQYQDATADEEDEYDEE 440
>At4g20890 tubulin beta-9 chain
Length = 444
Score = 394 bits (1012), Expect = e-110
Identities = 186/450 (41%), Positives = 286/450 (63%), Gaps = 13/450 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G WE+ C EHGI G DT + + + N +F+E GK
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVICGEHGIDQTGQSCGDTDLQL--ERINVYFNEASGGK 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
+VPRA+ +DLEP +D +R+G + Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 YVPRAVLMDLEPGTMDSLRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + + F+++PSP+VS
Sbjct: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMMTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + PT+ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLANPTFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +S+PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYSALSVPELTQQMWDAKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ ++RG + K+V+ + N++ K + FV+W P K +
Sbjct: 299 MMCAADPRHGRYLTASAVFRGKMSTKEVDEQMMNVQNKNSSYFVEWIPNNVKSSV----C 354
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
+ P G ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 355 DIAPTG----LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AR---EDLAALEKDYEEVGAEGEEGEEDEE 447
A DL A + Y++ EE EEDEE
Sbjct: 411 AESNMNDLVAEYQQYQDATVGEEEYEEDEE 440
>At1g20010 beta tubulin 1, putative
Length = 449
Score = 393 bits (1010), Expect = e-110
Identities = 186/455 (40%), Positives = 291/455 (63%), Gaps = 17/455 (3%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G+ WE+ C EHGI G DT+ + + N +++E G+
Sbjct: 1 MREILHIQGGQCGNQIGSKFWEVICDEHGIDSTGRYSGDTA-DLQLERINVYYNEASGGR 59
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
+VPRA+ +DLEP +D +R+G + Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 60 YVPRAVLMDLEPGTMDSIRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDAVLD 119
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 120 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 179
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 180 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTC 239
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
SLRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 240 SLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYISLTVPELTQQMWDSKN 299
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ ++RG + K+V+ + NI+ K + FV+W P K + PP
Sbjct: 300 MMCAADPRHGRYLTASAIFRGQMSTKEVDEQILNIQNKNSSYFVEWIPNNVKSSVCDIPP 359
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
++ A + N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 360 --------KGLKMAATFVGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 411
Query: 421 AR---EDLAALEKDYEEVGAEGEEG----EEDEEG 448
A DL A + Y++ A+ EEG EE+EEG
Sbjct: 412 AESNMNDLVAEYQQYQDATAD-EEGEYDVEEEEEG 445
>At5g62700 tubulin beta-2/beta-3 chain (sp|P29512)
Length = 450
Score = 392 bits (1008), Expect = e-109
Identities = 184/450 (40%), Positives = 285/450 (62%), Gaps = 13/450 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G WE+ C EHGI P G D+ + + + N +++E G+
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYTGDSDLQL--ERINVYYNEASCGR 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
VPRA+ +DLEP +D +R+G Y Q F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 FVPRAVLMDLEPGTMDSLRSGPYGQTFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRSLTVPELTQQMWDSKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ M+RG + K+V+ + N++ K + FV+W P K + PP
Sbjct: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMLNVQNKNSSYFVEWIPNNVKSTVCDIPP 358
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
T ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AR---EDLAALEKDYEEVGAEGEEGEEDEE 447
A DL + + Y++ A+ E EDEE
Sbjct: 411 AESNMNDLVSEYQQYQDATADEEGDYEDEE 440
>At5g62690 tubulin beta-2/beta-3 chain (sp|P29512)
Length = 450
Score = 392 bits (1008), Expect = e-109
Identities = 184/450 (40%), Positives = 285/450 (62%), Gaps = 13/450 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G WE+ C EHGI P G D+ + + + N +++E G+
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDPTGRYTGDSDLQL--ERINVYYNEASCGR 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
VPRA+ +DLEP +D +R+G Y Q F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 FVPRAVLMDLEPGTMDSLRSGPYGQTFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRSLTVPELTQQMWDSKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ M+RG + K+V+ + N++ K + FV+W P K + PP
Sbjct: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMLNVQNKNSSYFVEWIPNNVKSTVCDIPP 358
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
T ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AR---EDLAALEKDYEEVGAEGEEGEEDEE 447
A DL + + Y++ A+ E EDEE
Sbjct: 411 AESNMNDLVSEYQQYQDATADEEGDYEDEE 440
>At5g12250 tubulin beta-6 chain (sp|P29514)
Length = 449
Score = 392 bits (1006), Expect = e-109
Identities = 185/451 (41%), Positives = 289/451 (64%), Gaps = 14/451 (3%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G+ WE+ C EHGI P G ++ + + + N +++E G+
Sbjct: 1 MREILHIQGGQCGNQIGSKFWEVVCDEHGIDPTGRYVGNSDLQL--ERVNVYYNEASCGR 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
+VPRA+ +DLEP +D VRTG Y Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 YVPRAILMDLEPGTMDSVRTGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDAVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDSKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ M+RG + K+V+ + N++ K + FV+W P K +
Sbjct: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSSV----C 354
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
+ P G + A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 355 DIAPRG----LSMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AR---EDLAALEKDYEEVGAEGE-EGEEDEE 447
A DL + + Y++ A+ E E EEDE+
Sbjct: 411 AESNMNDLVSEYQQYQDATADDEGEYEEDED 441
>At5g23860 beta tubulin
Length = 449
Score = 391 bits (1004), Expect = e-109
Identities = 184/450 (40%), Positives = 285/450 (62%), Gaps = 13/450 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G WE+ C EHGI G + + + + N +++E G+
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVVCAEHGIDSTGRYQGENDLQL--ERVNVYYNEASCGR 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
VPRA+ +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 FVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + L F+++PSP+VS
Sbjct: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMLTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLTTPSFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRALTVPELTQQMWDAKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ M+RG + K+V+ + N++ K + FV+W P K + PP
Sbjct: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMINVQNKNSSYFVEWIPNNVKSTVCDIPP 358
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
T ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AR---EDLAALEKDYEEVGAEGEEGEEDEE 447
A DL + + Y++ A+ EEG E EE
Sbjct: 411 AESNMNDLVSEYQQYQDATADEEEGYEYEE 440
>At2g29550 tubulin beta-7 chain
Length = 449
Score = 391 bits (1004), Expect = e-109
Identities = 182/449 (40%), Positives = 287/449 (63%), Gaps = 12/449 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G+ WE+ LEHGI G D+ + + + N +++E G+
Sbjct: 1 MREILHIQGGQCGNQIGSKFWEVVNLEHGIDQTGRYVGDSELQL--ERVNVYYNEASCGR 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
+VPRA+ +DLEP +D VR+G Y Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 YVPRAVLMDLEPGTMDSVRSGPYGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+NC LQGF V +++GGGTGSG+G+LL+ ++ +Y + + F+++PSP+VS
Sbjct: 119 VVRKEAENCDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMMTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + P++ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLSTPSFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +++PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYRNLTVPELTQQMWDAKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ M+RG + K+V+ + N++ K + FV+W P K + PP
Sbjct: 299 MMCAADPRHGRYLTASAMFRGKMSTKEVDEQMLNVQNKNSSYFVEWIPNNVKSTVCDIPP 358
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
T ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 359 T--------GLKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AREDLAALEKDYEEV--GAEGEEGEEDEE 447
A ++ L +Y++ EEGE +EE
Sbjct: 411 AESNMNDLVSEYQQYQDATADEEGEYEEE 439
>At5g44340 tubulin beta-4 chain (sp|P24636)
Length = 444
Score = 388 bits (997), Expect = e-108
Identities = 185/452 (40%), Positives = 287/452 (62%), Gaps = 13/452 (2%)
Query: 1 MREVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGK 60
MRE++ I GQ G Q+G WE+ C EHGI G D+ + + + + +F+E GK
Sbjct: 1 MREILHIQGGQCGNQIGAKFWEVICDEHGIDHTGQYVGDSPLQL--ERIDVYFNEASGGK 58
Query: 61 HVPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKEDAANNFARGHYTVGREIVELCLD 120
+VPRA+ +DLEP +D +R+G + Q+F P+ + G+ A NN+A+GHYT G E+++ LD
Sbjct: 59 YVPRAVLMDLEPGTMDSLRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELIDSVLD 118
Query: 121 RTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSPQVSTA 180
RK A+N LQGF V +++GGGTGSG+G+LL+ ++ +Y + + F+++PSP+VS
Sbjct: 119 VVRKEAENSDCLQGFQVCHSLGGGTGSGMGTLLISKIREEYPDRMMMTFSVFPSPKVSDT 178
Query: 181 VVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSLTT 240
VVEPYN+ LS H L+E+ D ++LDNEA+YDIC R+L + PT+ +LN LIS +S +T
Sbjct: 179 VVEPYNATLSVHQLVENADECMVLDNEALYDICFRTLKLANPTFGDLNHLISATMSGVTC 238
Query: 241 SLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKAYHEQISIPEITSAVFEPSN 300
LRF G +N D+ + NL+P+PR+HF + +AP+ S + +S+PE+T +++ N
Sbjct: 239 CLRFPGQLNSDLRKLAVNLIPFPRLHFFMVGFAPLTSRGSQQYSALSVPELTQQMWDAKN 298
Query: 301 MMAKCDPRHGKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFKCGINYQPP 360
MM DPRHG+Y+ ++RG + K+V+ + NI+ K + FV+W P K +
Sbjct: 299 MMCAADPRHGRYLTASAVFRGKLSTKEVDEQMMNIQNKNSSYFVEWIPNNVKSSV----C 354
Query: 361 TVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEGMEEGEFSE 420
+ P G ++ A I N+T++ E+F R+ +F M+ ++AF+HWY GEGM+E EF+E
Sbjct: 355 DIAPKG----LKMASTFIGNSTSIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTE 410
Query: 421 AR---EDLAALEKDYEEVGAEGEEGEEDEEGY 449
A DL A + Y++ A EE EE+EE Y
Sbjct: 411 AESNMNDLVAEYQQYQDATAGEEEYEEEEEEY 442
>At5g05620 tubulin gamma-2 chain (sp|P38558)
Length = 474
Score = 248 bits (633), Expect = 5e-66
Identities = 141/445 (31%), Positives = 252/445 (55%), Gaps = 21/445 (4%)
Query: 2 REVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGKH 61
RE+I++ +GQ G Q+G W+ CLEHGI DG++ + G D + FF + +
Sbjct: 3 REIITLQVGQCGNQIGMEFWKQLCLEHGISKDGILEDFATQG--GDRKDVFFYQADDQHY 60
Query: 62 VPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKED--AANNFARGHYTVGREIVELCL 119
+PRAL +DLEP VI+ ++ GEYR L++ E + A NN+A G++ G+ + E +
Sbjct: 61 IPRALLIDLEPRVINGIQNGEYRNLYNHENIFLSDHGGGAGNNWASGYHQ-GKGVEEEIM 119
Query: 120 DRTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSP-QVS 178
D + AD L+GF++ +++ GGTGSG+GS LLE L+ Y KK ++++P+ + S
Sbjct: 120 DMIDREADGSDSLEGFVLCHSIAGGTGSGMGSYLLETLNDRYSKKLVQTYSVFPNQMETS 179
Query: 179 TAVVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSL 238
VV+PYNS+L+ L + D VV+LDN A+ I L + PT+ N L+S V+S+
Sbjct: 180 DVVVQPYNSLLTLKRLTLNADCVVVLDNTALNRIAVERLHLTNPTFAQTNSLVSTVMSAS 239
Query: 239 TTSLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKA-YHEQISIPEITSAVFE 297
TT+LR+ G +N D+ +L+P PR HF+++ Y P+ +A + ++ ++ + +
Sbjct: 240 TTTLRYPGYMNNDLVGLLASLIPTPRCHFLMTGYTPLTVERQANVIRKTTVLDVMRRLLQ 299
Query: 298 PSNMMAKCDPRH-----GKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFK 352
N+M R+ KY++ + +G+V P V+ ++ I+ ++ V F+DW P +
Sbjct: 300 TKNIMVSSYARNKEASQAKYISILNIIQGEVDPTQVHESLQRIRERKLVNFIDWGPASIQ 359
Query: 353 CGINYQPPTVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEG 412
++ + P V + ++ + M++++T++ +FSR ++D + K+AF+ Y
Sbjct: 360 VALSKKSPYVQTSHRVSGL-----MLASHTSIRHLFSRCLSQYDKLRKKQAFLDNYRKFP 414
Query: 413 M----EEGEFSEAREDLAALEKDYE 433
M + EF E+R+ + +L +Y+
Sbjct: 415 MFADNDLSEFDESRDIIESLVDEYK 439
>At3g61650 tubulin gamma-1 chain
Length = 474
Score = 245 bits (625), Expect = 4e-65
Identities = 139/445 (31%), Positives = 252/445 (56%), Gaps = 21/445 (4%)
Query: 2 REVISIHIGQAGIQVGNSCWELYCLEHGIQPDGMMPSDTSVGISNDAFNTFFSETGSGKH 61
RE+I++ +GQ G Q+G W+ CLEHGI DG++ + G D + FF + +
Sbjct: 3 REIITLQVGQCGNQIGMEFWKQLCLEHGISKDGILEDFATQG--GDRKDVFFYQADDQHY 60
Query: 62 VPRALFVDLEPSVIDEVRTGEYRQLFHPEQLISGKED--AANNFARGHYTVGREIVELCL 119
+PRAL +DLEP VI+ ++ G+YR L++ E + A NN+A G++ G+ + E +
Sbjct: 61 IPRALLIDLEPRVINGIQNGDYRNLYNHENIFVADHGGGAGNNWASGYHQ-GKGVEEEIM 119
Query: 120 DRTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKSKLGFTIYPSP-QVS 178
D + AD L+GF++ +++ GGTGSG+GS LLE L+ Y KK ++++P+ + S
Sbjct: 120 DMIDREADGSDSLEGFVLCHSIAGGTGSGMGSYLLETLNDRYSKKLVQTYSVFPNQMETS 179
Query: 179 TAVVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYTNLNRLISQVISSL 238
VV+PYNS+L+ L + D VV+LDN A+ I L + PT+ N L+S V+S+
Sbjct: 180 DVVVQPYNSLLTLKRLTLNADCVVVLDNTALGRIAVERLHLTNPTFAQTNSLVSTVMSAS 239
Query: 239 TTSLRFDGAINVDVTEFQTNLVPYPRIHFMLSSYAPVISAAKA-YHEQISIPEITSAVFE 297
TT+LR+ G +N D+ +L+P PR HF+++ Y P+ +A + ++ ++ + +
Sbjct: 240 TTTLRYPGYMNNDLVGLLASLIPTPRCHFLMTGYTPLTVERQANVIRKTTVLDVMRRLLQ 299
Query: 298 PSNMMAKCDPRH-----GKYMACCLMYRGDVVPKDVNAAVANIKTKRTVQFVDWCPTGFK 352
N+M R+ KY++ + +G+V P V+ ++ I+ ++ V F++W P +
Sbjct: 300 TKNIMVSSYARNKEASQAKYISILNIIQGEVDPTQVHESLQRIRERKLVNFIEWGPASIQ 359
Query: 353 CGINYQPPTVVPAGDLAKVQRAVCMISNNTAVAEVFSRIDYKFDLMYSKRAFVHWYVGEG 412
++ + P V A ++ + M++++T++ +FS+ ++D + K+AF+ Y
Sbjct: 360 VALSKKSPYVQTAHRVSGL-----MLASHTSIRHLFSKCLSQYDKLRKKQAFLDNYRKFP 414
Query: 413 M----EEGEFSEAREDLAALEKDYE 433
M + EF E+R+ + +L +Y+
Sbjct: 415 MFADNDLSEFDESRDIIESLVDEYK 439
>At4g26110 nucleosome assembly protein I-like protein
Length = 372
Score = 33.5 bits (75), Expect = 0.24
Identities = 14/43 (32%), Positives = 25/43 (57%)
Query: 405 VHWYVGEGMEEGEFSEAREDLAALEKDYEEVGAEGEEGEEDEE 447
V W+ GE ME +F ++ +++D +E E EE ++DE+
Sbjct: 292 VSWFTGEAMEAEDFEIDDDEEDDIDEDEDEEDEEDEEDDDDED 334
>At1g66660 hypothetical protein
Length = 412
Score = 33.1 bits (74), Expect = 0.32
Identities = 17/37 (45%), Positives = 24/37 (63%), Gaps = 3/37 (8%)
Query: 411 EGMEEGEFSEAREDLAALEKDYEEVGAEGEEGEEDEE 447
EG E+GEF E E+ E++ EE+ E +E EE+EE
Sbjct: 110 EGYEDGEFEEDEEE---FEEEEEELEEEEDEEEEEEE 143
>At5g55280 cell division protein FtsZ chloroplast homolog precursor
(sp|Q42545)
Length = 433
Score = 32.7 bits (73), Expect = 0.42
Identities = 29/149 (19%), Positives = 65/149 (43%), Gaps = 8/149 (5%)
Query: 106 GHYTVGREIVELCLDRTRKLADNCTGLQGFLVFNAVGGGTGSGLGSLLLERLSVDYGKKS 165
G+ +G + E D +A+ G + +GGGTGSG ++ + +S D G +
Sbjct: 135 GNPLLGEQAAEESKDA---IANALKGSDLVFITAGMGGGTGSGAAPVVAQ-ISKDAGYLT 190
Query: 166 KLGFTIYPSPQVSTAVVEPYNSVLSTHSLLEHTDVVVLLDNEAIYDICRRSLDIERPTYT 225
+G YP ++ + L ++ D ++++ N+ + DI ++ +
Sbjct: 191 -VGVVTYPFSFEGRK--RSLQALEAIEKLQKNVDTLIVIPNDRLLDIADEQTPLQ-DAFL 246
Query: 226 NLNRLISQVISSLTTSLRFDGAINVDVTE 254
+ ++ Q + ++ + G +NVD +
Sbjct: 247 LADDVLRQGVQGISDIITIPGLVNVDFAD 275
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,251,712
Number of Sequences: 26719
Number of extensions: 455893
Number of successful extensions: 1433
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1339
Number of HSP's gapped (non-prelim): 62
length of query: 449
length of database: 11,318,596
effective HSP length: 103
effective length of query: 346
effective length of database: 8,566,539
effective search space: 2964022494
effective search space used: 2964022494
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0371b.4


