

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0370.9
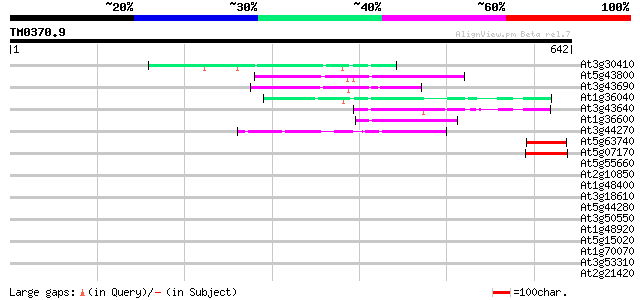
(642 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g30410 hypothetical protein 59 8e-09
At5g43800 putative protein 55 9e-08
At3g43690 unknown protein 53 6e-07
At1g36040 putative envelope-like protein gb|AAD20656 52 1e-06
At3g43640 putative protein 49 9e-06
At1g36600 hypothetical protein 47 3e-05
At3g44270 putative protein 46 6e-05
At5g63740 unknown protein 43 6e-04
At5g07170 putative protein 42 8e-04
At5g55660 putative protein 42 0.001
At2g10850 pseudogene 41 0.002
At1g48400 41 0.002
At3g18610 unknown protein 40 0.003
At5g44280 putative protein 40 0.004
At3g50550 unknown protein 40 0.005
At1g48920 unknown protein 39 0.007
At5g15020 transcriptional regulatory - like protein 39 0.009
At1g70070 Similar to Synechocystis antiviral protein (At1g70070) 39 0.009
At3g53310 unknown protein 38 0.015
At2g21420 Mutator-like transposase 38 0.015
>At3g30410 hypothetical protein
Length = 421
Score = 58.9 bits (141), Expect = 8e-09
Identities = 70/303 (23%), Positives = 119/303 (39%), Gaps = 29/303 (9%)
Query: 160 SRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAGSVRKS 219
S G+ SD V + + E + K + V + ++ + AA S +S
Sbjct: 12 SGAGQPSAATPSDPPPPSSVPMFIPKTEPGIYMSKKTKSKVSAPSQSKRQCTAATSKTRS 71
Query: 220 KG------ETTSKGKGLNVSNKKRKHVSDHESDQDGEPTM--PDISTT---TRKRVKGKR 268
K + + + + + +HV + QDG+ T P + + +R+ R
Sbjct: 72 KAPALRRLSSKKSTRSASAAGQTEEHVFESSDLQDGDVTQIAPPLFLSGYQENRRLLSAR 131
Query: 269 VPMNVADVPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKT 328
P ++ +N F A+ R+K + R R LS + + LLS A L+ T
Sbjct: 132 DPQVSSETSYNNC-FLTAEGLGRYKLIGGRYFNDMRFLSLDGNNTASTLQLLSTAGLLLT 190
Query: 329 VKGIGRCYEKLVREFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEF--------- 379
V I +++V EF N+ + A F VF R SP +IN+
Sbjct: 191 VTEIDSYVQEVVMEFYANLPDGEEGDQLAYF--VFVRGNMYEFSPAIINQIFQLPNPPYP 248
Query: 380 LGRSTTAVAEGESDLNEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATN 439
L S V E ++E+ S + W +L S L+ + +L+KI NW+ T
Sbjct: 249 LNMSKIKVPE---SMDEVDVARSNSKANSWK---MLTSQLLSPEIALLNKIYCHNWMPTV 302
Query: 440 HTS 442
+ S
Sbjct: 303 NRS 305
>At5g43800 putative protein
Length = 515
Score = 55.5 bits (132), Expect = 9e-08
Identities = 57/247 (23%), Positives = 105/247 (42%), Gaps = 13/247 (5%)
Query: 281 VSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLV 340
+ F + +R+K R +R + E ++++ ++ L+ TV I ++++
Sbjct: 170 IQFLTSQGVERYKDFKLRDPYEQRYVPRGPPEMRDMMKVIEDNHLLDTVAEIDPFVKEVI 229
Query: 341 REFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTT---AVAEGES---DL 394
EF N ++ + KVF R K SP +IN+ A AE E
Sbjct: 230 WEFYANFHIFNEETKKS---KVFVRGKMYEFSPQMINKTFRSPAIKWHAKAEFERVTMSK 286
Query: 395 NEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFW 454
+ +AK +S +V+ W L + + L I NW+ + + S +T+ AK+++
Sbjct: 287 HALAKYLSDGKVVTWKD---LRTPVMPTHLAGLYLICCINWIPSKNNSNVTVDRAKMLYL 343
Query: 455 IGTGAMMDFGQHVFEHTFKHVGSYAVKLSIM-FPCLITELILQQHPSILQADEAPSKKGL 513
+ +FG+ V++ K S +I+ FP LI +++ Q L+ E P
Sbjct: 344 LNERIPFNFGKLVYDQVEKAARSPGQIQNILPFPNLIYRVLMFQRVITLENYEKPGPHPK 403
Query: 514 PLNFDYR 520
N D R
Sbjct: 404 LFNPDVR 410
>At3g43690 unknown protein
Length = 539
Score = 52.8 bits (125), Expect = 6e-07
Identities = 48/203 (23%), Positives = 86/203 (41%), Gaps = 13/203 (6%)
Query: 276 VPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRC 335
VP F A A+R+K ++ R ++ L + L + + +TV + +
Sbjct: 235 VPPHTKRFLSAAAAERYKHIAKRDFIFQKTLPLDPEVLTATKYFLEHSGMAQTVVAVEQF 294
Query: 336 YEKLVREFIVNISS-DYDDAGSAEFHKVFFREKCVHLSPVVINEFLGRSTTA------VA 388
++VREF N+ +Y + G V+ R K SP +IN +A V
Sbjct: 295 VPEVVREFYANLPEMEYRECG---LDLVYVRGKMYEFSPALINHMFSIDDSALDPEAPVT 351
Query: 389 EGESDLNEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPL 448
+ +++A +++G +W + L P+ L +L K+ NW T +TS + +
Sbjct: 352 LSTASRDDLALMMTGGTTRRWLR--LQPADHLDT-MKMLHKVCCGNWFPTTNTSTLRVDR 408
Query: 449 AKLIFWIGTGAMMDFGQHVFEHT 471
+LI G + G+ V HT
Sbjct: 409 LRLIDMGTHGKSFNLGKLVVTHT 431
>At1g36040 putative envelope-like protein gb|AAD20656
Length = 444
Score = 51.6 bits (122), Expect = 1e-06
Identities = 72/340 (21%), Positives = 121/340 (35%), Gaps = 79/340 (23%)
Query: 291 RWKFVSHRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREFIVNISSD 350
R+K + +R R L + LL L+ TV I ++V EF N+
Sbjct: 173 RYKVIGNRCFNDMRFLPLDGNNTASTQQLLFNGGLLPTVTEIDSYVHEVVMEFYANLPDG 232
Query: 351 YDDAGSAEFHKVFFREKCVHLSPVVINEFL----------GRSTTAVAEGESDLNEIAKV 400
+ G + VF R S +IN+ G S V E ++E+A
Sbjct: 233 --EEGDNLAYSVFVRRNMYEFSLAIINQMFQLPNPSYPLDGMSEIQVPES---MDEVAIA 287
Query: 401 ISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWIGTGAM 460
+S + W +L S L+ + +L+KI NW T + S + L++ +
Sbjct: 288 LSNGKANSWK---MLTSRLLSPELALLNKICCHNWSPTVNRSVLKPERMTLLYMVAKALP 344
Query: 461 MDFGQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSKKGLPLNFDYR 520
+FG+ +F+ I+++DE + PL F
Sbjct: 345 FNFGKLIFDENLGR--------------------------IVRSDEGDTTSSAPLKFTME 378
Query: 521 LFAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKVRKHNV 580
+ VP + ++P L +TL A++ VR
Sbjct: 379 V---KPVPLLQSDSP----------------------TLDAALETLIASLTTMHVR---- 409
Query: 581 DQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDD 620
LA +VS D+ + D++EEDDD+ L D+
Sbjct: 410 ------LAGGEYSDVSYSVPDDVGDEDVEEEDDDDELDDN 443
>At3g43640 putative protein
Length = 428
Score = 48.9 bits (115), Expect = 9e-06
Identities = 53/229 (23%), Positives = 98/229 (42%), Gaps = 43/229 (18%)
Query: 394 LNEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIF 453
++E+A +S + W +L S L+ + +L+KI NW T + S + L++
Sbjct: 238 MDEVAIALSNGKANSWK---MLTSRLLSPELALLNKICCHNWSPTVNHSVLKPERMTLLY 294
Query: 454 WIGTGAMMDFGQHVFEHTF---KHVGSYAVKLSIMFPCLITELILQQHPSILQADEAPSK 510
+ +FG+ +F+ + V + + L ++ P LI +++ Q I+ +DE +
Sbjct: 295 MVAKALPFNFGKLIFDEIWACSNAVANPSSTLRLVLPNLIDQMLRFQR--IVPSDEGDTT 352
Query: 511 KGLPLNFDYRLFAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATI 570
PL F + P VL + D+P+ L +TL A++
Sbjct: 353 SSAPLKFTMEV-----KPVPVLQS--DSPT------------------LDAALETLIASL 387
Query: 571 QASKVRKHNVDQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSD 619
+VR LA +VS D+ + +++EEDDD+ L D
Sbjct: 388 TTMRVR----------LAGGEYSDVSYSVPDDVGDENVEEEDDDDELDD 426
>At1g36600 hypothetical protein
Length = 403
Score = 47.4 bits (111), Expect = 3e-05
Identities = 32/119 (26%), Positives = 53/119 (43%), Gaps = 5/119 (4%)
Query: 396 EIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNHTSGITIPLAKLIFWI 455
E+A+ ++ +V + K + L KI NW + +I +L++ I
Sbjct: 174 EVARYLTDGRVQNLQN---IAMNKFSENCKSLFKISCRNWSPQTNEGYASIDRGRLVYRI 230
Query: 456 GTGAMMDFGQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHP--SILQADEAPSKKG 512
+DFG+ V++H + K +MFP LI LI QHP IL+ A + +G
Sbjct: 231 AHKMPIDFGEMVYDHVLQLALMSVSKFFLMFPSLIYRLIQTQHPDIQILRRTSASASQG 289
>At3g44270 putative protein
Length = 324
Score = 46.2 bits (108), Expect = 6e-05
Identities = 58/239 (24%), Positives = 102/239 (42%), Gaps = 32/239 (13%)
Query: 261 RKRVKGKRVPMNVADVPLDNVSFHYADNAQRWKFVSHRRIAIERELSEEALEFKEIIDLL 320
+K+ KGK V A + F + +R+ + SH+ I +E LS + + ++ + +
Sbjct: 93 QKKGKGKGV---AASSTFEETRFVSREAEKRYAYFSHKNIIVEEGLSRKTDD--KVREFI 147
Query: 321 SKAELMKTVKGIGRCYEKLVREFIVNISSDYDDAGSAEFHKVFFREKCVHLSPVVINEFL 380
A L++TV + E+LV EF N+ + D+ S +S + +
Sbjct: 148 YDAGLIRTVTELKPFEEELVFEFWANLPTVKVDSTS--------------VSVMQQMDMA 193
Query: 381 GRSTTAVAEGESDLNEIAKVISGKQVMQWPKKGLLPSGKLTAKFTVLSKIGAANWLATNH 440
G VA+ +D GK ++ K+ LL + L + L K+ NW T++
Sbjct: 194 GLLEDEVAQFLTD---------GK--VKLLKRLLLSAFSLPGR--ELFKLCCTNWSPTSN 240
Query: 441 TSGITIPLAKLIFWIGTGAMMDFGQHVFEHTFKHVGSYAVKLSIMFPCLITELILQQHP 499
S AKL++ I +F + F+H + K + FP L+ +L QHP
Sbjct: 241 NSYAQPDRAKLVYMIMHKMPFNFVRLAFDHILQLALKPETKFFLPFPNLVYQLTQIQHP 299
>At5g63740 unknown protein
Length = 226
Score = 42.7 bits (99), Expect = 6e-04
Identities = 20/46 (43%), Positives = 28/46 (60%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTS 637
DD+ D ADD D + D D+EDDDE DDD+ ++E + D + S
Sbjct: 96 DDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDEECDDEYDS 141
Score = 41.6 bits (96), Expect = 0.001
Identities = 19/51 (37%), Positives = 28/51 (54%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
+DE D DD+D + D D+ DDDE D+D+ E+E + D D E +
Sbjct: 87 EDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDEECDD 137
Score = 41.2 bits (95), Expect = 0.002
Identities = 18/50 (36%), Positives = 30/50 (60%)
Query: 593 DEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
D D +DEDA+AD DE++D++ DDD+ +++ + DAD + E
Sbjct: 66 DGDGDEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDE 115
Score = 38.9 bits (89), Expect = 0.009
Identities = 18/51 (35%), Positives = 27/51 (52%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
+DE D DED + D DE+DDD+ DDD+ ++ + D D + E
Sbjct: 71 EDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDE 121
Score = 37.7 bits (86), Expect = 0.020
Identities = 20/58 (34%), Positives = 32/58 (54%), Gaps = 4/58 (6%)
Query: 581 DQLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSE 638
D+ IK+ DD +D D D + D DE++D++A +D+D E+E E D D +
Sbjct: 46 DEYIKVY----DDHNNDGEGDGDGDGDGDEDEDEDADADEDEDEDEDEDDDDDDDDDD 99
Score = 37.7 bits (86), Expect = 0.020
Identities = 19/51 (37%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Query: 592 DDEVSDPADDEDAE-ADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTS 641
DD+ D DD+DA+ AD DE+DDDE +D++ +++ + D + E S
Sbjct: 91 DDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDEECDDEYDS 141
Score = 37.4 bits (85), Expect = 0.026
Identities = 16/54 (29%), Positives = 30/54 (54%)
Query: 589 APADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
A AD++ + D++D + D D++DDD +DDD +++ + D D + E
Sbjct: 77 ADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDE 130
Score = 36.6 bits (83), Expect = 0.044
Identities = 15/54 (27%), Positives = 31/54 (56%)
Query: 589 APADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
A D++ + DD+D + D D++D D+A D+D+ +E+ + D D + ++
Sbjct: 79 ADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDEND 132
Score = 36.2 bits (82), Expect = 0.058
Identities = 17/51 (33%), Positives = 27/51 (52%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
+DE D DD+D + D D++ DD +DD+ E++ E D D + E
Sbjct: 83 EDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDE 133
Score = 35.8 bits (81), Expect = 0.076
Identities = 17/50 (34%), Positives = 25/50 (50%)
Query: 593 DEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
DE D D D + D DE++DD+ DDD+ ++ + D D E E
Sbjct: 70 DEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDE 119
Score = 35.4 bits (80), Expect = 0.099
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
+DE +D +DED + D D++DDD+ DDD+ ++ + D D + +
Sbjct: 73 EDEDADADEDEDEDEDEDDDDDDD-DDDDDDADDADDDEDDDDEDDDEDED 122
Score = 34.7 bits (78), Expect = 0.17
Identities = 15/51 (29%), Positives = 28/51 (54%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
D++ + DD+D + D D+ DD + DDD+ +++ + D D E+ E
Sbjct: 84 DEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDDDDDDENDEE 134
Score = 34.7 bits (78), Expect = 0.17
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query: 593 DEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
D D +DED +AD DE D+DE +DD+ +++ + DAD + +
Sbjct: 64 DGDGDGDEDEDEDADADE-DEDEDEDEDDDDDDDDDDDDDADDADDDEDD 112
Score = 33.9 bits (76), Expect = 0.29
Identities = 14/51 (27%), Positives = 28/51 (54%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
+D +D +DED + D D++DDD+ D D+ +++ + + D E +
Sbjct: 75 EDADADEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDDD 125
Score = 33.1 bits (74), Expect = 0.49
Identities = 15/39 (38%), Positives = 25/39 (63%), Gaps = 3/39 (7%)
Query: 588 AAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEE 626
A ADD+ D +D+D + D D++DDDE +D+ ++E
Sbjct: 103 ADDADDDEDDDDEDDDEDEDDDDDDDDE---NDEECDDE 138
>At5g07170 putative protein
Length = 542
Score = 42.4 bits (98), Expect = 8e-04
Identities = 18/48 (37%), Positives = 30/48 (62%)
Query: 591 ADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSE 638
ADD+ D DDED + D D++DDD+ DDD+ +++ + D++ E
Sbjct: 106 ADDDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDSEVEEE 153
Score = 38.1 bits (87), Expect = 0.015
Identities = 16/37 (43%), Positives = 24/37 (64%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGE 628
DD+ D DD+D + D D++DDD+ D + EEEG+
Sbjct: 120 DDDDDDDDDDDDDDDDDDDDDDDDESKDSEVEEEEGD 156
Score = 37.0 bits (84), Expect = 0.034
Identities = 17/46 (36%), Positives = 26/46 (55%)
Query: 597 DPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
D ADD D + D D++DDD+ DDD+ +++ + D SE E
Sbjct: 108 DDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDSEVEEE 153
Score = 35.4 bits (80), Expect = 0.099
Identities = 16/44 (36%), Positives = 25/44 (56%)
Query: 599 ADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
ADD+ + D DE+DDD+ DDD+ +++ + D D SE
Sbjct: 106 ADDDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDESKDSE 149
Score = 35.0 bits (79), Expect = 0.13
Identities = 20/58 (34%), Positives = 29/58 (49%), Gaps = 10/58 (17%)
Query: 592 DDEVSDPADD----------EDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSES 639
DD SD DD +DA+ D+EDDD+ DDD+ +++ + D D ES
Sbjct: 88 DDSSSDEEDDSESTHCYAADDDADDTDDDEDDDDDDDDDDDDDDDDDDDDDDDDDDES 145
>At5g55660 putative protein
Length = 759
Score = 42.0 bits (97), Expect = 0.001
Identities = 49/202 (24%), Positives = 70/202 (34%), Gaps = 16/202 (7%)
Query: 38 TRSSSGIAAQSSSPGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSP------------ISE 85
T S AA SSS S +K + T KK++ S + + E
Sbjct: 469 TPKKSSPAAGSSSSKRSAKSQKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEE 528
Query: 86 SDTENVADIPDLDIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDA--PPMK 143
+ EN IPD A L + SE + +K S S D + A K
Sbjct: 529 EEEENENGIPDKSEDEAPQLSESEENVESEEESEEETKKKKRGSRTSSDKKESAGKSRSK 588
Query: 144 KSVIKTGSAKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKST 203
K+ + T S+ T R + D D DTS K E +++ A P+KS
Sbjct: 589 KTAVPTKSSPPKKATQKRSAGKRKKSDDDSDTSPKASSKRKKTEKPAKEQA--AAPLKSV 646
Query: 204 IRTPKDSKAAGSVRKSKGETTS 225
+ G K K + S
Sbjct: 647 SKEKPVIGKRGGKGKDKNKEPS 668
Score = 38.1 bits (87), Expect = 0.015
Identities = 42/165 (25%), Positives = 68/165 (40%), Gaps = 7/165 (4%)
Query: 119 QSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKEHVILDSDGDTSDD 178
Q E++T ++ SV DD +K + + E + E+ I D D +
Sbjct: 489 QKKTEEATRTNKKSVAHSDDESEEEKEDDEEEEKEQEVEEEEEEN-ENGIPDKSEDEAPQ 547
Query: 179 VDVTVKNIESRLRKKMKHATPVKSTIRTPKDSK-AAGSVRKSKGETTSKGKGLNVSNKKR 237
+ + +N+ES + + K RT D K +AG R K +K + +KR
Sbjct: 548 LSESEENVESE-EESEEETKKKKRGSRTSSDKKESAGKSRSKKTAVPTKSSPPKKATQKR 606
Query: 238 KHVSDHESDQDGEPTMPDISTTTRKRVKGKRVPMNVADVPLDNVS 282
+SD D + T P S+ KR K ++ A PL +VS
Sbjct: 607 SAGKRKKSDDDSD-TSPKASS---KRKKTEKPAKEQAAAPLKSVS 647
Score = 32.3 bits (72), Expect = 0.84
Identities = 21/86 (24%), Positives = 43/86 (49%), Gaps = 11/86 (12%)
Query: 560 KEISKTLQATIQASKVRKHNVDQLIKMLAAPADDEVSDPADD---EDAEADLDEEDDDEA 616
K +K+ + T +A++ K +V A +DDE + +D E+ E +++EE+++
Sbjct: 483 KRSAKSQKKTEEATRTNKKSV--------AHSDDESEEEKEDDEEEEKEQEVEEEEEENE 534
Query: 617 LSDDDNVEEEGEYPTDADSTSESTSE 642
D E+E ++++ ES E
Sbjct: 535 NGIPDKSEDEAPQLSESEENVESEEE 560
Score = 32.0 bits (71), Expect = 1.1
Identities = 23/98 (23%), Positives = 45/98 (45%), Gaps = 6/98 (6%)
Query: 537 DAPSASGTKGVTSTGKDDILAELKEISKTLQA-TIQASKVRKHNVDQLIKMLAAPADDEV 595
D A + V K++ L E E + T + +V++ N + ++ A+ EV
Sbjct: 176 DVDEAEKVENVDEDDKEEALKEKNEAELAEEEETNKGEEVKEANKEDDVEADTKVAEPEV 235
Query: 596 SDPA-----DDEDAEADLDEEDDDEALSDDDNVEEEGE 628
D ++ED E + ++E +DE +D+ E++ E
Sbjct: 236 EDKKTESKDENEDKEEEKEDEKEDEKEESNDDKEDKKE 273
>At2g10850 pseudogene
Length = 285
Score = 41.2 bits (95), Expect = 0.002
Identities = 42/175 (24%), Positives = 71/175 (40%), Gaps = 19/175 (10%)
Query: 246 DQDGEPTMPDISTTTRKRVKG------KRVPMN---VADVPLDNVSFHYADNAQRWKFVS 296
D + P+ P +++ RKR KR+ D + F + +R+ S
Sbjct: 71 DPEATPSQPSVASRLRKRKSSAADPRIKRMKQGKGVTGSSSFDGIRFVSTEAEKRYAQFS 130
Query: 297 HRRIAIERELSEEALEFKEIIDLLSKAELMKTVKGIGRCYEKLVREFIVNISSDYDDAGS 356
R E ELS + E + + +A L++TV + LV EF N+ + D
Sbjct: 131 QRNFIEEVELSRKTNEAAR--EFIKQAGLIRTVTKFNPFTQNLVFEFWANLPTMKVDTYM 188
Query: 357 AEFHKVFFREKCVHLSPVVINEFLGRSTTAVAEGESDL-----NEIAKVISGKQV 406
KV R + LSP INE G + + D+ ++A+ ++G +V
Sbjct: 189 V---KVLVRNREYELSPGKINEMYGLPSVDARQQRMDIAGLVDEQVAEFLTGGKV 240
>At1g48400
Length = 513
Score = 41.2 bits (95), Expect = 0.002
Identities = 16/38 (42%), Positives = 27/38 (70%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEY 629
DD+ D DD+D + D D++DDD+ DDD+ +++G+Y
Sbjct: 291 DDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDGDY 328
Score = 38.9 bits (89), Expect = 0.009
Identities = 16/44 (36%), Positives = 28/44 (63%)
Query: 591 ADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDAD 634
+DD+ D DD+D + D D++DDD+ DDD+ +++ + D D
Sbjct: 284 SDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDGD 327
Score = 37.4 bits (85), Expect = 0.026
Identities = 16/51 (31%), Positives = 31/51 (60%)
Query: 584 IKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDAD 634
+++ + D + SD DD+D + D D++DDD+ DDD+ +++ + D D
Sbjct: 272 LRLWVSTNDYDYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 322
Score = 34.7 bits (78), Expect = 0.17
Identities = 14/46 (30%), Positives = 27/46 (58%)
Query: 597 DPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
D +DD+D + D D++DDD+ DDD+ +++ + D D + +
Sbjct: 282 DYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDGD 327
Score = 34.7 bits (78), Expect = 0.17
Identities = 15/46 (32%), Positives = 26/46 (55%)
Query: 593 DEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSE 638
D D DD+D + D D++DDD+ DDD+ +++ + D D +
Sbjct: 282 DYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDGD 327
Score = 33.5 bits (75), Expect = 0.37
Identities = 17/45 (37%), Positives = 24/45 (52%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADST 636
DD+ D DD+D + D D++DDD+ D D E + P D T
Sbjct: 298 DDDDDDDDDDDDDDDDDDDDDDDDDDDDGDYYIVEPKAPIFGDVT 342
Score = 28.9 bits (63), Expect = 9.2
Identities = 12/43 (27%), Positives = 23/43 (52%)
Query: 600 DDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
+D D D D++DDD+ DDD+ +++ + D D + +
Sbjct: 279 NDYDYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 321
>At3g18610 unknown protein
Length = 636
Score = 40.4 bits (93), Expect = 0.003
Identities = 47/184 (25%), Positives = 73/184 (39%), Gaps = 14/184 (7%)
Query: 39 RSSSGIAAQSSSPGHSPTPKKMRTKQVVTKKKTLRFSPPQDTSPISESDTENVADIP-DL 97
++ S + SS PTP K + VV K + S ++T + + T V D +
Sbjct: 207 KAESSSSDDGSSSDEEPTPAK-KEPIVVKKDSSDESSSDEETPVVKKKPTTVVKDAKAES 265
Query: 98 DIVHAEDLLDLASARVSEPPVQSNVE---KSTSFSEDSVDVH---DDAPPMKKSVIKTGS 151
E D +P V N + K +S SE+ D D+ PP KK+ K S
Sbjct: 266 SSSEEESSSDDEPTPAKKPTVVKNAKPAAKDSSSSEEDSDEEESDDEKPPTKKA--KVSS 323
Query: 152 AKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSK 211
+ E+SS + + +SD + S D VT K +S + +TP +
Sbjct: 324 KTSKQESSSDESSD----ESDKEESKDEKVTPKKKDSDVEMVDAEQKSNAKQPKTPTNQT 379
Query: 212 AAGS 215
GS
Sbjct: 380 QGGS 383
Score = 38.5 bits (88), Expect = 0.012
Identities = 66/273 (24%), Positives = 97/273 (35%), Gaps = 63/273 (23%)
Query: 47 QSSSPGHSPTPKKMRTKQVVTKKKTLRFSPPQDTS--------------PI-------SE 85
+SSS S + ++ +TK+ +K K S +D S PI S
Sbjct: 63 ESSSSDASDSDEEEKTKETPSKLKDESSSEEEDDSSSDEEIAPAKKRPEPIKKAKVESSS 122
Query: 86 SDTENVADIPDLDIVHAEDLLDLASARVS-----------------EPPVQSNVEKSTSF 128
SD ++ +D + +L+ A S +P V + +S
Sbjct: 123 SDDDSTSDEETAPVKKQPAVLEKAKVESSSSDDDSSSDEETVPVKKQPAVLEKAKIESSS 182
Query: 129 SEDSVDVHDDAPPMKKSVIKTGSAKAPPETSSRKGKEHVILDSDGDTSDDVDVTVKNIES 188
S+D ++ PMKK AKA E+SS DG +SD+ K
Sbjct: 183 SDDDSSSDEETVPMKKQTAVLEKAKA--ESSS---------SDDGSSSDEEPTPAKKEPI 231
Query: 189 RLRK------KMKHATPV--KSTIRTPKDSKAAGSVRK----SKGETTSKGKGLNVSNKK 236
++K TPV K KD+KA S + S E T K V N K
Sbjct: 232 VVKKDSSDESSSDEETPVVKKKPTTVVKDAKAESSSSEEESSSDDEPTPAKKPTVVKNAK 291
Query: 237 --RKHVSDHESDQDGEPTMPDISTTTRKRVKGK 267
K S E D D E + + T + +V K
Sbjct: 292 PAAKDSSSSEEDSDEEESDDEKPPTKKAKVSSK 324
>At5g44280 putative protein
Length = 486
Score = 40.0 bits (92), Expect = 0.004
Identities = 31/121 (25%), Positives = 58/121 (47%), Gaps = 16/121 (13%)
Query: 522 FAGTHVPNIVLNAPKDAPSASGTKGVTSTGKDDILAELKEISKTLQATIQASKVRKHNVD 581
F+ +P++ + P+D + T+ +L+E +T + +V KH+
Sbjct: 8 FSSAEIPDVA-DQPRDRFNPEATQ------------DLQEKDETKEEKEGDEEV-KHDEA 53
Query: 582 QLIKMLAAPADDEVSDPADD--EDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSES 639
+ + + P D E D DD ED E +++ E+D+EA +++ EEE E D+ S S
Sbjct: 54 EEDQEVVKPNDAEEDDDGDDAEEDEEEEVEAEEDEEAEEEEEEEEEEEEEEEDSKERSPS 113
Query: 640 T 640
+
Sbjct: 114 S 114
Score = 35.8 bits (81), Expect = 0.076
Identities = 22/65 (33%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Query: 581 DQLIKMLAAPADDEVSDPAD-DEDAEADLDEEDDDEALS--DDDNVEEEGEYPTDADSTS 637
D+ +K A D EV P D +ED + D EED++E + +D+ EEE E + +
Sbjct: 45 DEEVKHDEAEEDQEVVKPNDAEEDDDGDDAEEDEEEEVEAEEDEEAEEEEEEEEEEEEEE 104
Query: 638 ESTSE 642
E + E
Sbjct: 105 EDSKE 109
>At3g50550 unknown protein
Length = 95
Score = 39.7 bits (91), Expect = 0.005
Identities = 22/51 (43%), Positives = 31/51 (60%), Gaps = 2/51 (3%)
Query: 593 DEVSDPADDEDAEA-DLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
DE D DD D E+ D D+E++D LS DD+ E +Y TD++S S+ E
Sbjct: 18 DEDDDDDDDTDGESSDEDDEEEDRNLSGDDSESSEDDY-TDSNSDSDDDDE 67
Score = 31.2 bits (69), Expect = 1.9
Identities = 13/37 (35%), Positives = 23/37 (62%)
Query: 596 SDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTD 632
S+ ++D+ +++ D +DDDE DD+ EEE + D
Sbjct: 48 SESSEDDYTDSNSDSDDDDEEDDDDEEEEEEEDSLVD 84
Score = 30.4 bits (67), Expect = 3.2
Identities = 15/40 (37%), Positives = 23/40 (57%)
Query: 599 ADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSE 638
+D+E E D D++D D SD+D+ EE+ D +SE
Sbjct: 13 SDEEYDEDDDDDDDTDGESSDEDDEEEDRNLSGDDSESSE 52
Score = 29.6 bits (65), Expect = 5.4
Identities = 14/39 (35%), Positives = 23/39 (58%)
Query: 599 ADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTS 637
+ DE+ + D D++DD + S D++ EEE + DS S
Sbjct: 12 SSDEEYDEDDDDDDDTDGESSDEDDEEEDRNLSGDDSES 50
>At1g48920 unknown protein
Length = 557
Score = 39.3 bits (90), Expect = 0.007
Identities = 54/211 (25%), Positives = 76/211 (35%), Gaps = 43/211 (20%)
Query: 40 SSSGIAAQSSSPGHSPTPKKM--RTKQVVTKKKTLRFSPPQDTSPISESDTENVADIPDL 97
SSS ++ SS P PKK T V KK S +S +S E VA
Sbjct: 87 SSSDESSDDSSSDDEPAPKKAVAATNGTVAKK-----SKDDSSSSDDDSSDEEVA----- 136
Query: 98 DIVHAEDLLDLASARVSEPPVQSNVEKSTSFSEDSVDVHDDAPPMKKSVIKTGSAKAPPE 157
+ +A V++ E S SED D+ P KK K A
Sbjct: 137 -------VTKKPAAAAKNGSVKAKKESS---SEDDSSSEDE--PAKKPAAKIAKPAAKDS 184
Query: 158 TSSRKGKEHVILDSDGDTSDDVDVTVKNIESRLRKKMKHATPVKSTIRTPKDSKAAGSVR 217
+SS DSD D+ D+ T K A P + + DS S
Sbjct: 185 SSSDD-------DSDEDSEDEKPAT------------KKAAPAAAKAASSSDSSDEDSDE 225
Query: 218 KSKGETTSKGKGLNVSNKKRKHVSDHESDQD 248
+S+ E ++ K ++KK ES++D
Sbjct: 226 ESEDEKPAQKKADTKASKKSSSDESSESEED 256
>At5g15020 transcriptional regulatory - like protein
Length = 1343
Score = 38.9 bits (89), Expect = 0.009
Identities = 22/52 (42%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query: 591 ADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSESTSE 642
ADDE D A + +A E DE DD+ VEEEGE+ + D +ES E
Sbjct: 976 ADDEDGDDASEAGEDASGTESIGDECSQDDNGVEEEGEH-DEIDGKAESEGE 1026
>At1g70070 Similar to Synechocystis antiviral protein (At1g70070)
Length = 1171
Score = 38.9 bits (89), Expect = 0.009
Identities = 18/51 (35%), Positives = 30/51 (58%), Gaps = 1/51 (1%)
Query: 589 APADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSES 639
+P+ ++ + DDE+ E + DE+DDDEA + DN+ +E D D E+
Sbjct: 56 SPSQSQLYEEEDDEEEEEE-DEDDDDEAADEYDNISDEIRNSDDDDDDEET 105
Score = 31.6 bits (70), Expect = 1.4
Identities = 17/48 (35%), Positives = 27/48 (55%), Gaps = 2/48 (4%)
Query: 592 DDEVSDPADDEDAEA--DLDEEDDDEALSDDDNVEEEGEYPTDADSTS 637
D+E + +D+D EA + D D+ SDDD+ +EE E+ D + S
Sbjct: 68 DEEEEEEDEDDDDEAADEYDNISDEIRNSDDDDDDEETEFSVDLPTES 115
>At3g53310 unknown protein
Length = 286
Score = 38.1 bits (87), Expect = 0.015
Identities = 20/63 (31%), Positives = 33/63 (51%), Gaps = 5/63 (7%)
Query: 582 QLIKMLAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGE-----YPTDADST 636
Q I + + +D V D D D D D+ED+DE DD + +E+ E YP D ++
Sbjct: 112 QAIPLSDSDSDSVVEDEKDSTDVVEDDDDEDEDEDEDDDGSFDEDEEISQSLYPIDEETA 171
Query: 637 SES 639
+++
Sbjct: 172 TDA 174
>At2g21420 Mutator-like transposase
Length = 468
Score = 38.1 bits (87), Expect = 0.015
Identities = 18/48 (37%), Positives = 30/48 (62%), Gaps = 2/48 (4%)
Query: 587 LAAPADDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDAD 634
L+ DD+ D DD+D + D D++DDD+ DD++ E++G +D D
Sbjct: 386 LSPGCDDDDDDDDDDDDDDDDDDDDDDDD--DDDEDDEDDGYIDSDDD 431
Score = 36.6 bits (83), Expect = 0.044
Identities = 15/49 (30%), Positives = 26/49 (52%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSEST 640
DD+ D DD+D + D D++DDD+ +DD + + D D + +
Sbjct: 394 DDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDDVDGDDNDDGS 442
Score = 33.1 bits (74), Expect = 0.49
Identities = 15/39 (38%), Positives = 24/39 (61%)
Query: 600 DDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSE 638
DD+D + D D++DDD+ DDD+ +E+ E DS +
Sbjct: 393 DDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDD 431
Score = 32.3 bits (72), Expect = 0.84
Identities = 20/47 (42%), Positives = 27/47 (56%), Gaps = 6/47 (12%)
Query: 592 DDEVSDPADDEDAEADLDEEDDDEALSDDDNVEEEGEYPTDADSTSE 638
DD+ D DD+D + D D+EDD SDDD+V+ + D D SE
Sbjct: 403 DDDDDDD-DDDDDDDDEDDEDDGYIDSDDDDVDGD-----DNDDGSE 443
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.128 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,637,132
Number of Sequences: 26719
Number of extensions: 678190
Number of successful extensions: 7022
Number of sequences better than 10.0: 339
Number of HSP's better than 10.0 without gapping: 156
Number of HSP's successfully gapped in prelim test: 191
Number of HSP's that attempted gapping in prelim test: 4338
Number of HSP's gapped (non-prelim): 1433
length of query: 642
length of database: 11,318,596
effective HSP length: 106
effective length of query: 536
effective length of database: 8,486,382
effective search space: 4548700752
effective search space used: 4548700752
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0370.9


