

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0368c.9
(764 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
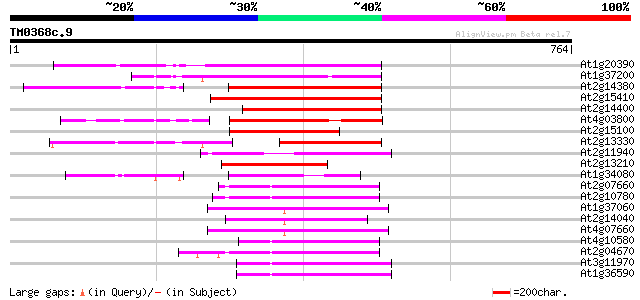
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 312 5e-85
At1g37200 hypothetical protein 254 2e-67
At2g14380 putative retroelement pol polyprotein 252 6e-67
At2g15410 putative retroelement pol polyprotein 251 1e-66
At2g14400 putative retroelement pol polyprotein 216 4e-56
At4g03800 189 7e-48
At2g15100 putative retroelement pol polyprotein 164 1e-40
At2g13330 F14O4.9 163 3e-40
At2g11940 putative retroelement gag/pol polyprotein 156 4e-38
At2g13210 pseudogene 154 2e-37
At1g34080 putative protein 139 5e-33
At2g07660 putative retroelement pol polyprotein 127 2e-29
At2g10780 pseudogene 124 2e-28
At1g37060 Athila retroelment ORF 1, putative 122 1e-27
At2g14040 putative retroelement pol polyprotein 120 4e-27
At4g07660 putative athila transposon protein 116 5e-26
At4g10580 putative reverse-transcriptase -like protein 113 3e-25
At2g04670 putative retroelement pol polyprotein 113 5e-25
At3g11970 hypothetical protein 111 2e-24
At1g36590 hypothetical protein 111 2e-24
>At1g20390 hypothetical protein
Length = 1791
Score = 312 bits (799), Expect = 5e-85
Identities = 181/448 (40%), Positives = 255/448 (56%), Gaps = 38/448 (8%)
Query: 60 ITISSADFFGVEQHLYDPIVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDERDLT 119
I+ + D G++ DP+VV L I+ V RVL+D GSS D+I+ D + + +R +
Sbjct: 553 ISFTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIK 612
Query: 120 PYTGSLVGFAGEQVWVRGILDLDTTFCERENIKMLKVRYLVLRAMGSYNIIIGRNTLNRL 179
P + L GF G+ V G + L + V+++V+ YN+I+G ++++
Sbjct: 613 PVSKPLAGFDGDFVMTIGTIKLPIFV----GGLIAWVKFVVIGKPAVYNVILGTPWIHQM 668
Query: 180 CAVISTAHTAVKYLLASGRIG-RISVDQKIARKCYCNAIDRYGRKSALTGHRCHEVNVPE 238
A+ ST H VK+ +G R + K + Y +S L R VN+ E
Sbjct: 669 QAIPSTYHQCVKFPTHNGIFTLRAPKEAKTPSRSY--------EESELC--RTEMVNIDE 718
Query: 239 ENLDSRGEGRVNRPTPIEETKELKFGDKVLKIGTRLTESQEQRLSKLLEENLDLFAWSHK 298
+ PT + + +G ++ S L LL+ N FAWS +
Sbjct: 719 SD-----------PT------------RCVGVGAEISPSIRLELIALLKRNSKTFAWSIE 755
Query: 299 DMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWL 358
DM GIDP H L +DP K V Q RR++G E+ + + +EV KLL A I E+KYP WL
Sbjct: 756 DMKGIDPAITAHELNVDPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWL 815
Query: 359 ANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQ 418
AN V+VKK NGK +CVDYTDLNKACPKDSYPLP ID+LV+ S N LLS MDA+SGY+Q
Sbjct: 816 ANPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQ 875
Query: 419 IKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMI 478
I MH D++KT+F+T R Y Y+ M FGLKN GATYQR ++++ ADQ+GR +EVY+ DM+
Sbjct: 876 ILMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDML 935
Query: 479 VKSVLEANHHEELLEAFGRLRKHNMRLN 506
VKS+ +H E L + F L + M+LN
Sbjct: 936 VKSLKPEDHVEHLSKCFDVLNTYGMKLN 963
>At1g37200 hypothetical protein
Length = 1564
Score = 254 bits (648), Expect = 2e-67
Identities = 147/348 (42%), Positives = 208/348 (59%), Gaps = 20/348 (5%)
Query: 167 YNIIIGRNTLNRLCAVISTAHTAVKYLLASGRIGRISVDQKIARKCYCNAIDRYGRKSAL 226
+N I+GR L+ + AV ST H +K+ G I I Q+ +R+CY + + +K+ L
Sbjct: 532 FNAILGRPWLHAMKAVPSTYHQCIKFPSEKG-IAVIYGSQRSSRRCYMGSYELI-KKAEL 589
Query: 227 TGHRCHEVNVPEENLDSRGEGRVNRPTPIEETKE------LKFGD--KVLKIGTRLTESQ 278
V + E+ L R + P+ K + D + + +G L +
Sbjct: 590 V------VLMIEDKLAEMKTVRSSDPSQCGPQKSSITQVYIDESDPKQCVGVGQDLDPAI 643
Query: 279 EQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQ 338
+ L+EN D FAWS ++ GI H L +DP + + Q RR++G E+ K +Q
Sbjct: 644 REDFITFLKENKDSFAWSSANLQGISLEVTSHELNVDPTYRPIKQKRRKLGPERAKAVQD 703
Query: 339 EVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLV 398
EV++LL I E+KYP WLAN V+VK NGK +C+D+ DLNKACPKDS+PLP ID+LV
Sbjct: 704 EVDRLLKIGSIREVKYPDWLANPVVVKNKNGKWRVCIDFMDLNKACPKDSFPLPHIDRLV 763
Query: 399 DGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLM 458
+ +ELLS MDA+SGY+QI M D++KTAF+T Y+ MPFGLKNTGATYQRL+
Sbjct: 764 KATAGHELLSFMDAFSGYNQILMRPDDQEKTAFITD----CYKVMPFGLKNTGATYQRLV 819
Query: 459 DRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
+R+FADQ+G+ ME+Y+ DM+VKS E +H +L E F L K M+LN
Sbjct: 820 NRMFADQLGKTMELYIDDMLVKSAHEKDHLPQLRECFKILNKFEMKLN 867
Score = 32.7 bits (73), Expect = 0.78
Identities = 21/86 (24%), Positives = 43/86 (49%), Gaps = 2/86 (2%)
Query: 18 PKEVGAIAGGFGGGGVTSAARKRYVRAVNAVTEVPFGFLHPD--ITISSADFFGVEQHLY 75
PK + I GG + + K + R ++ T+ + PD IT+ ++ +++
Sbjct: 443 PKRIDFIMGGSQLCNDSINSIKTHQRKADSYTKGKSLMMGPDHKITLWESETTDLDKPHD 502
Query: 76 DPIVVLLRINQFNVQRVLLDQGSSAD 101
D IV+ + + + + R+++D GSS D
Sbjct: 503 DAIVIRIDVGNYKLSRIMIDTGSSVD 528
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 252 bits (643), Expect = 6e-67
Identities = 118/208 (56%), Positives = 153/208 (72%)
Query: 299 DMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWL 358
DM GIDP CH L +DP KLV Q RR++G E+ K + EV+KLL A I E+KYP WL
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 359 ANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQ 418
AN V+VKK N K +C+D+TDLNKACPKDS+PLP ID++V+ + NELLS MDA+SGY+Q
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 419 IKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMI 478
I MH D++KT+F+ R Y Y+ MPFGLKN GA YQRL++++FA Q+G+ MEVY+ DM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 479 VKSVLEANHHEELLEAFGRLRKHNMRLN 506
VKS A+H + L F L K+NM+LN
Sbjct: 656 VKSTRSADHIDHLKACFETLNKYNMKLN 683
Score = 69.3 bits (168), Expect = 8e-12
Identities = 57/219 (26%), Positives = 98/219 (44%), Gaps = 16/219 (7%)
Query: 19 KEVGAIAGGFGGGGVTSAARKRYVRAVNAVT-EVPFGFLHPDITISSADFFGVEQHLYDP 77
+ V I GG + + K Y+++ A P +T +S D FGV+ DP
Sbjct: 288 RRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEMTPLTFTSEDLFGVDLPHNDP 347
Query: 78 IVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDERDLTPYTGSLVGFAGEQVWVRG 137
+V+ L I + V R+L+D GSS ++++ D ++ + +R + P L GF G + G
Sbjct: 348 LVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTNG 407
Query: 138 ILDLDTTFCERENIKMLKVRYLVLRAMGSYNIIIGRNTLNRLCAVISTAHTAVKYLLASG 197
+ L +++V+ YNII+G ++ + A+ S+ H +K + G
Sbjct: 408 TIKLPIYLGGAATWH----KFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG 463
Query: 198 RIGRISVDQKIARKCYCNAIDRYGRKSALTGHRCHEVNV 236
I I +Q +A D G + CHE+NV
Sbjct: 464 -IETIRGNQNLAH-------DMVGIDPEVA---CHELNV 491
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 251 bits (640), Expect = 1e-66
Identities = 119/233 (51%), Positives = 161/233 (69%)
Query: 274 LTESQEQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKE 333
L + L L +N FAWS +DM GID CH L +DP K + Q RR++G ++
Sbjct: 736 LPSELQNELVNFLRQNAATFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRT 795
Query: 334 KVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPS 393
K + +EV KLL A I E++YP WL N V+VKK NGK +C+D+TDLNKACPKDS+PLP
Sbjct: 796 KDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPH 855
Query: 394 IDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGAT 453
ID+LV+ + NELLS MDA+SGY+QI MH +D++KT F+T + Y Y+ MPFGLKN GAT
Sbjct: 856 IDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGAT 915
Query: 454 YQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNMRLN 506
Y RL++++F DQ+ +MEVY+ DM+VKS+ H L + F L ++NM+LN
Sbjct: 916 YPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCFQVLNRYNMKLN 968
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 216 bits (550), Expect = 4e-56
Identities = 100/190 (52%), Positives = 140/190 (73%)
Query: 317 GIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVD 376
GI RR++G E+ K + EV+KLL I E++YP WLAN V+VKK NGK +C+D
Sbjct: 461 GISRADVNRRKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCID 520
Query: 377 YTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARA 436
+TDLNKACPKDS+PLP ID+LV+ + NELLS MDA+SGY+QI M+ D++KT F+T R
Sbjct: 521 FTDLNKACPKDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRG 580
Query: 437 NYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFG 496
Y Y+ MPFGL+N GATY RL++++F++ VG+ MEVY+ DM++KS+ + +H + L E F
Sbjct: 581 IYCYKVMPFGLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFA 640
Query: 497 RLRKHNMRLN 506
L ++ M+LN
Sbjct: 641 ILNQYQMKLN 650
Score = 39.3 bits (90), Expect = 0.008
Identities = 16/43 (37%), Positives = 27/43 (62%)
Query: 76 DPIVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDERDL 118
D +VV L + F V R+L+D GSS D+I+ +++G+ D+
Sbjct: 425 DALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRADV 467
>At4g03800
Length = 637
Score = 189 bits (479), Expect = 7e-48
Identities = 94/208 (45%), Positives = 139/208 (66%), Gaps = 15/208 (7%)
Query: 300 MSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLA 359
M IDP+ ICH L +DP K + Q RR++G E+ K + +++KLL I E++YP W+A
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 360 NVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQI 419
V+VKK NGK +C+D+TDLNKACPKDS+PLP ID+LV+ + NELL+ MDA+ GY+QI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 420 KMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIV 479
M+ D++KT+F+T R GATYQ L++++F + + + MEV + D +V
Sbjct: 441 MMNPEDQEKTSFITDR---------------GATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 480 KSVLEANHHEELLEAFGRLRKHNMRLNL 507
KS+ + +H + L E F L ++ M+LNL
Sbjct: 486 KSLKKEDHVKHLGECFEILNQYQMKLNL 513
Score = 48.1 bits (113), Expect = 2e-05
Identities = 55/205 (26%), Positives = 89/205 (42%), Gaps = 32/205 (15%)
Query: 70 VEQHLYDPIVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDERDLTPYTGSLVGFA 129
+E+ D +++ L + F + R+L+D GSS D+I+ L P LV F
Sbjct: 195 IERPHDDALIITLDVANFKISRILVDTGSSVDLIF-------------LGP-PSPLVAFT 240
Query: 130 GEQVWVRGILDLDTTFCERENIKMLKVRYLVLRAMGSYNIIIGRNTLNRLCAVISTAHTA 189
E G + L + I V ++V +YNII+G + ++ AV ST H
Sbjct: 241 SESAMSLGTIKLPVLAGKMSKI----VDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQW 296
Query: 190 VKYLLASGRIGRISVDQKIAR-KCYCNAIDRYGRKSALTGHRCHEVNVPEENLDSRGEGR 248
+K+ ++G + I D + +R Y ID CHE+NV + +
Sbjct: 297 LKFPTSNG-VETIWGDLEGSRTSAYMPDID--------PSIICHELNVDPRFKPLK---Q 344
Query: 249 VNRPTPIEETKELKFG-DKVLKIGT 272
R +E K + DK+LKIG+
Sbjct: 345 KRRKLGVERAKAVNGKIDKLLKIGS 369
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 164 bits (416), Expect = 1e-40
Identities = 79/150 (52%), Positives = 106/150 (70%)
Query: 300 MSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLA 359
M+GI I H L +DP K V Q RR++G ++ + + EV +LL I E+KYP WLA
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 360 NVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQI 419
N V+VKK NGK +CVD+TDLNKAC KD +PLP ID+LV+ + +E+LS MDA+SGY+QI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 420 KMHSSDKDKTAFMTARANYSYQTMPFGLKN 449
M+ D++KT+F+T Y Y+ MPFGLKN
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKN 561
Score = 29.6 bits (65), Expect = 6.6
Identities = 16/50 (32%), Positives = 25/50 (50%)
Query: 156 VRYLVLRAMGSYNIIIGRNTLNRLCAVISTAHTAVKYLLASGRIGRISVD 205
V + V +YN+I+G L + V ST H VK+ G++ IS +
Sbjct: 369 VEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKFPTPVGKMTGISTE 418
>At2g13330 F14O4.9
Length = 889
Score = 163 bits (413), Expect = 3e-40
Identities = 77/139 (55%), Positives = 101/139 (72%)
Query: 368 NGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKD 427
NGK +C+D+ DLNKACPKDS+PLP ID+LV+ +E LS MDA+ GY+QI M ++
Sbjct: 291 NGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQE 350
Query: 428 KTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANH 487
KTAF+T R Y Y+ MPFGLKN G TYQRL++R+F DQ+G+ +EVY+ DM+VKS E +H
Sbjct: 351 KTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDH 410
Query: 488 HEELLEAFGRLRKHNMRLN 506
+L E F L K M+LN
Sbjct: 411 VPQLRECFKILIKFEMKLN 429
Score = 75.5 bits (184), Expect = 1e-13
Identities = 69/263 (26%), Positives = 118/263 (44%), Gaps = 26/263 (9%)
Query: 55 FLH----PD--ITISSADFFGVEQHLYDPIVVLLRINQFNVQRVLLDQGSSADIIYGDAF 108
FLH PD IT ++ +++ D +V+ + + + + +++D GSS D+++ DAF
Sbjct: 36 FLHKRQKPDDGITFWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAF 95
Query: 109 DQLGLDERDLTPYTGSLVGFAGEQVWVRGILDLDTTFCERENIKMLKVRYLVLRAMGSYN 168
+ G + L L GFAG+ + G + L T ++ L +LV+ +N
Sbjct: 96 KRTGHLDSKLQGRKTPLTGFAGDTTFSIGTIQLPTI---ARGVRQL-TNFLVVDKKAPFN 151
Query: 169 IIIGRNTLNRLCAVISTAHTAVKYLLASGRIGRISVDQKIARKCYCNAIDRYGRKSALTG 228
I+GR L+ + AV ST H +K+ G I + Q+ +RKCY + + + +
Sbjct: 152 AILGRPWLHVMKAVPSTYHQCIKFPSYKG-IAVVYGSQRSSRKCYMGSYEDIKKADPV-- 208
Query: 229 HRCHEVNVPEENLDSRGEGRVNRPTPIEETKEL--------KFGDKVLKIGTRLTESQEQ 280
V + E+ L R P+ K L + + IG L + +
Sbjct: 209 -----VLMIEDELAEMKTVRSLDPSQRGTRKSLITQVCIDESDPKRCVGIGHDLDLTVRE 263
Query: 281 RLSKLLEENLDLFAWSHKDMSGI 303
L L+EN D FAWS ++ GI
Sbjct: 264 DLITFLKENKDSFAWSSANLQGI 286
>At2g11940 putative retroelement gag/pol polyprotein
Length = 1212
Score = 156 bits (395), Expect = 4e-38
Identities = 90/261 (34%), Positives = 139/261 (52%), Gaps = 45/261 (17%)
Query: 260 ELKFGDKVLKIGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIK 319
+L F D + +G ++ + L + LE+N FAW+ ++M+GI I H L +DP K
Sbjct: 346 DLIFLDTLANLG----QNFRRELIQFLEKNKSTFAWTIREMTGISTEVISHELNVDPTFK 401
Query: 320 LVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTD 379
V Q R++ G ++ + + +V +LL A
Sbjct: 402 PVKQKRQKHGPDRAEAVNVKVVRLLKA--------------------------------- 428
Query: 380 LNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYS 439
D +PLP ID+ ++ + E+LS MDA+SGY+QI M+ D++KT+F+T R Y
Sbjct: 429 -------DCFPLPHIDRPIESTTGQEMLSFMDAFSGYNQIMMNPDDQEKTSFITERGTYY 481
Query: 440 YQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLR 499
Y+ MPFGLKN GATYQ L++ +F D +G+ MEVY+ DM+VK+ A+H + + F L
Sbjct: 482 YKVMPFGLKNVGATYQLLVNIMFKDLLGKTMEVYIDDMLVKTAFSASHLDHFQQCFDILN 541
Query: 500 KHNMRLN-LKSSLLVYKEESL 519
+ M+LN LK + V E L
Sbjct: 542 EFGMKLNPLKCTFAVPSREFL 562
Score = 32.7 bits (73), Expect = 0.78
Identities = 18/41 (43%), Positives = 23/41 (55%)
Query: 76 DPIVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDER 116
D V+ L I VQRVL+ GSS D+I+ D LG + R
Sbjct: 321 DAFVITLDIANCKVQRVLIVTGSSVDLIFLDTLANLGQNFR 361
>At2g13210 pseudogene
Length = 152
Score = 154 bits (389), Expect = 2e-37
Identities = 71/145 (48%), Positives = 101/145 (68%)
Query: 289 NLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEF 348
++ F WS +D+ G+ + H L +DP K V Q RR++G E+ + ++ EV KLL
Sbjct: 2 DVSTFVWSPEDLPGVSVEIVAHELNVDPTFKPVKQKRRKLGRERAEAVKAEVEKLLRIGS 61
Query: 349 I*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLS 408
I + KYP WLAN V+VKK NGK +C+D+TDLNKACPKDS+P+ I +LV+ S ++LLS
Sbjct: 62 ITKAKYPEWLANPVVVKKKNGKWRVCIDFTDLNKACPKDSFPMSHIGRLVESTSGDKLLS 121
Query: 409 LMDAYSGYHQIKMHSSDKDKTAFMT 433
MDA++GY+QI M+S D++KT F T
Sbjct: 122 FMDAFAGYNQIMMNSEDQEKTTFYT 146
>At1g34080 putative protein
Length = 811
Score = 139 bits (351), Expect = 5e-33
Identities = 77/179 (43%), Positives = 101/179 (56%), Gaps = 26/179 (14%)
Query: 299 DMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWL 358
+M+GI I H L +DP +K V Q RR++G ++ + + EV +LL A I E+KYP WL
Sbjct: 627 EMTGISTEVISHDLNVDPTLKPVKQKRRKLGPDRAEAVNAEVVRLLKAGLIREVKYPEWL 686
Query: 359 ANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQ 418
AN V+VKK NGK +CVD+TDLNKACPKD +PLP ID+LV+
Sbjct: 687 ANPVVVKKKNGKWRVCVDFTDLNKACPKDFFPLPHIDRLVESR----------------- 729
Query: 419 IKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDM 477
K F N Q K GATYQRL++R+F D +G+ MEVY+ DM
Sbjct: 730 ---------KDFFHYGTRNILLQGHALRTKKWGATYQRLVNRMFKDFLGKTMEVYIDDM 779
Score = 63.2 bits (152), Expect = 5e-10
Identities = 48/172 (27%), Positives = 83/172 (47%), Gaps = 15/172 (8%)
Query: 76 DPIVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDERDLTPYTGSLVGFAGEQVWV 135
D +V+ L + VQR+L+D GSS D+I+ D ++G+ ++D+ LV F E
Sbjct: 475 DALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETSMS 534
Query: 136 RGILDLDTTFCERENIKMLKVRYLVLRAMGSYNIIIGRNTLNRLCAVISTAHTAVKYLLA 195
G + L T + +KM + + V +YNII+G L + AV ST H VK+ L
Sbjct: 535 LGTITLPVT--AQGVVKM--IEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKFPLP 590
Query: 196 S-------GRIGRISVDQKIARKCYCNAIDRYGRKSALTGHR----CHEVNV 236
+ ++G V++ + + + + + G +TG H++NV
Sbjct: 591 TRAAPNPFKKVGNDLVERVVINEEHPDQLVGIGANLEMTGISTEVISHDLNV 642
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 127 bits (319), Expect = 2e-29
Identities = 75/220 (34%), Positives = 120/220 (54%), Gaps = 6/220 (2%)
Query: 285 LLEENLDLFAWSHKDMSGIDPNFIC-HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKL 343
++ E D+FA +SG+ P+ + L+PG +++ RM + +++++ +L
Sbjct: 122 IVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAELKKQLEEL 177
Query: 344 LAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASN 403
LA FI P W A V+ VKK +G +C+DY LNK K+ YPLP ID+L+D
Sbjct: 178 LAKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGG 236
Query: 404 NELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFA 463
+ S +D SGYHQI + +D KTAF T ++ + MPFGL N A + ++M+ VF
Sbjct: 237 AQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFR 296
Query: 464 DQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNM 503
D + + +++ D++V S H E L RLR+H +
Sbjct: 297 DFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHEL 336
>At2g10780 pseudogene
Length = 1611
Score = 124 bits (312), Expect = 2e-28
Identities = 76/228 (33%), Positives = 122/228 (53%), Gaps = 6/228 (2%)
Query: 277 SQEQRLSKLLEENLDLFAWSHKDMSGIDPNFIC-HRLALDPGIKLVTQTRRRMGDEKEKV 335
S E + ++ E D+FA +SG+ P+ + L+PG +++ RM +
Sbjct: 619 SAELKDIPIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAK 674
Query: 336 IQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSID 395
+++++ +LL FI P W A V+ VKK +G +C+DY LNK K+ YPLP ID
Sbjct: 675 LKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRID 733
Query: 396 KLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQ 455
+L+D + S +D SGYHQI + +D KTAF T ++ + MPFGL N A +
Sbjct: 734 ELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFM 793
Query: 456 RLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNM 503
++M+ VF D + + +++ D++V S H E L RLR+H +
Sbjct: 794 KMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLREHEL 841
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 122 bits (305), Expect = 1e-27
Identities = 77/265 (29%), Positives = 126/265 (47%), Gaps = 18/265 (6%)
Query: 270 IGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMG 329
+ LT Q L L + +S D+ GI P HR+ L+ + +RR+
Sbjct: 849 VNDELTADQVNLLITELMKYRKAIGYSLDDIKGISPTLCTHRIHLENESYSSIEPQRRLN 908
Query: 330 DEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*P----------------- 372
++V+++E+ KLL A I I TW++ V V K G
Sbjct: 909 PNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGH 968
Query: 373 -MCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAF 431
MC++Y LN A K+ +PLP ID +++ +N+ +D+YSG+ QI +H +D+ KT F
Sbjct: 969 RMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTF 1028
Query: 432 MTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEEL 491
++Y+ MPFGL N AT+QR M +F+D + +EV++ D V ++ L
Sbjct: 1029 TCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNL 1088
Query: 492 LEAFGRLRKHNMRLNLKSSLLVYKE 516
R + N+ LN + + +E
Sbjct: 1089 CRVLKRCEETNLVLNWEKCHFMVRE 1113
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 120 bits (300), Expect = 4e-27
Identities = 68/211 (32%), Positives = 105/211 (49%), Gaps = 18/211 (8%)
Query: 295 WSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKY 354
+S D++GI P H + L+ + +RR+ V+++E+ KLL A I I
Sbjct: 349 YSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLDAGIIYPISD 408
Query: 355 PTWLANVVMVKKANGK*P------------------MCVDYTDLNKACPKDSYPLPSIDK 396
TW++ V +V K G MC+DY LN A KD++PL ID+
Sbjct: 409 STWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKDNFPLSFIDQ 468
Query: 397 LVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQR 456
+++ SN +D Y G+ QI +H D++KT F ++Y+ MPFGL N AT+Q
Sbjct: 469 MLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGLCNAPATFQH 528
Query: 457 LMDRVFADQVGRNMEVYVGDMIVKSVLEANH 487
M +F+D + MEV++ D +V E H
Sbjct: 529 CMKYIFSDMIEDFMEVFIDDFLVLQRCEDKH 559
>At4g07660 putative athila transposon protein
Length = 724
Score = 116 bits (290), Expect = 5e-26
Identities = 74/265 (27%), Positives = 127/265 (47%), Gaps = 18/265 (6%)
Query: 270 IGTRLTESQEQRLSKLLEENLDLFAWSHKDMSGIDPNFICHRLALDPGIKLVTQTRRRMG 329
I L +++ L L + +S D+ I P+ HR+ L+ + +RR+
Sbjct: 299 INVELNDNEVNLLLSELRKYKRAIGYSLSDIKRISPSLCNHRIHLENESYSSIEPQRRLN 358
Query: 330 DEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PM---------------- 373
++V+++E+ KLL A I I TW+ V V K G +
Sbjct: 359 LNLKEVVKKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGH 418
Query: 374 --CVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAF 431
C+DY LN A KD +PLP +++++G +N+ +D YSG+ QI +H +D++KT F
Sbjct: 419 RVCIDYRKLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTF 478
Query: 432 MTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEEL 491
++Y+ MPFGL N T+QR M +F+D + + +EV++ D V ++ L
Sbjct: 479 TCPYGTFAYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNL 538
Query: 492 LEAFGRLRKHNMRLNLKSSLLVYKE 516
+ + N+ LN + + KE
Sbjct: 539 GRVLTKWEETNLVLNWEKCYFMVKE 563
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 113 bits (283), Expect = 3e-25
Identities = 63/192 (32%), Positives = 102/192 (52%), Gaps = 1/192 (0%)
Query: 312 LALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANGK* 371
+ L+PG +++ RM + +++++ LL FI P W A V+ VKK +G
Sbjct: 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSP-WGAPVLFVKKKDGSF 541
Query: 372 PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAF 431
+C+DY +LN+ K+ YPLP ID+L+D S +D SGYHQI + +D KTAF
Sbjct: 542 RLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAF 601
Query: 432 MTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHEEL 491
T ++ + MPFGL N A + RLM+ VF + + + +++ D++V S L
Sbjct: 602 RTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHL 661
Query: 492 LEAFGRLRKHNM 503
+LR+ +
Sbjct: 662 RRVMEKLREQKL 673
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 113 bits (282), Expect = 5e-25
Identities = 81/284 (28%), Positives = 136/284 (47%), Gaps = 16/284 (5%)
Query: 231 CHEVNVPEENLDSRGEGRVNRPTP-------IEETKELKFGDKVLKIGTRLTESQEQRLS 283
CH V E + R + RPT ++ K ++ G + + + ES Q
Sbjct: 421 CHRGRVSFERPEGRLVYQGVRPTSGSLVISAVQAKKMIEKGCEAYLVTISMPESLGQVAV 480
Query: 284 ---KLLEENLDLFAWSHKDMSGIDPNFIC-HRLALDPGIKLVTQTRRRMGDEKEKVIQQE 339
++++E D+F + + G+ P+ + L+PG +++ RM + ++++
Sbjct: 481 SDIRVIQEFEDVF----QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQ 536
Query: 340 VNKLLAAEFI*EIKYPTWLANVVMVKKANGK*PMCVDYTDLNKACPKDSYPLPSIDKLVD 399
+ LL FI P W A V+ VKK +G +C+DY LN K+ YPLP ID+L+D
Sbjct: 537 LEDLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLD 595
Query: 400 GASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMD 459
S +D SGYHQI + +D KTAF T ++ + MPF L N A + RLM+
Sbjct: 596 QLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMN 655
Query: 460 RVFADQVGRNMEVYVGDMIVKSVLEANHHEELLEAFGRLRKHNM 503
VF + + + +++ D++V S H L +LR+ +
Sbjct: 656 SVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKL 699
>At3g11970 hypothetical protein
Length = 1499
Score = 111 bits (277), Expect = 2e-24
Identities = 71/211 (33%), Positives = 109/211 (51%), Gaps = 2/211 (0%)
Query: 310 HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANG 369
H++ L G V Q R ++ I + V LL + P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 370 K*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKT 429
+CVDY +LN KDS+P+P I+ L+D + S +D +GYHQ++M D KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 430 AFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHE 489
AF T ++ Y MPFGL N AT+Q LM+ +F + + + V+ D++V S H +
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 490 ELLEAFGRLRKHNMRLNL-KSSLLVYKEESL 519
L + F +R + + L K + V K E L
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYL 800
>At1g36590 hypothetical protein
Length = 1499
Score = 111 bits (277), Expect = 2e-24
Identities = 71/211 (33%), Positives = 109/211 (51%), Gaps = 2/211 (0%)
Query: 310 HRLALDPGIKLVTQTRRRMGDEKEKVIQQEVNKLLAAEFI*EIKYPTWLANVVMVKKANG 369
H++ L G V Q R ++ I + V LL + P + + VV+VKK +G
Sbjct: 591 HKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSP-YASPVVLVKKKDG 649
Query: 370 K*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKT 429
+CVDY +LN KDS+P+P I+ L+D + S +D +GYHQ++M D KT
Sbjct: 650 TWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKT 709
Query: 430 AFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANHHE 489
AF T ++ Y MPFGL N AT+Q LM+ +F + + + V+ D++V S H +
Sbjct: 710 AFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSSLEEHRQ 769
Query: 490 ELLEAFGRLRKHNMRLNL-KSSLLVYKEESL 519
L + F +R + + L K + V K E L
Sbjct: 770 HLKQVFEVMRANKLFAKLSKCAFAVPKVEYL 800
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.145 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,733,856
Number of Sequences: 26719
Number of extensions: 583615
Number of successful extensions: 2588
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 2505
Number of HSP's gapped (non-prelim): 70
length of query: 764
length of database: 11,318,596
effective HSP length: 107
effective length of query: 657
effective length of database: 8,459,663
effective search space: 5557998591
effective search space used: 5557998591
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0368c.9


