

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0368b.6
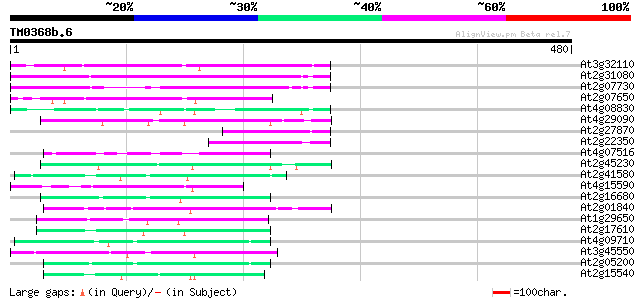
(480 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g32110 non-LTR reverse transcriptase, putative 131 9e-31
At2g31080 putative non-LTR retroelement reverse transcriptase 123 3e-28
At2g07730 putative non-LTR retroelement reverse transcriptase 101 8e-22
At2g07650 putative non-LTR retrolelement reverse transcriptase 90 3e-18
At4g08830 putative protein 89 5e-18
At4g29090 putative protein 81 1e-15
At2g27870 putative non-LTR retroelement reverse transcriptase 77 3e-14
At2g22350 putative non-LTR retroelement reverse transcriptase 74 2e-13
At4g07516 putative protein 67 2e-11
At2g45230 putative non-LTR retroelement reverse transcriptase 67 2e-11
At2g41580 putative non-LTR retroelement reverse transcriptase 62 9e-10
At4g15590 reverse transcriptase like protein 61 2e-09
At2g16680 putative non-LTR retroelement reverse transcriptase 56 4e-08
At2g01840 putative non-LTR retroelement reverse transcriptase 55 7e-08
At1g29650 reverse transcriptase, putative 54 2e-07
At2g17610 putative non-LTR retroelement reverse transcriptase 53 3e-07
At4g09710 RNA-directed DNA polymerase -like protein 52 6e-07
At3g45550 putative protein 52 6e-07
At2g05200 putative non-LTR retroelement reverse transcriptase 52 9e-07
At2g15540 putative non-LTR retroelement reverse transcriptase 51 1e-06
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 131 bits (329), Expect = 9e-31
Identities = 83/285 (29%), Positives = 136/285 (47%), Gaps = 21/285 (7%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIR--------WQIGDGK 52
LW V+R KY G+ H + + + +TW S G+R W IGDG+
Sbjct: 1419 LWAQVVRSKYKVGD------VHDRNWTVAKSNWSSTWRSVGVGLRDVIWREQHWVIGDGR 1472
Query: 53 VVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEI 112
+RF DRWL S +A+ ++ P+ + + W+ + + + ++
Sbjct: 1473 QIRFWTDRWL-SETPIADDSIVPLSLAQMLCTARDLWRDGTGWDMSQIAPFVTDNKRLDL 1531
Query: 113 MESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDPMWCL---LWRWKGAP 169
+ + ++ A WG T+ G + SA+ +LT +D+ M L +W+ +
Sbjct: 1532 LAVIVDSVTGAHDRLAWGMTSDGRFTVKSAFAMLTN--DDSPRQDMSSLYGRVWKVQAPE 1589
Query: 170 CVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLP 229
V++FLW V+N +M N +R HL DSD+C +C ES++H LRDCP + + +P
Sbjct: 1590 RVRVFLWLVVNQAIMTNSERKRRHLCDSDVCQVCRGGIESILHVLRDCPAMSGIWDRIVP 1649
Query: 230 TGGESHFFSMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
+ FF+M L WL N+R GL D W+ FA+ VW W+
Sbjct: 1650 RRLQQSFFTMSLLEWLYSNLRQGLMTEGSD-WSTMFAMAVWWGWK 1693
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 123 bits (308), Expect = 3e-28
Identities = 81/277 (29%), Positives = 124/277 (44%), Gaps = 9/277 (3%)
Query: 1 LWVHVLRHKYGCGN-NVMSIVSHRASESSIWNGVR-NTWESFSCGIRWQIGDGKVVRF*Q 58
LW V+R KY G S + + SS W V E G+ W GDG +RF
Sbjct: 743 LWARVVRKKYKVGGVQDTSWLKPQPRWSSTWRSVAVGLREVVVKGVGWVPGDGCTIRFWL 802
Query: 59 DRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
DRWL L+ E+ IP G +V ++ W +P V + ++ + +
Sbjct: 803 DRWLLQEPLV-ELGTDMIPEGERIKVAADYWLPGSGWNLEILGLYLPETVKRRLLSVVVQ 861
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTENIEDT-GPDPMWCLLWRWKGAPCVKMFLWK 177
W T G + SAY LL ++ D + +W+ V++F+W
Sbjct: 862 VFLGNGDEISWKGTQDGAFTVRSAYSLLQGDVGDRPNMGSFFNRIWKLITPERVRVFIWL 921
Query: 178 VLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFF 237
V N +M NV+RV HL+++ +C +C E+++H LRDCP ++P+ LP FF
Sbjct: 922 VSQNVIMTNVERVRRHLSENAICSVCNGAEETILHVLRDCPAMEPIWRRLLPLRRHHEFF 981
Query: 238 SMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
S L WL NM D ++G W F + +W W+
Sbjct: 982 SQSLLEWLFTNM-DPVKG----IWPTLFGMGIWWAWK 1013
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 101 bits (252), Expect = 8e-22
Identities = 83/277 (29%), Positives = 117/277 (41%), Gaps = 52/277 (18%)
Query: 1 LWVHVLRHKYGCGNNVMSI-VSHRASESSIWNGVR-NTWESFSCGIRWQIGDGKVVRF*Q 58
LW VL KY G S + ++S SS W V E + G+RW GDGK + F
Sbjct: 525 LWARVLSKKYKVGEMQNSNWLKPKSSWSSTWRSVTVGLREVVAKGVRWVPGDGKTISFWS 584
Query: 59 DRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPR 118
DRWL L A VA + I +L L
Sbjct: 585 DRWLLQEQLSA-VAKESISADAL----------------------------------LSD 609
Query: 119 HLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDPMWCL-LWRWKGAPCVKMFLWK 177
LS W T +G + SAYELL E+ + +W+ V++F+W
Sbjct: 610 ELS-------WKGTQNGDFTVRSAYELLKPEAEERPLIGSFLKQIWKLVAPERVRVFIWL 662
Query: 178 VLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFF 237
V + +M NV+RV HL+D C +C ES++H LRDCP + P+ LP ++ FF
Sbjct: 663 VSHMVIMTNVERVRRHLSDIATCSVCNGADESILHVLRDCPAMTPIWQRLLPQRRQNEFF 722
Query: 238 SMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
S WL N+ D +G W F++ +W W+
Sbjct: 723 SQ--FEWLFTNL-DPAKG----DWPTLFSMGIWWAWK 752
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 89.7 bits (221), Expect = 3e-18
Identities = 64/241 (26%), Positives = 102/241 (41%), Gaps = 30/241 (12%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRN----TWESFSCGIR--------WQI 48
LW ++R C V + W VR+ TW S + G+R W I
Sbjct: 405 LWARIMR----CNYRVQDV------RDGAWTKVRSVCSSTWRSVALGMREVVIPGLSWVI 454
Query: 49 GDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIV 108
GDG+ + F D+WL + I LAE+ VQ P G + + W+ A + +
Sbjct: 455 GDGREILFWMDKWLTN-IPLAELLVQEPPAGWKGMRARDLRRNGIGWDMATIAPYISSYN 513
Query: 109 FQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPDP----MWCLLWR 164
++ + ++ A WG + G ++ Y LT D P + +W
Sbjct: 514 RLQLQSFVLDDITGARDRISWGESQDGRFKVSTTYSFLTR---DEAPRQDMKRFFDRVWS 570
Query: 165 WKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVR 224
V++FLW V +M N +R HL+ + +C +C ES+IH LRDCP ++ +
Sbjct: 571 VTAPERVRLFLWLVAQQAIMTNQERHRRHLSTTTICQVCKGAKESIIHVLRDCPAMEGIW 630
Query: 225 L 225
L
Sbjct: 631 L 631
>At4g08830 putative protein
Length = 947
Score = 89.0 bits (219), Expect = 5e-18
Identities = 74/281 (26%), Positives = 113/281 (39%), Gaps = 53/281 (18%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
LW +LR KY G E S G RW +G+G+ + F D
Sbjct: 557 LWARILRSKYRVGLR----------------------EVVSRGSRWVVGNGRDILFWSDN 594
Query: 61 WLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPRHL 120
WL+ L+ ++ IP V + + W+ ++ P I + +E +
Sbjct: 595 WLSHEALINRAVIE-IPNSEKELRVKDLWANGLGWKLD---KIEPYISYHTRLELAAVVV 650
Query: 121 SSAPGLP---VWGRTTSGINSTASAYELLTENIEDTGPD--PMWCLLWRWKGAPCVKMFL 175
S G WG + G+ + SAY LLTE+ D P+ + LWR VK FL
Sbjct: 651 DSVTGARDRLSWGYSADGVFTVKSAYRLLTED-HDPRPNMAAFFDRLWRVVALERVKTFL 709
Query: 176 WKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESH 235
W H+ D+ +C +C E+++H L+DCP + + +
Sbjct: 710 W----------------HIGDTSVCQVCKGGDETILHVLKDCPSIAGIWRRLVQVQRSYD 753
Query: 236 FFSMELQAWLLRN--MRDGLQGWDRDAWTKRFAINVWCLWR 274
FF+ L WL N M++ G+ AW FAI VW W+
Sbjct: 754 FFNGSLFGWLYVNLGMKNAETGY---AWATLFAIVVWWSWK 791
>At4g29090 putative protein
Length = 575
Score = 81.3 bits (199), Expect = 1e-15
Identities = 68/270 (25%), Positives = 115/270 (42%), Gaps = 31/270 (11%)
Query: 27 SSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPP------GS 80
S +W + + E G R +G+G+ + + +WL S A + +Q +PP S
Sbjct: 108 SFVWKSIHASQEILRQGARAVVGNGEDIIIWRHKWLDSKPASAALRMQRVPPQEYASVSS 167
Query: 81 LNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLP---RHLSSAPGLPVWGRTTSGIN 137
+ +V D S W L P + + I E P R L S W T+SG
Sbjct: 168 ILKVSDLIDESGREWRKDVIEMLFPEVERKLIGELRPGGRRILDSY----TWDYTSSGDY 223
Query: 138 STASAYELLTE---------NIEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVK 188
+ S Y +LT+ + + +P++ +W+ + +P ++ FLWK L+N L V
Sbjct: 224 TVKSGYWVLTQIINKRSSPQEVSEPSLNPIYQKIWKSQTSPKIQHFLWKCLSNSLPVAGA 283
Query: 189 RVHNHLADSDLCPLCGTLSESVIHALRDCPMVK---PVRLANLPTGGESHFFSMELQAWL 245
+ HL+ C C + E+V H L C + + +P GGE S+ + +
Sbjct: 284 LAYRHLSKESACIRCPSCKETVNHLLFKCTFARLTWAISSIPIPLGGE-WADSIYVNLYW 342
Query: 246 LRNMRDGLQGWDRDAWTKRFAINVWCLWRM 275
+ N+ +G W K + W LWR+
Sbjct: 343 VFNLGNG-----NPQWEKASQLVPWLLWRL 367
>At2g27870 putative non-LTR retroelement reverse transcriptase
Length = 314
Score = 76.6 bits (187), Expect = 3e-14
Identities = 32/92 (34%), Positives = 53/92 (56%), Gaps = 1/92 (1%)
Query: 183 LMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPTGGESHFFSMELQ 242
LM N +R HL+DSD+C +C +++IH LRDCP ++ + + +P G FF+ L
Sbjct: 5 LMTNAERRRRHLSDSDICQICKGAEKTIIHILRDCPAMEGIWIRLVPAGKRREFFTQSLL 64
Query: 243 AWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
WL N+ D + + W+ FA+++W W+
Sbjct: 65 EWLFANLGDRRKTCE-STWSTLFALSIWWAWK 95
>At2g22350 putative non-LTR retroelement reverse transcriptase
Length = 321
Score = 73.9 bits (180), Expect = 2e-13
Identities = 33/104 (31%), Positives = 57/104 (54%), Gaps = 6/104 (5%)
Query: 171 VKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANLPT 230
V++FLW V+ ++ NV+R HL+D+ +C +C E+++H LRDCP + + +P
Sbjct: 6 VRVFLWLVVQQVIITNVERYRRHLSDTRVCQICQGGEETILHVLRDCPAMAGIWSRLVPR 65
Query: 231 GGESHFFSMELQAWLLRNMRDGLQGWDRDAWTKRFAINVWCLWR 274
FF+ L W+ +N+R +R +W F + VW W+
Sbjct: 66 DQIRQFFTASLLEWIYKNLR------ERGSWPTVFVMAVWWGWK 103
>At4g07516 putative protein
Length = 740
Score = 67.0 bits (162), Expect = 2e-11
Identities = 53/194 (27%), Positives = 83/194 (42%), Gaps = 37/194 (19%)
Query: 30 WNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFL 89
W+G+R+ GI W IGDGK +RF D WL L V Q L V+ +F+
Sbjct: 398 WSGLRDV---VLPGISWVIGDGKEIRFWSDTWLTDRPLSKMVMRQ------LPAVIQQFV 448
Query: 90 GSNGTWETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTEN 149
+ + L + V + + +S WG + + + AY LT
Sbjct: 449 SQH--------IRLQLSSVVLDCILGAKDRVS-------WGGSINRDFTVKLAYMFLTR- 492
Query: 150 IEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSES 209
+W V+ F+W + +M NV+R HL+D+DL +C E+
Sbjct: 493 ------------VWSIVVPERVRAFIWLPVIQVVMTNVERKRRHLSDTDLYSVCKQGEET 540
Query: 210 VIHALRDCPMVKPV 223
+IH LRDCP ++ +
Sbjct: 541 IIHILRDCPAMRGI 554
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 67.0 bits (162), Expect = 2e-11
Identities = 68/271 (25%), Positives = 100/271 (36%), Gaps = 33/271 (12%)
Query: 27 SSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQ------PIPPGS 80
S W + GIR IG+G+ + D W+ + A AV+ S
Sbjct: 905 SFAWKSIYEAQVLIKQGIRAVIGNGETINVWTDPWIGAKPAKAAQAVKRSHLVSQYAANS 964
Query: 81 LNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTA 140
++ V D L W L P+ + I+ P + W + SG S
Sbjct: 965 IHVVKDLLLPDGRDWNWNLVSLLFPDNTQENILALRPGGKETRDRF-TWEYSRSGHYSVK 1023
Query: 141 SAYELLTENIEDTGP---------DPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVH 191
S Y ++TE I DP++ +W+ P + FLW+ +NN L V +
Sbjct: 1024 SGYWVMTEIINQRNNPQEVLQPSLDPIFQQIWKLDVPPKIHHFLWRCVNNCLSVASNLAY 1083
Query: 192 NHLADSDLCPLCGTLSESVIHALRDCPMVK---PVRLANLPTGGESHFFSMELQAW---L 245
HLA C C + E+V H L CP + + P GGE W L
Sbjct: 1084 RHLAREKSCVRCPSHGETVNHLLFKCPFARLTWAISPLPAPPGGE----------WAESL 1133
Query: 246 LRNMRDGLQ-GWDRDAWTKRFAINVWCLWRM 275
RNM L + + A+ W LWR+
Sbjct: 1134 FRNMHHVLSVHKSQPEESDHHALIPWILWRL 1164
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 61.6 bits (148), Expect = 9e-10
Identities = 58/249 (23%), Positives = 99/249 (39%), Gaps = 28/249 (11%)
Query: 5 VLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLAS 64
+L+ +Y ++ +S + S W + E G++ IG+G+ D W+
Sbjct: 613 ILKSRYYMNSDFLS-ATKGTRPSYAWQSILYGRELLVSGLKKIIGNGENTYVWMDNWIFD 671
Query: 65 GILLAEVAVQPIPPGSLNRVVDEFLGSNG-------TWETAGFVELMPNIVFQEIMESLP 117
+P P SL +VD L + W +L P Q I + P
Sbjct: 672 D--------KPRRPESLQIMVDIQLKVSQLIDPFSRNWNLNMLRDLFPWKEIQIICQQRP 723
Query: 118 RHLSSAPGLPVWGRTTSGINSTASAYELLTENI---------EDTGPDPMWCLLWRWKGA 168
++S W T G+ + S Y+L + + E +P++ +W A
Sbjct: 724 --MASRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWNLNSA 781
Query: 169 PCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPVRLANL 228
P +K+FLWKVL + V + + D C +C +E++ H L CP+ + V A
Sbjct: 782 PKIKVFLWKVLKGAVAVEDRLRTRGVLIEDGCSMCPEKNETLNHILFQCPLARQV-WALT 840
Query: 229 PTGGESHFF 237
P +H F
Sbjct: 841 PMQSPNHGF 849
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 60.8 bits (146), Expect = 2e-09
Identities = 54/203 (26%), Positives = 86/203 (41%), Gaps = 21/203 (10%)
Query: 1 LWVHVLRHKYGCGN-NVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QD 59
LW VLR KY + + S + +A+ SS TW S G+R + G
Sbjct: 678 LWARVLRRKYKVTDVHDSSWLVPKATWSS-------TWRSIGVGLREGVAKG-------- 722
Query: 60 RWLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPRH 119
W+ L A + P LN V+EF W+ + +P V + + +
Sbjct: 723 -WILHEPLCTR-ATCLLSPEELNARVEEFWTEGVGWDMVKLGQCLPRSVTDRLHAVVIKG 780
Query: 120 LSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGP--DPMWCLLWRWKGAPCVKMFLWK 177
+ W T+ G + SAY LLT+ E++ P + + +W V++FLW
Sbjct: 781 VLGLRDRISWQGTSDGDFTVGSAYVLLTQE-EESKPCMESFFKRIWGVIAPERVRVFLWL 839
Query: 178 VLNNGLMVNVKRVHNHLADSDLC 200
V +M NV+RV H+ D ++C
Sbjct: 840 VGQQVIMTNVERVRRHIGDIEVC 862
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 56.2 bits (134), Expect = 4e-08
Identities = 48/207 (23%), Positives = 80/207 (38%), Gaps = 14/207 (6%)
Query: 27 SSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVD 86
S W + E + G++ IG+G+ D+WL G + + + +++ V
Sbjct: 857 SYTWRSILYGRELLNGGLKRLIGNGEQTNVWIDKWLFDGHSRRPMNLHSLM--NIHMKVS 914
Query: 87 EFLGS-NGTWETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYE- 144
+ W EL Q IM P L S+ W T +G+ + S YE
Sbjct: 915 HLIDPLTRNWNLKKLTELFHEKDVQLIMHQRP--LISSEDSYCWAGTNNGLYTVKSGYER 972
Query: 145 --------LLTENIEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLAD 196
L E +P++ +W + P +K+F+WK L L V + +
Sbjct: 973 SSRETFKNLFKEADVYPSVNPLFDKVWSLETVPKIKVFMWKALKGALAVEDRLRSRGIRT 1032
Query: 197 SDLCPLCGTLSESVIHALRDCPMVKPV 223
+D C C E++ H L CP + V
Sbjct: 1033 ADGCLFCKEEIETINHLLFQCPFARQV 1059
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 55.5 bits (132), Expect = 7e-08
Identities = 60/256 (23%), Positives = 109/256 (42%), Gaps = 19/256 (7%)
Query: 30 WNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFL 89
WN ++ G+R ++GDG+ + +D WL + L A PI + +V D +
Sbjct: 1252 WNSIQEGKLLLQQGLRVRLGDGQTTKIWEDPWLPT--LPPRPARGPILDEDM-KVADLWR 1308
Query: 90 GSNGTWETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLTE- 148
+ W+ F ++ N Q++ +SL +A W T + + S Y + T
Sbjct: 1309 ENKREWDPVIFEGVL-NPEDQQLAKSLYLSNYAARDSYKWAYTRNTQYTVRSGYWVATHV 1367
Query: 149 NIEDT-------GPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCP 201
N+ + G P+ +WR K P +K F+W+ L+ L + + ++ C
Sbjct: 1368 NLTEEEIINPLEGDVPLKQEIWRLKITPKIKHFIWRCLSGALSTTTQLRNRNIPADPTCQ 1427
Query: 202 LCGTLSESVIHALRDCPMVKPV-RLANLPTGGESHFFSMELQAWLLRNMRDGLQG-WDRD 259
C E++ H + C + V R AN +G F+ L+ N+R LQG +++
Sbjct: 1428 RCCNADETINHIIFTCSYAQVVWRSANF-SGSNRLCFTDNLE----ENIRLILQGKKNQN 1482
Query: 260 AWTKRFAINVWCLWRM 275
+ W +WR+
Sbjct: 1483 LPILNGLMPFWIMWRL 1498
>At1g29650 reverse transcriptase, putative
Length = 1557
Score = 53.9 bits (128), Expect = 2e-07
Identities = 48/210 (22%), Positives = 87/210 (40%), Gaps = 18/210 (8%)
Query: 24 ASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNR 83
A S W V + + + G+R +IG+G + D W+ + +Q SLN
Sbjct: 1176 ARPSYAWRSVLHGRDLLTKGLRKEIGNGNSISVWMDNWIYDNG--PRIPLQKHFSVSLNL 1233
Query: 84 VVDEFLGSNGT-WETAGFVELMPNIVFQEIMESL--PRHLSSAPGLPVWGRTTSGINSTA 140
V + + +G W +L+ ++ FQ ++ + + + VW + SG S
Sbjct: 1234 RVKDLINLDGRCWNR----DLLQDLFFQVDIDLICKKKPVVDMDDFWVWLHSKSGEYSVK 1289
Query: 141 SAY---------ELLTENIEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVH 191
S Y ELL + + + +W P +K+FLW++L+ L V + +
Sbjct: 1290 SGYWLVFQSNKPELLFKACRQPSTNGLKEKIWSTSTFPKIKLFLWRILSAALPVADQILR 1349
Query: 192 NHLADSDLCPLCGTLSESVIHALRDCPMVK 221
+ C +CG ES+ H L C + +
Sbjct: 1350 RGMNVDPCCQICGQEGESINHVLFTCSVAR 1379
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 53.1 bits (126), Expect = 3e-07
Identities = 45/214 (21%), Positives = 82/214 (38%), Gaps = 22/214 (10%)
Query: 24 ASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPP-GSLN 82
+ S W + + + S G+++ G+G ++ +D WL P PP G+ +
Sbjct: 357 SQSSYAWKSILHGTKLISRGLKYIAGNGNNIQLWKDNWLPLN--------PPRPPVGTCD 408
Query: 83 RVVDEFLGSNGTWETAGFVELMPNIVFQEIM---ESLPRHLSSAPGLPVWGRTTSGINST 139
+ + S+ E +L+ ++ Q + ++ ++ A W T G S
Sbjct: 409 SIYSQLKVSDLLIEGRWNEDLLCKLIHQNDIPHIRAIRPSITGANDAITWIYTHDGNYSV 468
Query: 140 ASAYELLTE----------NIEDTGPDPMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKR 189
S Y LL + + + ++ +W+ P +K F W+ +N L
Sbjct: 469 KSGYHLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQNAPPKIKHFWWRSAHNALPTAGNL 528
Query: 190 VHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
L D C CG SE V H L C + K +
Sbjct: 529 KRRRLITDDTCQRCGEASEDVNHLLFQCRVSKEI 562
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 52.4 bits (124), Expect = 6e-07
Identities = 52/224 (23%), Positives = 84/224 (37%), Gaps = 12/224 (5%)
Query: 5 VLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLAS 64
VL KY ++ M + + S W G+ + G+ W IG G + + WL+
Sbjct: 818 VLLGKYCNTSSFMDCSASPSFASHGWRGILAGRDLLRKGLGWSIGQGDSINVWTEAWLSP 877
Query: 65 GILLAEVAVQPIPPGSLNR---VVDEFLGSNGTWETAGFVELMPNIVFQEIMESLPRHLS 121
+ PP N+ V D +W + +P +++ + + +
Sbjct: 878 SSPQTPIG----PPTETNKDLSVHDLICHDVKSWNVEAIRKHLPQ--YEDQIRKITINAL 931
Query: 122 SAPGLPVWGRTTSGINSTASAYELLTENIEDTGP-DPMWCL-LWRWKGAPCVKMFLWKVL 179
VW SG +T + Y L N D W +W+ +P VK FLWK +
Sbjct: 932 PLQDSLVWLPVKSGEYTTKTGYALAKLNSFPASQLDFNWQKNIWKIHTSPKVKHFLWKAM 991
Query: 180 NNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV 223
L V ++ C CG +ES +H + CP K V
Sbjct: 992 KGALPVGEALSRRNIEAEVTCKRCGQ-TESSLHLMLLCPYAKKV 1034
>At3g45550 putative protein
Length = 851
Score = 52.4 bits (124), Expect = 6e-07
Identities = 56/241 (23%), Positives = 100/241 (41%), Gaps = 18/241 (7%)
Query: 1 LWVHVLRHKYGCGNNVMSIVSHRASESSIWNGVRNTWESFSCGIRWQIGDGKVVRF*QDR 60
L+ V++ +Y N+++ + R+ +S W+ + + G R+ IGDGK +R D
Sbjct: 501 LFARVMKARYFKDNSIIDAKT-RSQQSYGWSSLLSGIALLRKGTRYVIGDGKTIRLGIDN 559
Query: 61 WLASGILLAEVAVQPIPPGSLNRVVDEFLGSNGTWETAG---FVELMPNIVFQEIMESLP 117
+ S + + SL+ + + G + W+ A FV+ + + I S
Sbjct: 560 VVDSHPPRPLLTDEQHNGLSLDNLF-QHRGHSRCWDNAKLQTFVDQSDHDYIKRIYLST- 617
Query: 118 RHLSSAPGLPVWGRTTSGINSTASAYELLTENIEDTGPD--------PMWCLLWRWKGAP 169
S +W ++G + S Y L T + +T P + +W P
Sbjct: 618 ---RSKTDRLIWSYNSTGDYTVRSGYWLSTHDPSNTIPTMAKPHGSVDLKTKIWNLPIMP 674
Query: 170 CVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTLSESVIHALRDCPMVKPV-RLANL 228
+K FLW++L+ L + + CP C +ES+ HAL CP RL++
Sbjct: 675 KLKHFLWRILSKALPTTDRLTTRGMRIDPGCPRCRRENESINHALFTCPFATMAWRLSDT 734
Query: 229 P 229
P
Sbjct: 735 P 735
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 51.6 bits (122), Expect = 9e-07
Identities = 47/197 (23%), Positives = 76/197 (37%), Gaps = 8/197 (4%)
Query: 30 WNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFL 89
W + E G+ W I +G+ V D WL+ L V + P + V +
Sbjct: 803 WRSIIAGREILKEGLGWLITNGEKVSIWNDPWLSISKPL--VPIGPALREHQDLRVSALI 860
Query: 90 GSNGT-WETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASAYELLT- 147
N W+ ++PN ++ +++ LP S W SG ++ S Y + +
Sbjct: 861 NQNTLQWDWNKIAVILPN--YENLIKQLPAPSSRGVDKLAWLPVKSGQYTSRSGYGIASV 918
Query: 148 ENIEDTGPDPMW-CLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHLADSDLCPLCGTL 206
+I W LW+ + P +K +WK L V ++ V H++ S C CG
Sbjct: 919 ASIPIPQTQFNWQSNLWKLQTLPKIKHLMWKAAMEALPVGIQLVRRHISPSAACHRCGA- 977
Query: 207 SESVIHALRDCPMVKPV 223
ES H C V
Sbjct: 978 PESTTHLFFHCEFAAQV 994
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 51.2 bits (121), Expect = 1e-06
Identities = 49/204 (24%), Positives = 71/204 (34%), Gaps = 24/204 (11%)
Query: 30 WNGVRNTWESFSCGIRWQIGDGKVVRF*QDRWLASGILLAEVAVQPIPPGSLNRVVDEFL 89
W + G+R IG G + R +D W+ S P P S+ + D L
Sbjct: 788 WRSIMAAKPLLLSGLRRTIGSGMLTRVWEDPWIPS--------FPPRPAKSILNIRDTHL 839
Query: 90 GSNGT-------WETAGFVELMPNIVFQEIMESLPRHLSSAPGLPVWGRTTSGINSTASA 142
N W+ EL+ I+ P + W T SG + S
Sbjct: 840 YVNDLIDPVTKQWKLGRLQELVDPSDIPLILGIRPSRTYKSDDFS-WSFTKSGNYTVKSG 898
Query: 143 YELLTENIEDT------GPD--PMWCLLWRWKGAPCVKMFLWKVLNNGLMVNVKRVHNHL 194
Y + T GP + +W+ K K F W+ L+ L N + H+
Sbjct: 899 YWAARDLSRPTCDLPFQGPSVSALQAQVWKIKTTRKFKHFEWQCLSGCLATNQRLFSRHI 958
Query: 195 ADSDLCPLCGTLSESVIHALRDCP 218
+CP CG ES+ H L CP
Sbjct: 959 GTEKVCPRCGAEEESINHLLFLCP 982
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.334 0.143 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,843,985
Number of Sequences: 26719
Number of extensions: 440345
Number of successful extensions: 1597
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1518
Number of HSP's gapped (non-prelim): 48
length of query: 480
length of database: 11,318,596
effective HSP length: 103
effective length of query: 377
effective length of database: 8,566,539
effective search space: 3229585203
effective search space used: 3229585203
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0368b.6


