

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.7
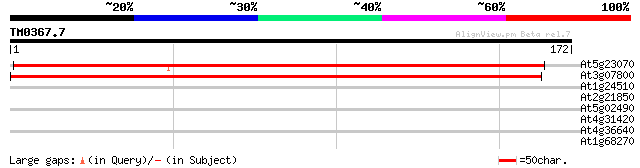
(172 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g23070 unknown protein 249 4e-67
At3g07800 putative thymidine kinase 243 3e-65
At1g24510 T-complex chaperonin protein , epsilon subunit 28 2.5
At2g21850 unknown protein 27 4.2
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 27 5.5
At4g31420 unknown protein with zinc finger 27 5.5
At4g36640 unknown protein 26 9.3
At1g68270 putative amp-binding protein 26 9.3
>At5g23070 unknown protein
Length = 282
Score = 249 bits (637), Expect = 4e-67
Identities = 117/164 (71%), Positives = 142/164 (86%), Gaps = 1/164 (0%)
Query: 2 VAIIKSSKDTRYGLDSIVTHDGTKLPCWALANLSSFKQKFGVDAYE-QLDVIGIDEAQFF 60
+AIIKS+KDTRY +SIVTHDG K PCW+L +LSSFK++FG D YE +LDVIGIDEAQFF
Sbjct: 108 IAIIKSNKDTRYCTESIVTHDGEKYPCWSLPDLSSFKERFGFDDYENRLDVIGIDEAQFF 167
Query: 61 DDLYDFCREAADHDGKTVVVAGLDGNYLRRNFGSVLDIIPLADSVTKLTARCEICGKHAF 120
DLY+FCREAAD +GKTV+VAGLDG+++RR FGSVLD+IP+AD+VTKLT+RCE+CGK A
Sbjct: 168 GDLYEFCREAADKEGKTVIVAGLDGDFMRRRFGSVLDLIPIADTVTKLTSRCEVCGKRAL 227
Query: 121 FTLRKTQETQVELIGGVDVYMPVCRQHYVNGQVAMETTRLVLES 164
FT+RKT+E + ELIGG +VYMPVCR HYV GQ +ET R VL+S
Sbjct: 228 FTMRKTEEKETELIGGAEVYMPVCRSHYVCGQNVLETARAVLDS 271
>At3g07800 putative thymidine kinase
Length = 238
Score = 243 bits (621), Expect = 3e-65
Identities = 112/163 (68%), Positives = 137/163 (83%)
Query: 1 NVAIIKSSKDTRYGLDSIVTHDGTKLPCWALANLSSFKQKFGVDAYEQLDVIGIDEAQFF 60
+VA++KSSKDTRY DS+VTHDG PCWAL +L SF +KFG+DAY +LDVIGIDEAQFF
Sbjct: 61 SVAMLKSSKDTRYAKDSVVTHDGIGFPCWALPDLMSFPEKFGLDAYNKLDVIGIDEAQFF 120
Query: 61 DDLYDFCREAADHDGKTVVVAGLDGNYLRRNFGSVLDIIPLADSVTKLTARCEICGKHAF 120
DLY+FC + AD DGK V+VAGLDG+YLRR+FG+VLDIIP+ADSVTKLTARCE+CG AF
Sbjct: 121 GDLYEFCCKVADDDGKIVIVAGLDGDYLRRSFGAVLDIIPIADSVTKLTARCEVCGHKAF 180
Query: 121 FTLRKTQETQVELIGGVDVYMPVCRQHYVNGQVAMETTRLVLE 163
FTLRK +T+ ELIGG DVYMPVCR+HY+ + ++ ++ VLE
Sbjct: 181 FTLRKNCDTRTELIGGADVYMPVCRKHYITNHIVIKASKKVLE 223
>At1g24510 T-complex chaperonin protein , epsilon subunit
Length = 535
Score = 28.1 bits (61), Expect = 2.5
Identities = 12/43 (27%), Positives = 23/43 (52%), Gaps = 4/43 (9%)
Query: 38 KQKFGVDAYEQLDVIGIDEAQFFDDLYDFCREAADHDGKTVVV 80
K K +D E+ + + E Q+FD++ C++ G T+V+
Sbjct: 259 KHKVDIDTVEKFETLRKQEQQYFDEMVQKCKDV----GATLVI 297
>At2g21850 unknown protein
Length = 772
Score = 27.3 bits (59), Expect = 4.2
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 7/63 (11%)
Query: 96 LDIIPLADSVTKLTARCEIC-----GKHAFFTLRKTQETQVELIGGVDVYMPVCRQHYVN 150
L ++P A + T +T C+IC G F + + V + DVY P+C + VN
Sbjct: 461 LTLLPDAPAGTDVTMNCDICRDDIKGLSTFVYRLINESSLVSCVVCKDVYHPLCLE--VN 518
Query: 151 GQV 153
Q+
Sbjct: 519 RQM 521
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 26.9 bits (58), Expect = 5.5
Identities = 18/55 (32%), Positives = 25/55 (44%), Gaps = 7/55 (12%)
Query: 97 DIIPLADSVTKLTARCEICGKHAFFTLRKTQETQVE---LIGGVDVYMPVCRQHY 148
DI ++ +L CE A TL T +T +E L GG D Y P+ R +
Sbjct: 258 DITGQPRALRRLRTACE----RAKRTLSSTAQTTIEIDSLYGGADFYSPITRARF 308
>At4g31420 unknown protein with zinc finger
Length = 404
Score = 26.9 bits (58), Expect = 5.5
Identities = 15/52 (28%), Positives = 25/52 (47%)
Query: 55 DEAQFFDDLYDFCREAADHDGKTVVVAGLDGNYLRRNFGSVLDIIPLADSVT 106
+E ++ YD+ D GK +VV+G N + GS L I +++ T
Sbjct: 267 EEDAELEEFYDYSSSYVDEAGKQIVVSGETDNTVELVGGSELLITEKSENTT 318
>At4g36640 unknown protein
Length = 294
Score = 26.2 bits (56), Expect = 9.3
Identities = 17/46 (36%), Positives = 24/46 (51%), Gaps = 1/46 (2%)
Query: 56 EAQFFDDLYDFCREAADHDGKTVVVAGLDG-NYLRRNFGSVLDIIP 100
EA D DF R+ + D KT GL+G +Y + N S D++P
Sbjct: 238 EATLEYDHEDFSRQMYEDDLKTAKYWGLEGKHYPKTNGFSPSDVVP 283
>At1g68270 putative amp-binding protein
Length = 535
Score = 26.2 bits (56), Expect = 9.3
Identities = 14/54 (25%), Positives = 28/54 (50%), Gaps = 3/54 (5%)
Query: 39 QKFGVDAYEQLDVIGIDEAQFFDDLYDFCREAADHDGKTVVVAGLDGNYLRRNF 92
Q+ + A + L ++ + E D Y+ +E+ HDGKT+ + GN + + +
Sbjct: 326 QQMELKARQGLGILSVAEV---DVKYNETQESVPHDGKTMGEIVMKGNNIMKGY 376
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,648,824
Number of Sequences: 26719
Number of extensions: 145031
Number of successful extensions: 299
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 294
Number of HSP's gapped (non-prelim): 8
length of query: 172
length of database: 11,318,596
effective HSP length: 92
effective length of query: 80
effective length of database: 8,860,448
effective search space: 708835840
effective search space used: 708835840
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0367.7


