

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.12
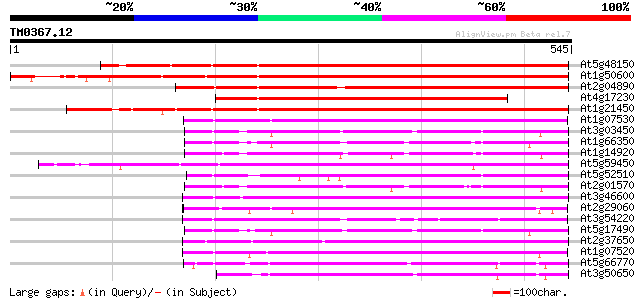
(545 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g48150 SCARECROW gene regulator-like 516 e-146
At1g50600 scarecrow-like 5 (SCL5) 504 e-143
At2g04890 putative SCARECROW gene regulator 460 e-129
At4g17230 scarecrow-like 13 (SCL13) 422 e-118
At1g21450 scarecrow-like 1 (SCL1) 381 e-106
At1g07530 transcription factor scarecrow-like 14, putative 216 2e-56
At3g03450 RGA1-like protein 215 6e-56
At1g66350 gibberellin regulatory protein like 214 1e-55
At1g14920 signal response protein (GAI) 213 2e-55
At5g59450 putative SCARECROW transcriptional regulatory protein ... 211 1e-54
At5g52510 scarecrow-like 8 (SCL8) 207 1e-53
At2g01570 putative RGA1, giberellin repsonse modulation protein 205 5e-53
At3g46600 scarecrow-like protein 204 1e-52
At2g29060 putative SCARECROW gene regulator 200 2e-51
At3g54220 SCARECROW1 197 2e-50
At5g17490 RGA-like protein 195 5e-50
At2g37650 scarecrow-like 9 (SCL9) 195 6e-50
At1g07520 transcription factor scarecrow-like 14, putative 184 1e-46
At5g66770 SCARECROW gene regulator 172 6e-43
At3g50650 scarecrow-like 7 (SCL7) 169 5e-42
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 516 bits (1329), Expect = e-146
Identities = 269/457 (58%), Positives = 338/457 (73%), Gaps = 11/457 (2%)
Query: 89 SQSYHSDQHQSSDNTYGSPSISAHSSDDGSYKLKHKLRELEISLLGPDS-DAVNSCNCCF 147
S Y++ + D+T GS D KHK+RE+E ++GPDS D + C F
Sbjct: 43 SPPYNALSTATYDDTCGS------CVTDELNDFKHKIREIETVMMGPDSLDLLVDCTDSF 96
Query: 148 KGGPSPIANYNWAQIEEMIPKLDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGD 207
S N W E I + DL+ L+ CA+A+S+ D+ A M L +MVSV+G+
Sbjct: 97 DSTASQEIN-GWRSTLEAISRRDLRADLVSCAKAMSENDLMMAHSMMEK-LRQMVSVSGE 154
Query: 208 PIQRLGAYMLEGLRARLESSGSLIYKAL-KCEQPITSKELMSYMDILYQVCPYWKFTYIS 266
PIQRLGAY+LEGL A+L SSGS IYKAL +C +P S EL+SYM ILY+VCPY+KF Y+S
Sbjct: 155 PIQRLGAYLLEGLVAQLASSGSSIYKALNRCPEP-ASTELLSYMHILYEVCPYFKFGYMS 213
Query: 267 SNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARG 326
+N I EAM+ E+R+HIIDFQI QG+QW+ LIQA A+RPGGPP IR+TG+DD S ARG
Sbjct: 214 ANGAIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPGGPPRIRITGIDDMTSAYARG 273
Query: 327 GGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDE 386
GGL IVG RL+ AK VPFEF+S ++S EV+ +NL +RPGEALAVNFAF LHHMPDE
Sbjct: 274 GGLSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLGVRPGEALAVNFAFVLHHMPDE 333
Query: 387 SVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALP 446
SVSTENHRDRLLR+VKSLSPKVVTLVEQESNTNT+ FF RF+ET++YY+AM+ESIDV LP
Sbjct: 334 SVSTENHRDRLLRMVKSLSPKVVTLVEQESNTNTAAFFPRFMETMNYYAAMFESIDVTLP 393
Query: 447 RDDKNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTAS 506
RD K RI+ EQHC+ARD+VN+IACEGA+RVERHEL GKWRSRF MAGF P PLS V ++
Sbjct: 394 RDHKQRINVEQHCLARDVVNIIACEGADRVERHELLGKWRSRFGMAGFTPYPLSPLVNST 453
Query: 507 VRNILNEFNENYRLEHKDVALYLTWKNRAMCTASAWR 543
++++L +++ YRLE +D ALYL W +R + + AW+
Sbjct: 454 IKSLLRNYSDKYRLEERDGALYLGWMHRDLVASCAWK 490
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 504 bits (1297), Expect = e-143
Identities = 282/555 (50%), Positives = 369/555 (65%), Gaps = 41/555 (7%)
Query: 1 MQASQKHSTSSGVHL-YHQP--VQDIDAYTHYQILQSNSSHGNNNSHGAAASFETCKENY 57
M+A+QKH G + YHQP V+ +D S +T ++Y
Sbjct: 72 MEATQKHMIQEGSSMFYHQPSSVKQMDL-----------------------SVQTF-DSY 107
Query: 58 FTLESSSPAATDLIGC-----DSASDASVSSNYRSPSQSYHSD----QHQSSDNTYGSPS 108
TLESSS T C +S+S S SSN SQ+ +++ + S + SP
Sbjct: 108 CTLESSS--GTKSHPCLNNKNNSSSTTSFSSNESPISQANNNNLSRFNNHSPEENNNSPL 165
Query: 109 ISAHSSDDGSYKLKHKLRELEISLLGPDSDAVNSCNCCFKGGPSPIANYNWAQIEEMIPK 168
+ +++ +L L++LE +++ PD D + F G + + + EMI +
Sbjct: 166 SGSSATNTNETELSLMLKDLETAMMEPDVDNSYNNQGGF-GQQHGVVSSAMYRSMEMISR 224
Query: 169 LDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSG 228
DLK VL CA+AV + D++ W+ + L +MVSV+G+P+QRLGAYMLEGL ARL SSG
Sbjct: 225 GDLKGVLYECAKAVENYDLEMT-DWLISQLQQMVSVSGEPVQRLGAYMLEGLVARLASSG 283
Query: 229 SLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQI 288
S IYKAL+C+ P T EL++YM ILY+ CPY+KF Y S+N I EA++NES +HIIDFQI
Sbjct: 284 SSIYKALRCKDP-TGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQI 342
Query: 289 TQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFE 348
+QG QW+ LI+AL +RPGGPP +R+TG+DD S AR GGL +VG+RL A+ CGVPFE
Sbjct: 343 SQGGQWVSLIRALGARPGGPPNVRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFE 402
Query: 349 FHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKV 408
FH AA+ EVE+E L +R GEALAVNF LHHMPDESV+ ENHRDRLLRLVK LSP V
Sbjct: 403 FHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNV 462
Query: 409 VTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMI 468
VTLVEQE+NTNT+PF RFVET+++Y A++ESIDV L RD K RI+ EQHC+AR++VN+I
Sbjct: 463 VTLVEQEANTNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLI 522
Query: 469 ACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVALY 528
ACEG ER ERHE GKWRSRF MAGF P PLSS V A+++ +L ++E Y LE +D ALY
Sbjct: 523 ACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLLESYSEKYTLEERDGALY 582
Query: 529 LTWKNRAMCTASAWR 543
L WKN+ + T+ AWR
Sbjct: 583 LGWKNQPLITSCAWR 597
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 460 bits (1183), Expect = e-129
Identities = 233/382 (60%), Positives = 294/382 (75%), Gaps = 9/382 (2%)
Query: 162 IEEMIPKLDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLR 221
I E I + DLK VL+ CA+AVS+ ++ A M + G MVS++G+PIQRLGAYMLEGL
Sbjct: 41 IVEAISRGDLKLVLVACAKAVSENNLLMARWCMGELRG-MVSISGEPIQRLGAYMLEGLV 99
Query: 222 ARLESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRI 281
ARL +SGS IYK+L+ +P S E +SY+ +L++VCPY+KF Y+S+N I EAM++E RI
Sbjct: 100 ARLAASGSSIYKSLQSREP-ESYEFLSYVYVLHEVCPYFKFGYMSANGAIAEAMKDEERI 158
Query: 282 HIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAK 341
HIIDFQI QG+QW+ LIQA A+RPGG P IR+TGV D G L V KRL AK
Sbjct: 159 HIIDFQIGQGSQWIALIQAFAARPGGAPNIRITGVGD-------GSVLVTVKKRLEKLAK 211
Query: 342 SCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLV 401
VPF F++ + CEVE+ENL +R GEAL VNFA+ LHH+PDESVS ENHRDRLLR+V
Sbjct: 212 KFDVPFRFNAVSRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMV 271
Query: 402 KSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVA 461
KSLSPKVVTLVEQE NTNTSPF RF+ETLSYY+AM+ESIDV LPR+ K RI+ EQHC+A
Sbjct: 272 KSLSPKVVTLVEQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMA 331
Query: 462 RDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYRLE 521
RD+VN+IACEGAER+ERHEL GKW+SRFSMAGF P PLSS ++A++R +L +++ Y +E
Sbjct: 332 RDVVNIIACEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNGYAIE 391
Query: 522 HKDVALYLTWKNRAMCTASAWR 543
+D ALYL W +R + ++ AW+
Sbjct: 392 ERDGALYLGWMDRILVSSCAWK 413
>At4g17230 scarecrow-like 13 (SCL13)
Length = 287
Score = 422 bits (1085), Expect = e-118
Identities = 205/283 (72%), Positives = 237/283 (83%), Gaps = 1/283 (0%)
Query: 201 MVSVAGDPIQRLGAYMLEGLRARLESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYW 260
MVSV+G PIQRLG YM EGLRARLE SGS IYK+LKC +P T +ELMSYM +LY++CPYW
Sbjct: 1 MVSVSGSPIQRLGTYMAEGLRARLEGSGSNIYKSLKCNEP-TGRELMSYMSVLYEICPYW 59
Query: 261 KFTYISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSL 320
KF Y ++NV I EA+ E+R+HIIDFQI QG+Q++ LIQ LA PGGPP +RVTGVDDS
Sbjct: 60 KFAYTTANVEILEAIAGETRVHIIDFQIAQGSQYMFLIQELAKHPGGPPLLRVTGVDDSQ 119
Query: 321 SFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFL 380
S ARGGGL +VG+RL+ A+SCGVPFEFH A MSGC+V+ E+L + PG A+ VNF + L
Sbjct: 120 STYARGGGLSLVGERLATLAQSCGVPFEFHDAIMSGCKVQREHLGLEPGFAVVVNFPYVL 179
Query: 381 HHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYES 440
HHMPDESVS ENHRDRLL L+KSLSPK+VTLVEQESNTNTSPF RFVETL YY+AM+ES
Sbjct: 180 HHMPDESVSVENHRDRLLHLIKSLSPKLVTLVEQESNTNTSPFLSRFVETLDYYTAMFES 239
Query: 441 IDVALPRDDKNRISAEQHCVARDIVNMIACEGAERVERHELFG 483
ID A PRDDK RISAEQHCVARDIVNMIACE +ERVERHE+ G
Sbjct: 240 IDAARPRDDKQRISAEQHCVARDIVNMIACEESERVERHEVLG 282
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 381 bits (978), Expect = e-106
Identities = 207/508 (40%), Positives = 316/508 (61%), Gaps = 29/508 (5%)
Query: 56 NYFTLESSSPAATDLIGCDSASDASVSSNYRSPSQSYHSDQHQSSDNTYGSPSISAHSSD 115
++ +L+S + ++GC + S S ++ Q S SPS+
Sbjct: 95 SFGSLDSFPYQSRPVLGCSMEFQLPLDSTSTSSTRLLGDYQAVSY-----SPSMDVVEEF 149
Query: 116 DGSYKLKHKLRELEISLLGPDSDAVNSCNCCF-------------------KGGPSPIAN 156
D +++ K++ELE +LLG + D + + K S +N
Sbjct: 150 DDE-QMRSKIQELERALLGDEDDKMVGIDNLMEIDSEWSYQNESEQHQDSPKESSSADSN 208
Query: 157 YNWAQIEEMIPKLDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYM 216
+ + +E++ + K +LI CA+A+S+G ++ AL +N L ++VS+ GDP QR+ AYM
Sbjct: 209 SHVSS-KEVVSQATPKQILISCARALSEGKLEEALSMVNE-LRQIVSIQGDPSQRIAAYM 266
Query: 217 LEGLRARLESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQ 276
+EGL AR+ +SG IY+ALKC++P S E ++ M +L++VCP +KF ++++N I EA++
Sbjct: 267 VEGLAARMAASGKFIYRALKCKEP-PSDERLAAMQVLFEVCPCFKFGFLAANGAILEAIK 325
Query: 277 NESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRL 336
E +HIIDF I QG Q++ LI+++A PG P +R+TG+DD S GGL I+G RL
Sbjct: 326 GEEEVHIIDFDINQGNQYMTLIRSIAELPGKRPRLRLTGIDDPESVQRSIGGLRIIGLRL 385
Query: 337 SDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDR 396
A+ GV F+F + V L +PGE L VNFAF LHHMPDESV+T N RD
Sbjct: 386 EQLAEDNGVSFKFKAMPSKTSIVSPSTLGCKPGETLIVNFAFQLHHMPDESVTTVNQRDE 445
Query: 397 LLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAE 456
LL +VKSL+PK+VT+VEQ+ NTNTSPFF RF+E YYSA++ES+D+ LPR+ + R++ E
Sbjct: 446 LLHMVKSLNPKLVTVVEQDVNTNTSPFFPRFIEAYEYYSAVFESLDMTLPRESQERMNVE 505
Query: 457 QHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNIL-NEFN 515
+ C+ARDIVN++ACEG ER+ER+E GKWR+R MAGF P P+S+ VT +++N++ ++
Sbjct: 506 RQCLARDIVNIVACEGEERIERYEAAGKWRARMMMAGFNPKPMSAKVTNNIQNLIKQQYC 565
Query: 516 ENYRLEHKDVALYLTWKNRAMCTASAWR 543
Y+L+ + L+ W+ +++ ASAWR
Sbjct: 566 NKYKLKEEMGELHFCWEEKSLIVASAWR 593
>At1g07530 transcription factor scarecrow-like 14, putative
Length = 769
Score = 216 bits (551), Expect = 2e-56
Identities = 129/375 (34%), Positives = 205/375 (54%), Gaps = 4/375 (1%)
Query: 170 DLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGS 229
DL+ +L+ CAQAVS D +TA + + + S G+ +RL Y L ARL +G+
Sbjct: 392 DLRTLLVLCAQAVSVDDRRTANEMLRQIR-EHSSPLGNGSERLAHYFANSLEARLAGTGT 450
Query: 230 LIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQIT 289
IY AL ++ + L +Y + VCP+ K I +N + N + IHIIDF I+
Sbjct: 451 QIYTALSSKKTSAADMLKAYQTYM-SVCPFKKAAIIFANHSMMRFTANANTIHIIDFGIS 509
Query: 290 QGTQWLLLIQALA-SRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFE 348
G QW LI L+ SRPGG P +R+TG++ G+ G RL+ + + VPFE
Sbjct: 510 YGFQWPALIHRLSLSRPGGSPKLRITGIELPQRGFRPAEGVQETGHRLARYCQRHNVPFE 569
Query: 349 FHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKV 408
+++ A +++E+L +R GE + VN F ++ DE+V + RD +L+L++ ++P V
Sbjct: 570 YNAIAQKWETIQVEDLKLRQGEYVVVNSLFRFRNLLDETVLVNSPRDAVLKLIRKINPNV 629
Query: 409 VTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMI 468
N N F RF E L +YSA+++ D L R+D+ R+ E+ R+IVN++
Sbjct: 630 FIPAILSGNYNAPFFVTRFREALFHYSAVFDMCDSKLAREDEMRLMYEKEFYGREIVNVV 689
Query: 469 ACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVR-NILNEFNENYRLEHKDVAL 527
ACEG ERVER E + +W++R AGF PL + +++ I N +++N+ ++ L
Sbjct: 690 ACEGTERVERPETYKQWQARLIRAGFRQLPLEKELMQNLKLKIENGYDKNFDVDQNGNWL 749
Query: 528 YLTWKNRAMCTASAW 542
WK R + +S W
Sbjct: 750 LQGWKGRIVYASSLW 764
>At3g03450 RGA1-like protein
Length = 547
Score = 215 bits (547), Expect = 6e-56
Identities = 133/381 (34%), Positives = 204/381 (52%), Gaps = 23/381 (6%)
Query: 171 LKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSL 230
L L+ CA+A+ ++ A + V G + + ++ Y + L R
Sbjct: 180 LVHALVACAEAIHQENLNLADALVKRV-GTLAGSQAGAMGKVATYFAQALARR------- 231
Query: 231 IYKALKCEQPITSKELMSYMDIL----YQVCPYWKFTYISSNVVIGEAMQNESRIHIIDF 286
IY+ E + + S+ ++L Y+ CPY KF + ++N I EA+ R+H+ID
Sbjct: 232 IYRDYTAETDVCAAVNPSFEEVLEMHFYESCPYLKFAHFTANQAILEAVTTARRVHVIDL 291
Query: 287 QITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVP 346
+ QG QW L+QALA RPGGPP R+TG+ + L +G +L+ FA++ GV
Sbjct: 292 GLNQGMQWPALMQALALRPGGPPSFRLTGIGPPQT--ENSDSLQQLGWKLAQFAQNMGVE 349
Query: 347 FEFHS-AAMSGCEVELENLVIRP-GEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSL 404
FEF AA S ++E E RP E L VN F LH + S S E +LL VK++
Sbjct: 350 FEFKGLAAESLSDLEPEMFETRPESETLVVNSVFELHRLLARSGSIE----KLLNTVKAI 405
Query: 405 SPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDI 464
P +VT+VEQE+N N F RF E L YYS++++S++ + ++R+ +E + + R I
Sbjct: 406 KPSIVTVVEQEANHNGIVFLDRFNEALHYYSSLFDSLEDSYSLPSQDRVMSEVY-LGRQI 464
Query: 465 VNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEF--NENYRLEH 522
+N++A EG++RVERHE +WR R AGF P L SS +L+ + + YR+E
Sbjct: 465 LNVVAAEGSDRVERHETAAQWRIRMKSAGFDPIHLGSSAFKQASMLLSLYATGDGYRVEE 524
Query: 523 KDVALYLTWKNRAMCTASAWR 543
D L + W+ R + T SAW+
Sbjct: 525 NDGCLMIGWQTRPLITTSAWK 545
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 214 bits (544), Expect = 1e-55
Identities = 135/381 (35%), Positives = 207/381 (53%), Gaps = 34/381 (8%)
Query: 171 LKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSL 230
L L+ CA+AV +++ A + +V G + S ++++ Y EGL R
Sbjct: 152 LVHALLACAEAVQQNNLKLADALVKHV-GLLASSQAGAMRKVATYFAEGLARR------- 203
Query: 231 IYKALKCEQPITSKELMSYMDIL----YQVCPYWKFTYISSNVVIGEAMQNESRIHIIDF 286
IY+ P L S+ D L Y+ CPY KF + ++N I E ++H+ID
Sbjct: 204 IYRIY----PRDDVALSSFSDTLQIHFYESCPYLKFAHFTANQAILEVFATAEKVHVIDL 259
Query: 287 QITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVP 346
+ G QW LIQALA RP GPP R+TG+ SL+ + VG +L A + GV
Sbjct: 260 GLNHGLQWPALIQALALRPNGPPDFRLTGIGYSLT------DIQEVGWKLGQLASTIGVN 313
Query: 347 FEFHSAAMSG-CEVELENLVIRPG-EALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSL 404
FEF S A++ +++ E L IRPG E++AVN F LH + ++ D+ L +KS+
Sbjct: 314 FEFKSIALNNLSDLKPEMLDIRPGLESVAVNSVFELHRL----LAHPGSIDKFLSTIKSI 369
Query: 405 SPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDI 464
P ++T+VEQE+N N + F RF E+L YYS++++S++ +D R+ +E + R I
Sbjct: 370 RPDIMTVVEQEANHNGTVFLDRFTESLHYYSSLFDSLEGPPSQD---RVMSELF-LGRQI 425
Query: 465 VNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSV--TASVRNILNEFNENYRLEH 522
+N++ACEG +RVERHE +WR+RF + GF P + S+ AS+ L + Y +E
Sbjct: 426 LNLVACEGEDRVERHETLNQWRNRFGLGGFKPVSIGSNAYKQASMLLALYAGADGYNVEE 485
Query: 523 KDVALYLTWKNRAMCTASAWR 543
+ L L W+ R + SAWR
Sbjct: 486 NEGCLLLGWQTRPLIATSAWR 506
>At1g14920 signal response protein (GAI)
Length = 533
Score = 213 bits (542), Expect = 2e-55
Identities = 132/380 (34%), Positives = 203/380 (52%), Gaps = 26/380 (6%)
Query: 171 LKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSL 230
L L+ CA+AV ++ A + + VS G ++++ Y E L R
Sbjct: 169 LVHALLACAEAVQKENLTVAEALVKQIGFLAVSQIG-AMRKVATYFAEALARR------- 220
Query: 231 IYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQITQ 290
IY+ + PI + Y+ CPY KF + ++N I EA Q + R+H+IDF ++Q
Sbjct: 221 IYRLSPSQSPIDHSLSDTLQMHFYETCPYLKFAHFTANQAILEAFQGKKRVHVIDFSMSQ 280
Query: 291 GTQWLLLIQALASRPGGPPFIRVTGVDDSL--SFDARGGGLHIVGKRLSDFAKSCGVPFE 348
G QW L+QALA RPGGPP R+TG+ +FD LH VG +L+ A++ V FE
Sbjct: 281 GLQWPALMQALALRPGGPPVFRLTGIGPPAPDNFDY----LHEVGCKLAHLAEAIHVEFE 336
Query: 349 FHS-AAMSGCEVELENLVIRPG--EALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLS 405
+ A + +++ L +RP E++AVN F LH + + D++L +V +
Sbjct: 337 YRGFVANTLADLDASMLELRPSEIESVAVNSVFELHKL----LGRPGAIDKVLGVVNQIK 392
Query: 406 PKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIV 465
P++ T+VEQESN N+ F RF E+L YYS +++S++ DK + +E + + + I
Sbjct: 393 PEIFTVVEQESNHNSPIFLDRFTESLHYYSTLFDSLEGVPSGQDK--VMSEVY-LGKQIC 449
Query: 466 NMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFN--ENYRLEHK 523
N++AC+G +RVERHE +WR+RF AGF + S+ +L FN E YR+E
Sbjct: 450 NVVACDGPDRVERHETLSQWRNRFGSAGFAAAHIGSNAFKQASMLLALFNGGEGYRVEES 509
Query: 524 DVALYLTWKNRAMCTASAWR 543
D L L W R + SAW+
Sbjct: 510 DGCLMLGWHTRPLIATSAWK 529
>At5g59450 putative SCARECROW transcriptional regulatory protein
(F2O15.13)
Length = 610
Score = 211 bits (536), Expect = 1e-54
Identities = 142/520 (27%), Positives = 248/520 (47%), Gaps = 19/520 (3%)
Query: 29 YQILQSNSSHGNNNSHGAAASFETCKENYFTLESSSPAATDLIGCDSASDASVSSNYRSP 88
Y+I+Q S + N+ +++S + + F S++ TD S A VSS
Sbjct: 93 YEIIQQQSPESDQNT--SSSSDQNSGDQDFCFPSTT---TD-------SSALVSSGESQR 140
Query: 89 SQSYHSDQHQSSDNTYGS--PSISAHSSDDGSYKLKHKLRELEISLLGPDSDAVNSCNCC 146
+ +D+ +N + P+I ++ + KL+H L + + + V +
Sbjct: 141 KYRHRNDEEDDLENNRRNKQPAIFVSEMEELAVKLEHVLLVCKTNQEEEEERTVITKQST 200
Query: 147 FKGGPSPIANYNWAQIEEMIPKLDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAG 206
+ N ++ + +DL+ +L +CAQAV+ D + A + + S G
Sbjct: 201 PNRAGRAKGSSNKSKTHKT-NTVDLRSLLTQCAQAVASFDQRRATDKLKEIRAHSSS-NG 258
Query: 207 DPIQRLGAYMLEGLRARLESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYIS 266
D QRL Y E L AR+ + S + +++ + CP + Y +
Sbjct: 259 DGTQRLAFYFAEALEARITGNISPPVSNPFPSSTTSMVDILKAYKLFVHTCPIYVTDYFA 318
Query: 267 SNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARG 326
+N I E +++HI+DF + G QW L++AL+ RPGGPP +RVTG++ +
Sbjct: 319 ANKSIYELAMKATKLHIVDFGVLYGFQWPCLLRALSKRPGGPPMLRVTGIELPQAGFRPS 378
Query: 327 GGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDE 386
+ G+RL F VPFEF+ A + L+ L+I PGE VN L + PDE
Sbjct: 379 DRVEETGRRLKRFCDQFNVPFEFNFIAKKWETITLDELMINPGETTVVNCIHRLQYTPDE 438
Query: 387 SVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALP 446
+VS ++ RD +L+L + ++P + E N+ F RF E L +YS++++ D +
Sbjct: 439 TVSLDSPRDTVLKLFRDINPDLFVFAEINGMYNSPFFMTRFREALFHYSSLFDMFDTTIH 498
Query: 447 RDD--KNRISAEQHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVT 504
+D KNR E+ + RD +++I+CEGAER R E + +WR R AGF P +S +
Sbjct: 499 AEDEYKNRSLLERELLVRDAMSVISCEGAERFARPETYKQWRVRILRAGFKPATISKQIM 558
Query: 505 ASVRNILNE-FNENYRLEHKDVALYLTWKNRAMCTASAWR 543
+ I+ + ++ ++ ++ + + WK R + S W+
Sbjct: 559 KEAKEIVRKRYHRDFVIDSDNNWMLQGWKGRVIYAFSCWK 598
>At5g52510 scarecrow-like 8 (SCL8)
Length = 525
Score = 207 bits (527), Expect = 1e-53
Identities = 130/387 (33%), Positives = 211/387 (53%), Gaps = 27/387 (6%)
Query: 172 KDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSLI 231
+ ++ A A+++G + A + V + ++ + ++L +M+ LR+R+ S + +
Sbjct: 151 RQTVMEIATAIAEGKTEIATEILARV-SQTPNLERNSEEKLVDFMVAALRSRIASPVTEL 209
Query: 232 YKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESR----IHIIDFQ 287
Y KE + +LY++ P +K + ++N+ I +A N H+IDF
Sbjct: 210 Y----------GKEHLISTQLLYELSPCFKLGFEAANLAILDAADNNDGGMMIPHVIDFD 259
Query: 288 ITQGTQWLLLIQALASRPGGP------PFIRVTGVDDS----LSFDARGGGLHIVGKRLS 337
I +G Q++ L++ L++R G P +++T V ++ L D L VG LS
Sbjct: 260 IGEGGQYVNLLRTLSTRRNGKSQSQNSPVVKITAVANNVYGCLVDDGGEERLKAVGDLLS 319
Query: 338 DFAKSCGVPFEFHSA-AMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDR 396
G+ F+ ++ ++ E+L P E LAVN AF L+ +PDESV TEN RD
Sbjct: 320 QLGDRLGISVSFNVVTSLRLGDLNRESLGCDPDETLAVNLAFKLYRVPDESVCTENPRDE 379
Query: 397 LLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAE 456
LLR VK L P+VVTLVEQE N+NT+PF R E+ + Y A+ ES++ +P + +R E
Sbjct: 380 LLRRVKGLKPRVVTLVEQEMNSNTAPFLGRVSESCACYGALLESVESTVPSTNSDRAKVE 439
Query: 457 QHCVARDIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNE 516
+ + R +VN +ACEG +R+ER E+FGKWR R SMAGF PLS + S+++ N +
Sbjct: 440 EG-IGRKLVNAVACEGIDRIERCEVFGKWRMRMSMAGFELMPLSEKIAESMKSRGNRVHP 498
Query: 517 NYRLEHKDVALYLTWKNRAMCTASAWR 543
+ ++ + + W RA+ ASAWR
Sbjct: 499 GFTVKEDNGGVCFGWMGRALTVASAWR 525
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 205 bits (522), Expect = 5e-53
Identities = 127/378 (33%), Positives = 204/378 (53%), Gaps = 22/378 (5%)
Query: 171 LKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSL 230
L L+ CA+A+ ++ A + + VS AG ++++ Y E L R
Sbjct: 221 LVHALMACAEAIQQNNLTLAEALVKQIGCLAVSQAG-AMRKVATYFAEALARR------- 272
Query: 231 IYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQITQ 290
IY+ + I + Y+ CPY KF + ++N I EA + + R+H+IDF + Q
Sbjct: 273 IYRLSPPQNQIDHCLSDTLQMHFYETCPYLKFAHFTANQAILEAFEGKKRVHVIDFSMNQ 332
Query: 291 GTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFEFH 350
G QW L+QALA R GGPP R+TG+ ++ LH VG +L+ A++ V FE+
Sbjct: 333 GLQWPALMQALALREGGPPTFRLTGIGPPAPDNS--DHLHEVGCKLAQLAEAIHVEFEYR 390
Query: 351 S-AAMSGCEVELENLVIRPG--EALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPK 407
A S +++ L +RP EA+AVN F LH + + +++L +VK + P
Sbjct: 391 GFVANSLADLDASMLELRPSDTEAVAVNSVFELHKL----LGRPGGIEKVLGVVKQIKPV 446
Query: 408 VVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNM 467
+ T+VEQESN N F RF E+L YYS +++S++ +P + ++++ +E + + + I N+
Sbjct: 447 IFTVVEQESNHNGPVFLDRFTESLHYYSTLFDSLE-GVP-NSQDKVMSEVY-LGKQICNL 503
Query: 468 IACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFN--ENYRLEHKDV 525
+ACEG +RVERHE +W +RF +G P L S+ +L+ FN + YR+E +
Sbjct: 504 VACEGPDRVERHETLSQWGNRFGSSGLAPAHLGSNAFKQASMLLSVFNSGQGYRVEESNG 563
Query: 526 ALYLTWKNRAMCTASAWR 543
L L W R + T SAW+
Sbjct: 564 CLMLGWHTRPLITTSAWK 581
>At3g46600 scarecrow-like protein
Length = 583
Score = 204 bits (518), Expect = 1e-52
Identities = 118/376 (31%), Positives = 194/376 (51%), Gaps = 4/376 (1%)
Query: 169 LDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSG 228
+D++++L++CAQAV+ D + A + + + S GD QRLG + E L AR+ +G
Sbjct: 207 VDMRNLLMQCAQAVASFDQRRAFEKLKEIR-EHSSRHGDATQRLGYHFAEALEARI--TG 263
Query: 229 SLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQI 288
++ + +++ Q CP Y ++N I E + +HIIDF I
Sbjct: 264 TMTTPISATSSRTSMVDILKAYKGFVQACPTLIMCYFTANRTINELASKATTLHIIDFGI 323
Query: 289 TQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFE 348
G QW LIQAL+ R GPP +RVTG++ S + G+RL F VPFE
Sbjct: 324 LYGFQWPCLIQALSKRDIGPPLLRVTGIELPQSGFRPSERVEETGRRLKRFCDKFNVPFE 383
Query: 349 FHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKV 408
+ A + + L++LVI GE VN L + PDE+VS + RD L+L + ++P +
Sbjct: 384 YSFIAKNWENITLDDLVINSGETTVVNCILRLQYTPDETVSLNSPRDTALKLFRDINPDL 443
Query: 409 VTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMI 468
E N+ F RF E L + S++++ + L DD R E+ + RD +++I
Sbjct: 444 FVFAEINGTYNSPFFLTRFREALFHCSSLFDMYETTLSEDDNCRTLVERELIIRDAMSVI 503
Query: 469 ACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNE-FNENYRLEHKDVAL 527
ACEG+ER R E + +W+ R AGF P LS + + I+ E +++++ +++ + +
Sbjct: 504 ACEGSERFARPETYKQWQVRILRAGFRPAKLSKQIVKDGKEIVKERYHKDFVIDNDNHWM 563
Query: 528 YLTWKNRAMCTASAWR 543
+ WK R + S W+
Sbjct: 564 FQGWKGRVLYAVSCWK 579
>At2g29060 putative SCARECROW gene regulator
Length = 1336
Score = 200 bits (508), Expect = 2e-51
Identities = 126/380 (33%), Positives = 199/380 (52%), Gaps = 12/380 (3%)
Query: 170 DLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGS 229
DL+ +L+ CAQAVS D +TA ++ + S GD +RL Y L ARL G+
Sbjct: 317 DLRTMLVSCAQAVSINDRRTADELLSRIRQHSSSY-GDGTERLAHYFANSLEARLAGIGT 375
Query: 230 LIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGE--AMQNESRIHIIDFQ 287
+Y AL ++ TS L +Y + VCP+ K I +N I + N IHIIDF
Sbjct: 376 QVYTALSSKKTSTSDMLKAYQTYI-SVCPFKKIAIIFANHSIMRLASSANAKTIHIIDFG 434
Query: 288 ITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPF 347
I+ G QW LI LA R G +R+TG++ G+ G+RL+ + + +PF
Sbjct: 435 ISDGFQWPSLIHRLAWRRGSSCKLRITGIELPQRGFRPAEGVIETGRRLAKYCQKFNIPF 494
Query: 348 EFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPK 407
E+++ A ++LE+L ++ GE +AVN F ++ DE+V+ + RD +L+L++ + P
Sbjct: 495 EYNAIAQKWESIKLEDLKLKEGEFVAVNSLFRFRNLLDETVAVHSPRDTVLKLIRKIKPD 554
Query: 408 VVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNM 467
V + N F RF E L +YS++++ D L R+D R+ E+ R+I+N+
Sbjct: 555 VFIPGILSGSYNAPFFVTRFREVLFHYSSLFDMCDTNLTREDPMRVMFEKEFYGREIMNV 614
Query: 468 IACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYRLEHKDVA- 526
+ACEG ERVER E + +W++R AGF PL + ++ ++ Y+ + DV
Sbjct: 615 VACEGTERVERPESYKQWQARAMRAGFRQIPLEKELVQKLKLMV---ESGYKPKEFDVDQ 671
Query: 527 ----LYLTWKNRAMCTASAW 542
L WK R + +S W
Sbjct: 672 DCHWLLQGWKGRIVYGSSIW 691
Score = 192 bits (487), Expect = 5e-49
Identities = 122/382 (31%), Positives = 199/382 (51%), Gaps = 9/382 (2%)
Query: 169 LDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLE-SS 227
+D + +L CAQA+S GD TAL ++ + + S GD QRL L ARL+ S+
Sbjct: 953 VDFRTLLTHCAQAISTGDKTTALEFLLQIR-QQSSPLGDAGQRLAHCFANALEARLQGST 1011
Query: 228 GSLI---YKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHII 284
G +I Y AL T+ + + + P+ Y S +I + ++ +HI+
Sbjct: 1012 GPMIQTYYNALTSSLKDTAADTIRAYRVYLSSSPFVTLMYFFSIWMILDVAKDAPVLHIV 1071
Query: 285 DFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCG 344
DF I G QW + IQ+++ R P +R+TG++ + G+RL+++ K
Sbjct: 1072 DFGILYGFQWPMFIQSISDRKDVPRKLRITGIELPQCGFRPAERIEETGRRLAEYCKRFN 1131
Query: 345 VPFEFHSAAMSGCE-VELENLVIRPGEALAVNFAFFLHHMPDESVSTEN-HRDRLLRLVK 402
VPFE+ + A E + +E+L IRP E LAVN L ++ DE+ S EN RD +L+L++
Sbjct: 1132 VPFEYKAIASQNWETIRIEDLDIRPNEVLAVNAGLRLKNLQDETGSEENCPRDAVLKLIR 1191
Query: 403 SLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVAR 462
+++P V + N F RF E + +YSA+++ D LPRD+K RI E+ R
Sbjct: 1192 NMNPDVFIHAIVNGSFNAPFFISRFKEAVYHYSALFDMFDSTLPRDNKERIRFEREFYGR 1251
Query: 463 DIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNE--FNENYRL 520
+ +N+IACE A+RVER E + +W+ R AGF + + R L + +++++ +
Sbjct: 1252 EAMNVIACEEADRVERPETYRQWQVRMVRAGFKQKTIKPELVELFRGKLKKWRYHKDFVV 1311
Query: 521 EHKDVALYLTWKNRAMCTASAW 542
+ L WK R + +S W
Sbjct: 1312 DENSKWLLQGWKGRTLYASSCW 1333
>At3g54220 SCARECROW1
Length = 653
Score = 197 bits (500), Expect = 2e-50
Identities = 131/377 (34%), Positives = 202/377 (52%), Gaps = 18/377 (4%)
Query: 169 LDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSG 228
L L +L++CA+AVS +++ A + + ++ + G QR+ AY E + ARL +S
Sbjct: 288 LHLLTLLLQCAEAVSADNLEEANKLLLEI-SQLSTPYGTSAQRVAAYFSEAMSARLLNSC 346
Query: 229 SLIYKALKCE-QPIT-SKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDF 286
IY AL P T S +++S + + P KF++ ++N I EA + E +HIID
Sbjct: 347 LGIYAALPSRWMPQTHSLKMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDL 406
Query: 287 QITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVP 346
I QG QW L LASRPGGPP +R+TG+ S+ L GKRLSDFA G+P
Sbjct: 407 DIMQGLQWPGLFHILASRPGGPPHVRLTGLGTSME------ALQATGKRLSDFADKLGLP 460
Query: 347 FEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSP 406
FEF A ++ E L +R EA+AV+ +L H + ++ H L L++ L+P
Sbjct: 461 FEFCPLAEKVGNLDTERLNVRKREAVAVH---WLQHSLYDVTGSDAH---TLWLLQRLAP 514
Query: 407 KVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVN 466
KVVT+VEQ+ ++ F RFVE + YYSA+++S+ + + + R EQ ++++I N
Sbjct: 515 KVVTVVEQDL-SHAGSFLGRFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRN 573
Query: 467 MIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEF-NENYRLEHKDV 525
++A G R F WR + GF L+ + +L F ++ Y L +
Sbjct: 574 VLAVGGPSR-SGEVKFESWREKMQQCGFKGISLAGNAATQATLLLGMFPSDGYTLVDDNG 632
Query: 526 ALYLTWKNRAMCTASAW 542
L L WK+ ++ TASAW
Sbjct: 633 TLKLGWKDLSLLTASAW 649
>At5g17490 RGA-like protein
Length = 523
Score = 195 bits (496), Expect = 5e-50
Identities = 128/381 (33%), Positives = 202/381 (52%), Gaps = 29/381 (7%)
Query: 171 LKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSGSL 230
L L+ CA+AV ++ A + V G + + + ++ Y E L R
Sbjct: 157 LVQALVACAEAVQLENLSLADALVKRV-GLLAASQAGAMGKVATYFAEALARR------- 208
Query: 231 IYKALKCEQPITSKELMSYMDIL----YQVCPYWKFTYISSNVVIGEAMQNESRIHIIDF 286
IY+ P + S+ +IL Y CPY KF + ++N I EA+ +H+ID
Sbjct: 209 IYRI----HPSAAAIDPSFEEILQMNFYDSCPYLKFAHFTANQAILEAVTTSRVVHVIDL 264
Query: 287 QITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVP 346
+ QG QW L+QALA RPGGPP R+TGV + + + G+ +G +L+ A++ GV
Sbjct: 265 GLNQGMQWPALMQALALRPGGPPSFRLTGVGNPSNRE----GIQELGWKLAQLAQAIGVE 320
Query: 347 FEFHSAAMSG-CEVELENLVIR-PGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSL 404
F+F+ ++E + R E L VN F LH + + S E +LL VK++
Sbjct: 321 FKFNGLTTERLSDLEPDMFETRTESETLVVNSVFELHPVLSQPGSIE----KLLATVKAV 376
Query: 405 SPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDI 464
P +VT+VEQE+N N F RF E L YYS++++S++ + ++R+ +E + + R I
Sbjct: 377 KPGLVTVVEQEANHNGDVFLDRFNEALHYYSSLFDSLEDGVVIPSQDRVMSEVY-LGRQI 435
Query: 465 VNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSV--TASVRNILNEFNENYRLEH 522
+N++A EG++R+ERHE +WR R AGF P L S AS+ L+ + YR+E
Sbjct: 436 LNLVATEGSDRIERHETLAQWRKRMGSAGFDPVNLGSDAFKQASLLLALSGGGDGYRVEE 495
Query: 523 KDVALYLTWKNRAMCTASAWR 543
D +L L W+ + + ASAW+
Sbjct: 496 NDGSLMLAWQTKPLIAASAWK 516
>At2g37650 scarecrow-like 9 (SCL9)
Length = 718
Score = 195 bits (495), Expect = 6e-50
Identities = 117/376 (31%), Positives = 194/376 (51%), Gaps = 5/376 (1%)
Query: 169 LDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLESSG 228
+DL+ +LI CAQAV+ D + A G + + + GD QRL GL ARL +G
Sbjct: 342 VDLRSLLIHCAQAVAADDRRCA-GQLLKQIRLHSTPFGDGNQRLAHCFANGLEARLAGTG 400
Query: 229 SLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIIDFQI 288
S IYK + +P ++ ++ + CP+ K +Y +N I + + N R+H+IDF I
Sbjct: 401 SQIYKGI-VSKPRSAAAVLKAHQLFLACCPFRKLSYFITNKTIRDLVGNSQRVHVIDFGI 459
Query: 289 TQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCGVPFE 348
G QW LI + G P +R+TG++ + G+RL+ +AK GVPFE
Sbjct: 460 LYGFQWPTLIHRFSMY--GSPKVRITGIEFPQPGFRPAQRVEETGQRLAAYAKLFGVPFE 517
Query: 349 FHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSLSPKV 408
+ + A ++LE+L I E VN + ++ DESV E+ RD +L L+ ++P +
Sbjct: 518 YKAIAKKWDAIQLEDLDIDRDEITVVNCLYRAENLHDESVKVESCRDTVLNLIGKINPDL 577
Query: 409 VTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDIVNMI 468
N F RF E L ++S++++ ++ +PR+D+ R+ E R+ +N+I
Sbjct: 578 FVFGIVNGAYNAPFFVTRFREALFHFSSIFDMLETIVPREDEERMFLEMEVFGREALNVI 637
Query: 469 ACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSV-TASVRNILNEFNENYRLEHKDVAL 527
ACEG ERVER E + +W R +G V P S+ S+ + +++++ ++ + L
Sbjct: 638 ACEGWERVERPETYKQWHVRAMRSGLVQVPFDPSIMKTSLHKVHTFYHKDFVIDQDNRWL 697
Query: 528 YLTWKNRAMCTASAWR 543
WK R + S W+
Sbjct: 698 LQGWKGRTVMALSVWK 713
>At1g07520 transcription factor scarecrow-like 14, putative
Length = 695
Score = 184 bits (466), Expect = 1e-46
Identities = 118/382 (30%), Positives = 197/382 (50%), Gaps = 10/382 (2%)
Query: 169 LDLKDVLIRCAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLE-SS 227
+D + +L CAQ+VS GD TA + + K S GD QRL + L ARLE S+
Sbjct: 313 VDFRTLLTLCAQSVSAGDKITADDLLRQIR-KQCSPVGDASQRLAHFFANALEARLEGST 371
Query: 228 GSLI---YKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHII 284
G++I Y ++ ++ ++ L SY + P+ Y SN +I +A ++ S +HI+
Sbjct: 372 GTMIQSYYDSISSKKRTAAQILKSY-SVFLSASPFMTLIYFFSNKMILDAAKDASVLHIV 430
Query: 285 DFQITQGTQWLLLIQALASRPGGPPFIRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCG 344
DF I G QW + IQ L+ G +R+TG++ + G+RL+++ K G
Sbjct: 431 DFGILYGFQWPMFIQHLSKSNPGLRKLRITGIEIPQHGLRPTERIQDTGRRLTEYCKRFG 490
Query: 345 VPFEFHSAAMSGCE-VELENLVIRPGEALAVNFAFFLHHMPDESVSTEN-HRDRLLRLVK 402
VPFE+++ A E +++E IRP E LAVN ++ D E+ RD L+L++
Sbjct: 491 VPFEYNAIASKNWETIKMEEFKIRPNEVLAVNAVLRFKNLRDVIPGEEDCPRDGFLKLIR 550
Query: 403 SLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVAR 462
++P V + N F RF E L +YSA+++ L +++ RI E R
Sbjct: 551 DMNPNVFLSSTVNGSFNAPFFTTRFKEALFHYSALFDLFGATLSKENPERIHFEGEFYGR 610
Query: 463 DIVNMIACEGAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNE--FNENYRL 520
+++N+IACEG +RVER E + +W+ R AGF P+ + + R + + +++++ L
Sbjct: 611 EVMNVIACEGVDRVERPETYKQWQVRMIRAGFKQKPVEAELVQLFREKMKKWGYHKDFVL 670
Query: 521 EHKDVALYLTWKNRAMCTASAW 542
+ WK R + ++S W
Sbjct: 671 DEDSNWFLQGWKGRILFSSSCW 692
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 172 bits (435), Expect = 6e-43
Identities = 133/386 (34%), Positives = 190/386 (48%), Gaps = 25/386 (6%)
Query: 170 DLKDVLIR----CAQAVSDGDMQTALGWMNNVLGKMVSVAGDPIQRLGAYMLEGLRARLE 225
DL+ L++ CA+ +SD D A + + + VS GDP +R+ Y E L RL
Sbjct: 212 DLEPPLLKAIYDCAR-ISDSDPNEASKTLLQIR-ESVSELGDPTERVAFYFTEALSNRLS 269
Query: 226 SSGSLIYKALKCEQPITSKELMSYMDILYQVCPYWKFTYISSNVVIGEAMQNESRIHIID 285
+ A T ++SY L CPY KF ++++N I EA + ++IHI+D
Sbjct: 270 PNSP----ATSSSSSSTEDLILSYKT-LNDACPYSKFAHLTANQAILEATEKSNKIHIVD 324
Query: 286 FQITQGTQWLLLIQALASRPGGPPF-IRVTGVDDSLSFDARGGGLHIVGKRLSDFAKSCG 344
F I QG QW L+QALA+R G P IRV+G+ ++ L G RL DFAK
Sbjct: 325 FGIVQGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLD 384
Query: 345 VPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFFLHHMPDESVSTENHRDRLLRLVKSL 404
+ F+F + + + P E LAVNF L+ + DE T D LRL KSL
Sbjct: 385 LNFDFIPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDE---TPTIVDTALRLAKSL 441
Query: 405 SPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYESIDVALPRDDKNRISAEQHCVARDI 464
+P+VVTL E E + N F R L +YSA++ES++ L RD + R+ E+ R I
Sbjct: 442 NPRVVTLGEYEVSLNRVGFANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRI 501
Query: 465 VNMIACE--GAERVERHELFGKWRSRFSMAGFVPCPLSSSVTASVRNILNEFNENYR--- 519
+I E G R ER E +WR AGF LS+ + + +L +N NY
Sbjct: 502 SGLIGPEKTGIHR-ERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILL--WNYNYSNLY 558
Query: 520 --LEHKDVALYLTWKNRAMCTASAWR 543
+E K + L W + + T S+WR
Sbjct: 559 SIVESKPGFISLAWNDLPLLTLSSWR 584
>At3g50650 scarecrow-like 7 (SCL7)
Length = 542
Score = 169 bits (427), Expect = 5e-42
Identities = 120/354 (33%), Positives = 177/354 (49%), Gaps = 25/354 (7%)
Query: 202 VSVAGDPIQRLGAYMLEGLRAR-LESSGSLIYKALKCEQPITSKELMSYMDILYQVCPYW 260
VS +GDPIQR+G Y E L + ES S +L+ ++SY L CPY
Sbjct: 202 VSESGDPIQRVGYYFAEALSHKETESPSSSSSSSLE-------DFILSYKT-LNDACPYS 253
Query: 261 KFTYISSNVVIGEAMQNESRIHIIDFQITQGTQWLLLIQALASRPGGPPF-IRVTGVDDS 319
KF ++++N I EA + IHI+DF I QG QW L+QALA+R G P IR++G+
Sbjct: 254 KFAHLTANQAILEATNQSNNIHIVDFGIFQGIQWSALLQALATRSSGKPTRIRISGIPAP 313
Query: 320 LSFDARGGGLHIVGKRLSDFAKSCGVPFEFHSAAMSGCEVELENLVIRPGEALAVNFAFF 379
D+ G L G RL DFA + FEF+ + + + P E L VNF
Sbjct: 314 SLGDSPGPSLIATGNRLRDFAAILDLNFEFYPVLTPIQLLNGSSFRVDPDEVLVVNFMLE 373
Query: 380 LHHMPDESVSTENHRDRLLRLVKSLSPKVVTLVEQESNTNTSPFFQRFVETLSYYSAMYE 439
L+ + DE+ +T LRL +SL+P++VTL E E + N F R +L +YSA++E
Sbjct: 374 LYKLLDETATTVG---TALRLARSLNPRIVTLGEYEVSLNRVEFANRVKNSLRFYSAVFE 430
Query: 440 SIDVALPRDDKNRISAEQHCVARDIVNMIACEG-----AERVERHELFGKWRSRFSMAGF 494
S++ L RD K R+ E+ R I++++ + R E +WR AGF
Sbjct: 431 SLEPNLDRDSKERLRVERVLFGRRIMDLVRSDDDNNKPGTRFGLMEEKEQWRVLMEKAGF 490
Query: 495 VPCPLSSSVTASVRNILNEFNENYR-----LEHKDVALYLTWKNRAMCTASAWR 543
P S+ + + +L +N NY +E + + L W N + T S+WR
Sbjct: 491 EPVKPSNYAVSQAKLLL--WNYNYSTLYSLVESEPGFISLAWNNVPLLTVSSWR 542
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,162,072
Number of Sequences: 26719
Number of extensions: 509608
Number of successful extensions: 1605
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1435
Number of HSP's gapped (non-prelim): 68
length of query: 545
length of database: 11,318,596
effective HSP length: 104
effective length of query: 441
effective length of database: 8,539,820
effective search space: 3766060620
effective search space used: 3766060620
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0367.12


