

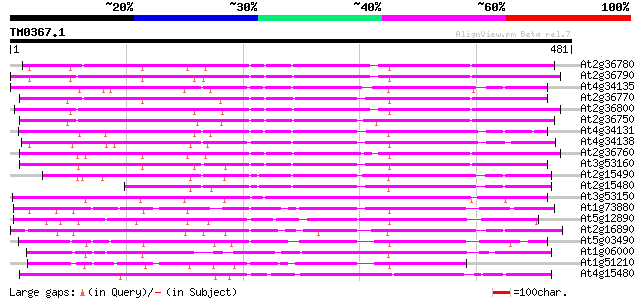
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0367.1
(481 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g36780 putative glucosyl transferase 321 5e-88
At2g36790 putative glucosyl transferase 318 5e-87
At4g34135 glucosyltransferase like protein 317 9e-87
At2g36770 putative glucosyl transferase 316 2e-86
At2g36800 putative glucosyl transferase 316 2e-86
At2g36750 putative glucosyl transferase 312 3e-85
At4g34131 glucosyltransferase -like protein 305 4e-83
At4g34138 glucosyltransferase -like protein 305 5e-83
At2g36760 putative glucosyl transferase 303 2e-82
At3g53160 glucosyltransferase - like protein 295 4e-80
At2g15490 putative glucosyltransferase 291 4e-79
At2g15480 glucosyltransferase like protein 266 1e-71
At3g53150 glucosyltransferase - like protein 258 4e-69
At1g73880 putative glucosyltransferase (At1g73880) 197 1e-50
At5g12890 glucosyltransferase -like protein 190 1e-48
At2g16890 glucosyltransferase like protein 188 6e-48
At5g03490 UDPG glucosyltransferase - like protein 183 2e-46
At1g06000 unknown protein 182 3e-46
At1g51210 putative glucosyl transferase 175 4e-44
At4g15480 indole-3-acetate beta-glucosyltransferase like protein 151 7e-37
>At2g36780 putative glucosyl transferase
Length = 496
Score = 321 bits (823), Expect = 5e-88
Identities = 185/490 (37%), Positives = 275/490 (55%), Gaps = 45/490 (9%)
Query: 12 LYFI--PYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTT--------TIDSSLPNLHL 61
L+F+ P++A GHMIP+ DIA L A RG +TI+TTP N I+S L L
Sbjct: 13 LHFVLFPFMAQGHMIPMIDIARLLAQRGVTITIVTTPHNAARFKNVLNRAIESGLAINIL 72
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPA--DCVVADY 119
H V FP ++ GLP+G E+ S + + + LL +P+ ME C+++D+
Sbjct: 73 H-VKFPYQEFGLPEGKENIDSLDSTELMVPFFKAVNLLEDPVMKLMEEMKPRPSCLISDW 131
Query: 120 CFPWIDDLTTELHIPRLSFNPSPLFALCAMRA-----------KRGSSSFLISGLPHG-- 166
C P+ + +IP++ F+ F L M K FL+ P
Sbjct: 132 CLPYTSIIAKNFNIPKIVFHGMGCFNLLCMHVLRRNLEILENVKSDEEYFLVPSFPDRVE 191
Query: 167 ----DIALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAW 222
+ + A+ ++ +++ E S+GV++N+F EL+ YVK Y+ G K W
Sbjct: 192 FTKLQLPVKANASGDWKEIMDEMVKAEYTSYGVIVNTFQELEP-PYVKDYKEAMDG-KVW 249
Query: 223 LLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHE 282
+GP SL +KAERG K+ + DE L WL +K+ SVLYVC GS+C+ QL E
Sbjct: 250 SIGPVSLCNKAGADKAERG-SKAAIDQDECLQWLDSKEEGSVLYVCLGSICNLPLSQLKE 308
Query: 283 MACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPE-GFEEK----GMILRGWAPQLLI 337
+ G+E S F+WV+ ++ KE E W+ E GFEE+ G++++GWAPQ+LI
Sbjct: 309 LGLGLEESRRSFIWVIRGSEKYKELFE------WMLESGFEERIKERGLLIKGWAPQVLI 362
Query: 338 LGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWS 397
L HPSVG FLTHCG NS LE +++G+P+ITWP+ GDQF +KL+ +V GV G EE
Sbjct: 363 LSHPSVGGFLTHCGWNSTLEGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEE-V 421
Query: 398 MGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNL 457
M G ++ V++ ++ ++KAV LMG ++A++ RRRV ELG LA+ AV+ GGSS+ N+
Sbjct: 422 MKWGEEDKIGVLVDKEGVKKAVEELMGDSDDAKERRRRVKELGELAHKAVEKGGSSHSNI 481
Query: 458 TALIAHLKTL 467
T L+ + L
Sbjct: 482 TLLLQDIMQL 491
>At2g36790 putative glucosyl transferase
Length = 495
Score = 318 bits (814), Expect = 5e-87
Identities = 183/506 (36%), Positives = 280/506 (55%), Gaps = 45/506 (8%)
Query: 1 MDVESEQRPLKLYFI--PYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTT-------- 50
M E P L+F+ P++A GHMIP+ DIA L A RG +TI+TTP N
Sbjct: 1 MAFEKNNEPFPLHFVLFPFMAQGHMIPMVDIARLLAQRGVLITIVTTPHNAARFKNVLNR 60
Query: 51 TIDSSLPNLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHH 110
I+S LP ++L V FP ++ GL +G E+ ++ + + LL+EP+Q +E
Sbjct: 61 AIESGLP-INLVQVKFPYQEAGLQEGQENMDLLTTMEQITSFFKAVNLLKEPVQNLIEEM 119
Query: 111 PA--DCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS----------- 157
C+++D C + ++ + IP++ F+ F L + R +
Sbjct: 120 SPRPSCLISDMCLSYTSEIAKKFKIPKILFHGMGCFCLLCVNVLRKNREILDNLKSDKEY 179
Query: 158 FLISGLP------HGDIALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKH 211
F++ P + + P +E ++ + S+GV++NSF EL+ Y K
Sbjct: 180 FIVPYFPDRVEFTRPQVPVETYVPAGWKEILEDMVEADKTSYGVIVNSFQELEP-AYAKD 238
Query: 212 YERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGS 271
++ G KAW +GP SL +KAERG+ KS + DE L WL +K+P SVLYVC GS
Sbjct: 239 FKEARSG-KAWTIGPVSLCNKVGVDKAERGN-KSDIDQDECLEWLDSKEPGSVLYVCLGS 296
Query: 272 MCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPE-GFEEK----GM 326
+C+ QL E+ G+E S F+WV+ ++ KE E W E GFE++ G+
Sbjct: 297 ICNLPLSQLLELGLGLEESQRPFIWVIRGWEKYKELVE------WFSESGFEDRIQDRGL 350
Query: 327 ILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRG 386
+++GW+PQ+LIL HPSVG FLTHCG NS LE ++AG+P++TWP+ DQF EKL+ ++
Sbjct: 351 LIKGWSPQMLILSHPSVGGFLTHCGWNSTLEGITAGLPMLTWPLFADQFCNEKLVVQILK 410
Query: 387 VGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAA 446
VGV +E M G E+ V++ ++ ++KAV LMG ++A++ RRR ELG A+ A
Sbjct: 411 VGVSAEVKE-VMKWGEEEKIGVLVDKEGVKKAVEELMGESDDAKERRRRAKELGESAHKA 469
Query: 447 VQPGGSSYQNLTALIAHLKTLRDSNS 472
V+ GGSS+ N+T L+ + L SN+
Sbjct: 470 VEEGGSSHSNITFLLQDIMQLAQSNN 495
>At4g34135 glucosyltransferase like protein
Length = 483
Score = 317 bits (812), Expect = 9e-87
Identities = 192/499 (38%), Positives = 283/499 (56%), Gaps = 59/499 (11%)
Query: 1 MDVESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN-- 58
M + R L + F P++A GHMIP D+A LF+SRG TI+TT N+ + +
Sbjct: 1 MGSDHHHRKLHVMFFPFMAYGHMIPTLDMAKLFSSRGAKSTILTTSLNSKILQKPIDTFK 60
Query: 59 -------LHLHTVPFPSRQVGLPDGIES---FSSSAD---LQTTFKVHQGIKLLREPIQL 105
+ + FP ++GLP+G E+ F+S+ + + K + ++ ++
Sbjct: 61 NLNPGLEIDIQIFNFPCVELGLPEGCENVDFFTSNNNDDKNEMIVKFFFSTRFFKDQLEK 120
Query: 106 FMEHHPADCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCA----------MRAKRGS 155
+ DC++AD FPW + + ++PRL F+ + F+LCA R S
Sbjct: 121 LLGTTRPDCLIADMFFPWATEAAGKFNVPRLVFHGTGYFSLCAGYCIGVHKPQKRVASSS 180
Query: 156 SSFLISGLPHGDIAL------NASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYV 209
F+I LP G+I + + + + + E+KS GV++NSF EL+ H+Y
Sbjct: 181 EPFVIPELP-GNIVITEEQIIDGDGESDMGKFMTEVRESEVKSSGVVLNSFYELE-HDYA 238
Query: 210 KHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCF 269
Y+ +AW +GP S+ +EKAERG +K+ + E L WL +KKP+SV+YV F
Sbjct: 239 DFYKSCVQ-KRAWHIGPLSVYNRGFEEKAERG-KKANIDEAECLKWLDSKKPNSVIYVSF 296
Query: 270 GSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE----KG 325
GS+ F NEQL E+A G+EAS F+WVV K +++E+WLPEGFEE KG
Sbjct: 297 GSVAFFKNEQLFEIAAGLEASGTSFIWVVRKTK--------DDREEWLPEGFEERVKGKG 348
Query: 326 MILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVR 385
MI+RGWAPQ+LIL H + G F+THCG NS LE V+AG+P++TWPV +QFY EKL+T+V
Sbjct: 349 MIIRGWAPQVLILDHQATGGFVTHCGWNSLLEGVAAGLPMVTWPVGAEQFYNEKLVTQVL 408
Query: 386 GVGVEVGAEEW---SMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNL 442
GV VGA + MG ISR+K++KAVR ++ GE AE+ RRR +L +
Sbjct: 409 RTGVSVGASKHMKVMMGD--------FISREKVDKAVREVL-AGEAAEERRRRAKKLAAM 459
Query: 443 ANAAVQPGGSSYQNLTALI 461
A AAV+ GGSS+ +L + +
Sbjct: 460 AKAAVEEGGSSFNDLNSFM 478
>At2g36770 putative glucosyl transferase
Length = 496
Score = 316 bits (810), Expect = 2e-86
Identities = 181/485 (37%), Positives = 273/485 (55%), Gaps = 43/485 (8%)
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSN--------TTTIDSSLPNLH 60
PL P++A GHMIP+ DIA L A RG VTI+TT N + ++S LP
Sbjct: 12 PLHFILFPFMAQGHMIPMIDIARLLAQRGATVTIVTTRYNAGRFENVLSRAMESGLPINI 71
Query: 61 LHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPA--DCVVAD 118
+H V FP ++ GLP+G E+ S ++ Q + +L +P+ ME C+++D
Sbjct: 72 VH-VNFPYQEFGLPEGKENIDSYDSMELMVPFFQAVNMLEDPVMKLMEEMKPRPSCIISD 130
Query: 119 YCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNASPPEAL 178
P+ + + IP++ F+ + F L M R + L + D L S P+ +
Sbjct: 131 LLLPYTSKIARKFSIPKIVFHGTGCFNLLCMHVLRRNLEILKNLKSDKDYFLVPSFPDRV 190
Query: 179 T-----------------ACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKA 221
A ++ ++ E S+GV++N+F EL+ YVK Y + G K
Sbjct: 191 EFTKPQVPVETTASGDWKAFLDEMVEAEYTSYGVIVNTFQELEP-AYVKDYTKARAG-KV 248
Query: 222 WLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLH 281
W +GP SL +KAERG++ + + DE L WL +K+ SVLYVC GS+C+ QL
Sbjct: 249 WSIGPVSLCNKAGADKAERGNQ-AAIDQDECLQWLDSKEDGSVLYVCLGSICNLPLSQLK 307
Query: 282 EMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPE-GFEEK----GMILRGWAPQLL 336
E+ G+E S F+WV+ + E+ E +W+ E GFEE+ G++++GW+PQ+L
Sbjct: 308 ELGLGLEKSQRSFIWVI------RGWEKYNELYEWMMESGFEERIKERGLLIKGWSPQVL 361
Query: 337 ILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEW 396
IL HPSVG FLTHCG NS LE +++G+P+ITWP+ GDQF +KL+ +V GV G EE
Sbjct: 362 ILSHPSVGGFLTHCGWNSTLEGITSGIPLITWPLFGDQFCNQKLVVQVLKAGVSAGVEE- 420
Query: 397 SMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQN 456
M G E+ V++ ++ ++KAV LMG ++A++ RRRV ELG A+ AV+ GGSS+ N
Sbjct: 421 VMKWGEEEKIGVLVDKEGVKKAVEELMGASDDAKERRRRVKELGESAHKAVEEGGSSHSN 480
Query: 457 LTALI 461
+T L+
Sbjct: 481 ITYLL 485
>At2g36800 putative glucosyl transferase
Length = 495
Score = 316 bits (809), Expect = 2e-86
Identities = 185/501 (36%), Positives = 276/501 (54%), Gaps = 44/501 (8%)
Query: 5 SEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTT--------TIDSSL 56
++ PL P++A GHMIP+ DIA L A RG +TI+TTP N I+S L
Sbjct: 6 TKSSPLHFVLFPFMAQGHMIPMVDIARLLAQRGVIITIVTTPHNAARFKNVLNRAIESGL 65
Query: 57 PNLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQ-LFMEHHPA-DC 114
P ++L V FP + GL +G E+ S ++ + + L EP+Q L E +P C
Sbjct: 66 P-INLVQVKFPYLEAGLQEGQENIDSLDTMERMIPFFKAVNFLEEPVQKLIEEMNPRPSC 124
Query: 115 VVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS-----------FLISGL 163
+++D+C P+ + + +IP++ F+ F L M R + F +
Sbjct: 125 LISDFCLPYTSKIAKKFNIPKILFHGMGCFCLLCMHVLRKNREILDNLKSDKELFTVPDF 184
Query: 164 PHG-DIALNASPPEALTAC------VEPLLRKELKSHGVLINSFVELDGHEYVKHYERTT 216
P + P E + ++ S+GV++NSF EL+ Y K Y+
Sbjct: 185 PDRVEFTRTQVPVETYVPAGDWKDIFDGMVEANETSYGVIVNSFQELEP-AYAKDYKEVR 243
Query: 217 GGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFS 276
G KAW +GP SL +KAERG+ KS + DE L WL +KK SVLYVC GS+C+
Sbjct: 244 SG-KAWTIGPVSLCNKVGADKAERGN-KSDIDQDECLKWLDSKKHGSVLYVCLGSICNLP 301
Query: 277 NEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPE-GFEEK----GMILRGW 331
QL E+ G+E S F+WV+ ++ KE E W E GFE++ G++++GW
Sbjct: 302 LSQLKELGLGLEESQRPFIWVIRGWEKYKELVE------WFSESGFEDRIQDRGLLIKGW 355
Query: 332 APQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEV 391
+PQ+LIL HPSVG FLTHCG NS LE ++AG+P++TWP+ DQF EKL+ EV GV
Sbjct: 356 SPQMLILSHPSVGGFLTHCGWNSTLEGITAGLPLLTWPLFADQFCNEKLVVEVLKAGVRS 415
Query: 392 GAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGG 451
G E+ M G E+ V++ ++ ++KAV LMG ++A++ RRR ELG+ A+ AV+ GG
Sbjct: 416 GVEQ-PMKWGEEEKIGVLVDKEGVKKAVEELMGESDDAKERRRRAKELGDSAHKAVEEGG 474
Query: 452 SSYQNLTALIAHLKTLRDSNS 472
SS+ N++ L+ + L + N+
Sbjct: 475 SSHSNISFLLQDIMELAEPNN 495
>At2g36750 putative glucosyl transferase
Length = 491
Score = 312 bits (799), Expect = 3e-85
Identities = 183/488 (37%), Positives = 273/488 (55%), Gaps = 38/488 (7%)
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSN--------TTTIDSSLPNLH 60
PL P++A GHMIP+ DIA L A RG +TI+TTP N + I S LP ++
Sbjct: 8 PLHFVLFPFMAQGHMIPMVDIARLLAQRGVTITIVTTPQNAGRFKNVLSRAIQSGLP-IN 66
Query: 61 LHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQ-LFMEHHPA-DCVVAD 118
L V FPS++ G P+G E+ L + + LL EP++ L E P +C++AD
Sbjct: 67 LVQVKFPSQESGSPEGQENLDLLDSLGASLTFFKAFSLLEEPVEKLLKEIQPRPNCIIAD 126
Query: 119 YCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFL-----------ISGLPHGD 167
C P+ + + L IP++ F+ F L + FL I P
Sbjct: 127 MCLPYTNRIAKNLGIPKIIFHGMCCFNLLCTHIMHQNHEFLETIESDKEYFPIPNFPDRV 186
Query: 168 IALNASPPEALTA-----CVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAW 222
+ P L A ++ + + S+GV++N+F EL+ YV+ Y++ G K W
Sbjct: 187 EFTKSQLPMVLVAGDWKDFLDGMTEGDNTSYGVIVNTFEELEP-AYVRDYKKVKAG-KIW 244
Query: 223 LLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHE 282
+GP SL +++AERG+ K+ + DE + WL +K+ SVLYVC GS+C+ QL E
Sbjct: 245 SIGPVSLCNKLGEDQAERGN-KADIDQDECIKWLDSKEEGSVLYVCLGSICNLPLSQLKE 303
Query: 283 MACGIEASSVEFVWVVPAEKRGKEEEEEEEK---EKWLPEGFEEKGMILRGWAPQLLILG 339
+ G+E S F+WV+ RG E+ E + E E +E+G+++ GW+PQ+LIL
Sbjct: 304 LGLGLEESQRPFIWVI----RGWEKYNELLEWISESGYKERIKERGLLITGWSPQMLILT 359
Query: 340 HPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMG 399
HP+VG FLTHCG NS LE +++GVP++TWP+ GDQF EKL ++ GV G EE SM
Sbjct: 360 HPAVGGFLTHCGWNSTLEGITSGVPLLTWPLFGDQFCNEKLAVQILKAGVRAGVEE-SMR 418
Query: 400 GGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTA 459
G E+ V++ ++ ++KAV LMG +A++ R+RV ELG LA+ AV+ GGSS+ N+T
Sbjct: 419 WGEEEKIGVLVDKEGVKKAVEELMGDSNDAKERRKRVKELGELAHKAVEEGGSSHSNITF 478
Query: 460 LIAHLKTL 467
L+ + L
Sbjct: 479 LLQDIMQL 486
>At4g34131 glucosyltransferase -like protein
Length = 481
Score = 305 bits (781), Expect = 4e-83
Identities = 187/489 (38%), Positives = 277/489 (56%), Gaps = 54/489 (11%)
Query: 8 RPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPN--------- 58
R L + F P++A GHMIP D+A LF+SRG TI+TTP N+ +
Sbjct: 7 RKLHVVFFPFMAYGHMIPTLDMAKLFSSRGAKSTILTTPLNSKIFQKPIERFKNLNPSFE 66
Query: 59 LHLHTVPFPSRQVGLPDGIESF-----SSSADLQ-TTFKVHQGIKLLREPIQLFMEHHPA 112
+ + FP +GLP+G E+ +++ D Q T K + + ++ ++ +E
Sbjct: 67 IDIQIFDFPCVDLGLPEGCENVDFFTSNNNDDRQYLTLKFFKSTRFFKDQLEKLLETTRP 126
Query: 113 DCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS----------FLISG 162
DC++AD FPW + + ++PRL F+ + F+LC+ R + F+I
Sbjct: 127 DCLIADMFFPWATEAAEKFNVPRLVFHGTGYFSLCSEYCIRVHNPQNIVASRYEPFVIPD 186
Query: 163 LPHGDIAL------NASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTT 216
LP G+I + + + + + ++KS GV++NSF EL+ +Y Y+
Sbjct: 187 LP-GNIVITQEQIADRDEESEMGKFMIEVKESDVKSSGVIVNSFYELEP-DYADFYKSVV 244
Query: 217 GGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFS 276
+AW +GP S+ +EKAERG +K+ ++ E L WL +KKP SV+Y+ FGS+ F
Sbjct: 245 L-KRAWHIGPLSVYNRGFEEKAERG-KKASINEVECLKWLDSKKPDSVIYISFGSVACFK 302
Query: 277 NEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEE----KGMILRGWA 332
NEQL E+A G+E S F+WVV + EKE+WLPEGFEE KGMI+RGWA
Sbjct: 303 NEQLFEIAAGLETSGANFIWVV-------RKNIGIEKEEWLPEGFEERVKGKGMIIRGWA 355
Query: 333 PQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVG 392
PQ+LIL H + F+THCG NS LE V+AG+P++TWPV +QFY EKL+T+V GV VG
Sbjct: 356 PQVLILDHQATCGFVTHCGWNSLLEGVAAGLPMVTWPVAAEQFYNEKLVTQVLRTGVSVG 415
Query: 393 AEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGS 452
A++ G ISR+K+ KAVR ++ GEEA++ R R +L +A AAV+ GGS
Sbjct: 416 AKKNVRTTGD------FISREKVVKAVREVL-VGEEADERRERAKKLAEMAKAAVE-GGS 467
Query: 453 SYQNLTALI 461
S+ +L + I
Sbjct: 468 SFNDLNSFI 476
>At4g34138 glucosyltransferase -like protein
Length = 488
Score = 305 bits (780), Expect = 5e-83
Identities = 192/494 (38%), Positives = 274/494 (54%), Gaps = 56/494 (11%)
Query: 11 KLYFI--PYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTI-----------DSSLP 57
KL+F+ P++A GHMIP D+A LFA++G TI+TTP N + L
Sbjct: 9 KLHFLLFPFMAHGHMIPTLDMAKLFATKGAKSTILTTPLNAKLFFEKPIKSFNQDNPGLE 68
Query: 58 NLHLHTVPFPSRQVGLPDGIESFS---SSADLQT---TFKVHQGIKLLREPIQLFMEHHP 111
++ + + FP ++GLPDG E+ S+ DL + K +K EP++ +
Sbjct: 69 DITIQILNFPCTELGLPDGCENTDFIFSTPDLNVGDLSQKFLLAMKYFEEPLEELLVTMR 128
Query: 112 ADCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKR-------GSSSFLISGLP 164
DC+V + FPW + + +PRL F+ + F+LCA R S F+I LP
Sbjct: 129 PDCLVGNMFFPWSTKVAEKFGVPRLVFHGTGYFSLCASHCIRLPKNVATSSEPFVIPDLP 188
Query: 165 HGDI------ALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGG 218
GDI + + ++ + E S GVL+NSF EL+ + Y ++
Sbjct: 189 -GDILITEEQVMETEEESVMGRFMKAIRDSERDSFGVLVNSFYELE--QAYSDYFKSFVA 245
Query: 219 HKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNE 278
+AW +GP SL +EKAERG +K+ + E L WL +KK SV+Y+ FG+M SF NE
Sbjct: 246 KRAWHIGPLSLGNRKFEEKAERG-KKASIDEHECLKWLDSKKCDSVIYMAFGTMSSFKNE 304
Query: 279 QLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQ 334
QL E+A G++ S +FVWVV + + EKE WLPEGFEEK G+I+RGWAPQ
Sbjct: 305 QLIEIAAGLDMSGHDFVWVV------NRKGSQVEKEDWLPEGFEEKTKGKGLIIRGWAPQ 358
Query: 335 LLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAE 394
+LIL H ++G FLTHCG NS LE V+AG+P++TWPV +QFY EKL+T+V GV VG +
Sbjct: 359 VLILEHKAIGGFLTHCGWNSLLEGVAAGLPMVTWPVGAEQFYNEKLVTQVLKTGVSVGVK 418
Query: 395 EWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSY 454
+ G ISR+K+E AVR +M G E+ R+R EL +A AV+ GGSS
Sbjct: 419 KMMQVVGD------FISREKVEGAVREVMVG----EERRKRAKELAEMAKNAVKEGGSSD 468
Query: 455 QNLTALIAHLKTLR 468
+ L+ L ++
Sbjct: 469 LEVDRLMEELTLVK 482
>At2g36760 putative glucosyl transferase
Length = 496
Score = 303 bits (775), Expect = 2e-82
Identities = 175/495 (35%), Positives = 268/495 (53%), Gaps = 41/495 (8%)
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSL-----PNLHLHT 63
PL P++A GHMIP+ DIA + A RG +TI+TTP N L LH+
Sbjct: 12 PLHFVLFPFMAQGHMIPMVDIARILAQRGVTITIVTTPHNAARFKDVLNRAIQSGLHIRV 71
Query: 64 --VPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPA--DCVVADY 119
V FP ++ GL +G E+ ++ + + +L P+ ME C+++D+
Sbjct: 72 EHVKFPFQEAGLQEGQENVDFLDSMELMVHFFKAVNMLENPVMKLMEEMKPKPSCLISDF 131
Query: 120 CFPWIDDLTTELHIPRLSFNPSPLFALCAMRA-----------KRGSSSFLISGLPHG-- 166
C P+ + +IP++ F+ F L +M K FL+ P
Sbjct: 132 CLPYTSKIAKRFNIPKIVFHGVSCFCLLSMHILHRNHNILHALKSDKEYFLVPSFPDRVE 191
Query: 167 ----DIALNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAW 222
+ + + ++ + + S+GV++N+F +L+ YVK+Y G K W
Sbjct: 192 FTKLQVTVKTNFSGDWKEIMDEQVDADDTSYGVIVNTFQDLES-AYVKNYTEARAG-KVW 249
Query: 223 LLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHE 282
+GP SL ++KAERG+ K+ + DE + WL +K SVLYVC GS+C+ QL E
Sbjct: 250 SIGPVSLCNKVGEDKAERGN-KAAIDQDECIKWLDSKDVESVLYVCLGSICNLPLAQLRE 308
Query: 283 MACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPE-GFEEK----GMILRGWAPQLLI 337
+ G+EA+ F+WV+ GK E E W+ E GFEE+ ++++GW+PQ+LI
Sbjct: 309 LGLGLEATKRPFIWVIRGG--GKYHELAE----WILESGFEERTKERSLLIKGWSPQMLI 362
Query: 338 LGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWS 397
L HP+VG FLTHCG NS LE +++GVP+ITWP+ GDQF +KL+ +V GV VG EE
Sbjct: 363 LSHPAVGGFLTHCGWNSTLEGITSGVPLITWPLFGDQFCNQKLIVQVLKAGVSVGVEE-V 421
Query: 398 MGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNL 457
M G E V++ ++ ++KAV +MG +EA++ R+RV ELG LA+ AV+ GGSS+ N+
Sbjct: 422 MKWGEEESIGVLVDKEGVKKAVDEIMGESDEAKERRKRVRELGELAHKAVEEGGSSHSNI 481
Query: 458 TALIAHLKTLRDSNS 472
L+ + +S S
Sbjct: 482 IFLLQDIMQQVESKS 496
>At3g53160 glucosyltransferase - like protein
Length = 490
Score = 295 bits (755), Expect = 4e-80
Identities = 179/486 (36%), Positives = 269/486 (54%), Gaps = 45/486 (9%)
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASR-GQHVTIITTPSNTTTIDSSLP------NLHL 61
PL IP++A GHMIPL DI+ L + R G V IITT N I +SL +++
Sbjct: 6 PLHFVVIPFMAQGHMIPLVDISRLLSQRQGVTVCIITTTQNVAKIKTSLSFSSLFATINI 65
Query: 62 HTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPA---DCVVAD 118
V F S+Q GLP+G ES A + K L E ++ ME C++ D
Sbjct: 66 VEVKFLSQQTGLPEGCESLDMLASMGDMVKFFDAANSLEEQVEKAMEEMVQPRPSCIIGD 125
Query: 119 YCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS----------FLISGLPHGDI 168
P+ L + IP+L F+ F+L +++ R S F + GLP D
Sbjct: 126 MSLPFTSRLAKKFKIPKLIFHGFSCFSLMSIQVVRESGILKMIESNDEYFDLPGLP--DK 183
Query: 169 ALNASPPEALTACVE--------PLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHK 220
P ++ VE ++ + S+GV++N+F EL+ +Y + Y + G K
Sbjct: 184 VEFTKPQVSVLQPVEGNMKESTAKIIEADNDSYGVIVNTFEELEV-DYAREYRKARAG-K 241
Query: 221 AWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQL 280
W +GP SL +KA+RG +K+ + D+ L WL +++ SVLYVC GS+C+ QL
Sbjct: 242 VWCVGPVSLCNRLGLDKAKRG-DKASIGQDQCLQWLDSQETGSVLYVCLGSLCNLPLAQL 300
Query: 281 HEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPE-GFEEK----GMILRGWAPQL 335
E+ G+EAS+ F+WV+ +E + + W+ + GFEE+ G++++GWAPQ+
Sbjct: 301 KELGLGLEASNKPFIWVI------REWGKYGDLANWMQQSGFEERIKDRGLVIKGWAPQV 354
Query: 336 LILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEE 395
IL H S+G FLTHCG NS LE ++AGVP++TWP+ +QF EKL+ ++ G+++G E+
Sbjct: 355 FILSHASIGGFLTHCGWNSTLEGITAGVPLLTWPLFAEQFLNEKLVVQILKAGLKIGVEK 414
Query: 396 WSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQ 455
M G+ E ++SR+ + KAV LMG EEAE+ RR+V EL +LAN A++ GGSS
Sbjct: 415 -LMKYGKEEEIGAMVSRECVRKAVDELMGDSEEAEERRRKVTELSDLANKALEKGGSSDS 473
Query: 456 NLTALI 461
N+T LI
Sbjct: 474 NITLLI 479
>At2g15490 putative glucosyltransferase
Length = 460
Score = 291 bits (746), Expect = 4e-79
Identities = 182/471 (38%), Positives = 258/471 (54%), Gaps = 50/471 (10%)
Query: 29 IAILFASRGQHVTIITTPSNTTTIDSSL-------PNLHL--HTVPFPSRQVGLPDGIE- 78
+A LFA RG T++TTP N ++ + P+L + + FP ++GLP+G E
Sbjct: 1 MAKLFARRGAKSTLLTTPINAKILEKPIEAFKVQNPDLEIGIKILNFPCVELGLPEGCEN 60
Query: 79 -----SFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYCFPWIDDLTTELHI 133
S+ S K K +++ ++ F+E +VAD FPW + ++ +
Sbjct: 61 RDFINSYQKSDSFDLFLKFLFSTKYMKQQLESFIETTKPSALVADMFFPWATESAEKIGV 120
Query: 134 PRLSFNPSPLFALCAMRAKR----------GSSSFLISGLPHGDIALNASPPEALTACV- 182
PRL F+ + FALC R S+ F+I GLP GDI +
Sbjct: 121 PRLVFHGTSSFALCCSYNMRIHKPHKKVASSSTPFVIPGLP-GDIVITEDQANVTNEETP 179
Query: 183 -----EPLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEK 237
+ + E S GVL+NSF EL+ Y Y R+ KAW +GP SL EK
Sbjct: 180 FGKFWKEVRESETSSFGVLVNSFYELES-SYADFY-RSFVAKKAWHIGPLSLSNRGIAEK 237
Query: 238 AERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWV 297
A RG +K+ + E L WL +K P SV+Y+ FGS NEQL E+A G+E S F+WV
Sbjct: 238 AGRG-KKANIDEQECLKWLDSKTPGSVVYLSFGSGTGLPNEQLLEIAFGLEGSGQNFIWV 296
Query: 298 VPAEKRGKEEEEEEEKEKWLPEGFEE----KGMILRGWAPQLLILGHPSVGAFLTHCGSN 353
V + + + E E WLP+GFEE KG+I+RGWAPQ+LIL H ++G F+THCG N
Sbjct: 297 V---SKNENQVGTGENEDWLPKGFEERNKGKGLIIRGWAPQVLILDHKAIGGFVTHCGWN 353
Query: 354 SALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRD 413
S LE ++AG+P++TWP+ +QFY EKL+T+V +GV VGA E G +ISR
Sbjct: 354 STLEGIAAGLPMVTWPMGAEQFYNEKLLTKVLRIGVNVGATELVKKG-------KLISRA 406
Query: 414 KIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
++EKAVR ++ GGE+AE+ R R ELG +A AAV+ GGSSY ++ + L
Sbjct: 407 QVEKAVREVI-GGEKAEERRLRAKELGEMAKAAVEEGGSSYNDVNKFMEEL 456
>At2g15480 glucosyltransferase like protein
Length = 372
Score = 266 bits (681), Expect = 1e-71
Identities = 155/386 (40%), Positives = 228/386 (58%), Gaps = 38/386 (9%)
Query: 99 LREPIQLFMEHHPADCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKR----- 153
+++ ++ F+E +VAD FPW + +L +PRL F+ + F+LC R
Sbjct: 1 MKQQLESFIETTKPSALVADMFFPWATESAEKLGVPRLVFHGTSFFSLCCSYNMRIHKPH 60
Query: 154 -----GSSSFLISGLPHGDIALN------ASPPEALTACVEPLLRKELKSHGVLINSFVE 202
S+ F+I GLP GDI + A + ++ + E S GVL+NSF E
Sbjct: 61 KKVATSSTPFVIPGLP-GDIVITEDQANVAKEETPMGKFMKEVRESETNSFGVLVNSFYE 119
Query: 203 LDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPS 262
L+ Y Y R+ +AW +GP SL EKA RG +K+ + E L WL +K P
Sbjct: 120 LES-AYADFY-RSFVAKRAWHIGPLSLSNRELGEKARRG-KKANIDEQECLKWLDSKTPG 176
Query: 263 SVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFE 322
SV+Y+ FGS +F+N+QL E+A G+E S F+WVV ++ E + + E+WLPEGF+
Sbjct: 177 SVVYLSFGSGTNFTNDQLLEIAFGLEGSGQSFIWVV------RKNENQGDNEEWLPEGFK 230
Query: 323 E----KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTE 378
E KG+I+ GWAPQ+LIL H ++G F+THCG NSA+E ++AG+P++TWP+ +QFY E
Sbjct: 231 ERTTGKGLIIPGWAPQVLILDHKAIGGFVTHCGWNSAIEGIAAGLPMVTWPMGAEQFYNE 290
Query: 379 KLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHE 438
KL+T+V +GV VGA E G +ISR ++EKAVR ++ GGE+AE+ R +
Sbjct: 291 KLLTKVLRIGVNVGATELVKKG-------KLISRAQVEKAVREVI-GGEKAEERRLWAKK 342
Query: 439 LGNLANAAVQPGGSSYQNLTALIAHL 464
LG +A AAV+ GGSSY ++ + L
Sbjct: 343 LGEMAKAAVEEGGSSYNDVNKFMEEL 368
>At3g53150 glucosyltransferase - like protein
Length = 507
Score = 258 bits (660), Expect = 4e-69
Identities = 160/499 (32%), Positives = 245/499 (49%), Gaps = 46/499 (9%)
Query: 3 VESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLH 62
+ S+ + L IP +A GH+IP+ DI+ + A +G VTI+TTP N + ++ L
Sbjct: 5 IVSKAKRLHFVLIPLMAQGHLIPMVDISKILARQGNIVTIVTTPQNASRFAKTVDRARLE 64
Query: 63 T--------VPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHH--PA 112
+ P P ++ GLP E+ + + + + L+EP++ F+E P
Sbjct: 65 SGLEINVVKFPIPYKEFGLPKDCETLDTLPSKDLLRRFYDAVDKLQEPMERFLEQQDIPP 124
Query: 113 DCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCA----------MRAKRGSSSFLISG 162
C+++D C W IPR+ F+ F+L + + F I G
Sbjct: 125 SCIISDKCLFWTSRTAKRFKIPRIVFHGMCCFSLLSSHNIHLHSPHLSVSSAVEPFPIPG 184
Query: 163 LPHGDIALNASPPEALTACV------EPLLRKELKSHGVLINSFVELDGHEYVKHYERTT 216
+PH A P A E + E ++ GV++NSF EL+ Y + Y
Sbjct: 185 MPHRIEIARAQLPGAFEKLANMDDVREKMRESESEAFGVIVNSFQELEPG-YAEAYAEAI 243
Query: 217 GGHKAWLLGPASLVRGTAKEKAERGHEKSV-VSTDELLSWLSTKKPSSVLYVCFGSMCSF 275
K W +GP SL + +RG ++ +S E L +L + +P SVLYV GS+C
Sbjct: 244 N-KKVWFVGPVSLCNDRMADLFDRGSNGNIAISETECLQFLDSMRPRSVLYVSLGSLCRL 302
Query: 276 SNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRGWAPQL 335
QL E+ G+E S F+WV+ E++ E +E K + E +G++++GW+PQ
Sbjct: 303 IPNQLIELGLGLEESGKPFIWVIKTEEKHMIELDEWLKRENFEERVRGRGIVIKGWSPQA 362
Query: 336 LILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAE- 394
+IL H S G FLTHCG NS +EA+ GVP+ITWP+ +QF EKL+ EV +GV VG E
Sbjct: 363 MILSHGSTGGFLTHCGWNSTIEAICFGVPMITWPLFAEQFLNEKLIVEVLNIGVRVGVEI 422
Query: 395 --EWSMGGGRRERERVVISRDKIEKAVRRLM----------GGGEEAEQIRRRVHELGNL 442
W G ER V++ + + KA++ LM E + RRR+ EL +
Sbjct: 423 PVRW----GDEERLGVLVKKPSVVKAIKLLMDQDCQRVDENDDDNEFVRRRRRIQELAVM 478
Query: 443 ANAAVQPGGSSYQNLTALI 461
A AV+ GSS N++ LI
Sbjct: 479 AKKAVEEKGSSSINVSILI 497
>At1g73880 putative glucosyltransferase (At1g73880)
Length = 473
Score = 197 bits (501), Expect = 1e-50
Identities = 149/507 (29%), Positives = 229/507 (44%), Gaps = 85/507 (16%)
Query: 4 ESEQRPLKLYFI--PYLAAGHMIPLCDIAILFASRGQ---HVTIITTPSNTTTID---SS 55
E +P K + + P+ A GHMIPL D A RG +T++ TP N + S+
Sbjct: 5 EENNKPTKTHVLIFPFPAQGHMIPLLDFTHRLALRGGAALKITVLVTPKNLPFLSPLLSA 64
Query: 56 LPNLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPAD-- 113
+ N+ +PFPS +P G+E+ +H + L P+ ++ HP+
Sbjct: 65 VVNIEPLILPFPSHP-SIPSGVENVQDLPPSGFPLMIH-ALGNLHAPLISWITSHPSPPV 122
Query: 114 CVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRA---------------------- 151
+V+D+ W T L IPR F+PS C +
Sbjct: 123 AIVSDFFLGW----TKNLGIPRFDFSPSAAITCCILNTLWIEMPTKINEDDDNEILHFPK 178
Query: 152 -------KRGSSSFLISGLPHGDIALNASPPEALTACVEPLLRKELKSHGVLINSFVELD 204
+ S L HGD A + R + S G+++NSF ++
Sbjct: 179 IPNCPKYRFDQISSLYRSYVHGDPAWEF---------IRDSFRDNVASWGLVVNSFTAME 229
Query: 205 GHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSV 264
G Y++H +R G + W +GP + G RG SV S D ++SWL ++ + V
Sbjct: 230 G-VYLEHLKREMGHDRVWAVGPIIPLSGD-----NRGGPTSV-SVDHVMSWLDAREDNHV 282
Query: 265 LYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK 324
+YVCFGS + EQ +A G+E S V F+W V KE E++ + +GF+++
Sbjct: 283 VYVCFGSQVVLTKEQTLALASGLEKSGVHFIWAV------KEPVEKDSTRGNILDGFDDR 336
Query: 325 ----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKL 380
G+++RGWAPQ+ +L H +VGAFLTHCG NS +EAV AGV ++TWP+ DQ+ L
Sbjct: 337 VAGRGLVIRGWAPQVAVLRHRAVGAFLTHCGWNSVVEAVVAGVLMLTWPMRADQYTDASL 396
Query: 381 MTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELG 440
+ + VGV E V D++ + + G + R + EL
Sbjct: 397 VVDELKVGVRA-----------CEGPDTVPDPDELARVFADSVTGNQTE---RIKAVELR 442
Query: 441 NLANAAVQPGGSSYQNLTALIAHLKTL 467
A A+Q GSS +L I H+ +L
Sbjct: 443 KAALDAIQERGSSVNDLDGFIQHVVSL 469
>At5g12890 glucosyltransferase -like protein
Length = 488
Score = 190 bits (483), Expect = 1e-48
Identities = 150/491 (30%), Positives = 234/491 (47%), Gaps = 64/491 (13%)
Query: 4 ESEQRPLKLYFIPYLAAGHMIPLCDIA-----ILFASRGQHVTI--ITTPSNTTTIDSSL 56
E++ R L++ P++ GH+IP +A I+ +R TI I TPSN I S+L
Sbjct: 3 EAKPRNLRIVMFPFMGQGHIIPFVALALRLEKIMIMNRANKTTISMINTPSNIPKIRSNL 62
Query: 57 P---NLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFM------ 107
P ++ L +PF S GLP E+F S + + + LREP + FM
Sbjct: 63 PPESSISLIELPFNSSDHGLPHDGENFDS-LPYSLVISLLEASRSLREPFRDFMTKILKE 121
Query: 108 EHHPADCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRA--------KRGSSSFL 159
E + V+ D+ WI + E+ + + F+ S F L R+ + FL
Sbjct: 122 EGQSSVIVIGDFFLGWIGKVCKEVGVYSVIFSASGAFGLGCYRSIWLNLPHKETKQDQFL 181
Query: 160 ISGLPH-GDIA--------LNASPPEALTACVEPLLRKELKSHGVLINSFVELDGHEYVK 210
+ P G+I L A + + ++ ++ G L N+ E+D +
Sbjct: 182 LDDFPEAGEIEKTQLNSFMLEADGTDDWSVFMKKIIPGWSDFDGFLFNTVAEID--QMGL 239
Query: 211 HYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFG 270
Y R G W +GP +++ K+ R E++V S WL +K SV+YVCFG
Sbjct: 240 SYFRRITGVPVWPVGP--VLKSPDKKVGSRSTEEAVKS------WLDSKPDHSVVYVCFG 291
Query: 271 SMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK------ 324
SM S + E+A +E+S F+WVV G E + E + + +LPEGFEE+
Sbjct: 292 SMNSILQTHMLELAMALESSEKNFIWVV-RPPIGVEVKSEFDVKGYLPEGFEERITRSER 350
Query: 325 GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEV 384
G++++ WAPQ+ IL H + FL+HCG NS LE++S GVP++ WP+ +QF+ LM +
Sbjct: 351 GLLVKKWAPQVDILSHKATCVFLSHCGWNSILESLSHGVPLLGWPMAAEQFFNSILMEKH 410
Query: 385 RGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLAN 444
GV VEV +R I D I ++ +M E ++IR++ E+ L
Sbjct: 411 IGVSVEVA-----------RGKRCEIKCDDIVSKIKLVMEETEVGKEIRKKAREVKELVR 459
Query: 445 AAVQPG--GSS 453
A+ G GSS
Sbjct: 460 RAMVDGVKGSS 470
>At2g16890 glucosyltransferase like protein
Length = 478
Score = 188 bits (477), Expect = 6e-48
Identities = 147/510 (28%), Positives = 235/510 (45%), Gaps = 69/510 (13%)
Query: 1 MDVESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQH-----VTIITTPSNTTTID-- 53
M V + + L+ P+++ GH+IPL L + VT+ TTP N I
Sbjct: 1 MSVSTHHHHVVLF--PFMSKGHIIPLLQFGRLLLRHHRKEPTITVTVFTTPKNQPFISDF 58
Query: 54 -SSLPNLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHP- 111
S P + + ++PFP G+P G+E+ + + KLL+ + ++ P
Sbjct: 59 LSDTPEIKVISLPFPENITGIPPGVENTEKLPSMSLFVPFTRATKLLQPFFEETLKTLPK 118
Query: 112 ADCVVADYCFPWIDDLTTELHIPR-LSFNPSPLFALCAM-----------RAKRGSSSFL 159
+V+D W + + +IPR +S+ + A ++ +K +
Sbjct: 119 VSFMVSDGFLWWTSESAAKFNIPRFVSYGMNSYSAAVSISVFKHELFTEPESKSDTEPVT 178
Query: 160 ISGLP-----HGDIALNASPPEALTACVE---PLLRKELKSHGVLINSFVELDGHEYVKH 211
+ P D + PE A +E ++ SHG L+NSF EL+ +V +
Sbjct: 179 VPDFPWIKVKKCDFDHGTTEPEESGAALELSMDQIKSTTTSHGFLVNSFYELES-AFVDY 237
Query: 212 YERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSS--VLYVCF 269
+ K+W +GP L + ++G K + WL K+ VLYV F
Sbjct: 238 NNNSGDKPKSWCVGPLCLT-----DPPKQGSAKPA-----WIHWLDQKREEGRPVLYVAF 287
Query: 270 GSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFE----EKG 325
G+ SN+QL E+A G+E S V F+WV ++ E+ + EGF E G
Sbjct: 288 GTQAEISNKQLMELAFGLEDSKVNFLWVT-----------RKDVEEIIGEGFNDRIRESG 336
Query: 326 MILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVR 385
MI+R W Q IL H SV FL+HCG NSA E++ GVP++ WP++ +Q K++ E
Sbjct: 337 MIVRDWVDQWEILSHESVKGFLSHCGWNSAQESICVGVPLLAWPMMAEQPLNAKMVVEEI 396
Query: 386 GVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANA 445
VGV V E+ S+ G ++R+++ ++ LM GE + R+ V E +A A
Sbjct: 397 KVGVRVETEDGSVKG--------FVTREELSGKIKELM-EGETGKTARKNVKEYSKMAKA 447
Query: 446 AVQPG-GSSYQNLTALIAHLKTLRDSNSVS 474
A+ G GSS++NL ++ L RDSN S
Sbjct: 448 ALVEGTGSSWKNLDMILKELCKSRDSNGAS 477
>At5g03490 UDPG glucosyltransferase - like protein
Length = 465
Score = 183 bits (465), Expect = 2e-46
Identities = 143/487 (29%), Positives = 227/487 (46%), Gaps = 76/487 (15%)
Query: 8 RPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTV--- 64
+P + P+ A GH++PL D+ RG +V++I TP N T + S L + H +V
Sbjct: 16 KPPHIVVFPFPAQGHLLPLLDLTHQLCLRGFNVSVIVTPGNLTYL-SPLLSAHPSSVTSV 74
Query: 65 --PFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPAD--CVVADYC 120
PFP L G+E+ + + ++ LREPI + + HP +++D+
Sbjct: 75 VFPFPPHP-SLSPGVENVKDVGN-SGNLPIMASLRQLREPIINWFQSHPNPPIALISDFF 132
Query: 121 FPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNASP---PEA 177
W DL ++ IPR +F F + ++ + + S P + L +P E
Sbjct: 133 LGWTHDLCNQIGIPRFAFFSISFFLVSVLQFCFENIDLIKSTDPIHLLDLPRAPIFKEEH 192
Query: 178 LTACVEPLLRK-------------ELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLL 224
L + V L+ L S+G + NS L+ +Y+++ ++ G + +++
Sbjct: 193 LPSIVRRSLQTPSPDLESIKDFSMNLLSYGSVFNSSEILED-DYLQYVKQRMGHDRVYVI 251
Query: 225 GPASLVRGTAKEKAERGHEKSVVSTD-ELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEM 283
GP + K + S D LLSWL SVLYVCFGS + + +Q +
Sbjct: 252 GPLCSIGSGLKSNSG--------SVDPSLLSWLDGSPNGSVLYVCFGSQKALTKDQCDAL 303
Query: 284 ACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILG 339
A G+E S FVWVV K+ +P+GFE++ G+++RGW QL +L
Sbjct: 304 ALGLEKSMTRFVWVV--------------KKDPIPDGFEDRVSGRGLVVRGWVSQLAVLR 349
Query: 340 HPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMG 399
H +VG FL+HCG NS LE +++G I+ WP+ DQF +L+ E GV V V
Sbjct: 350 HVAVGGFLSHCGWNSVLEGITSGAVILGWPMEADQFVNARLLVEHLGVAVRV-------- 401
Query: 400 GGRRERERVVISRDKIEKAVRRLMG-GGEE----AEQIRRRVHELGNLANAAVQPGGSSY 454
E V D++ + + MG GG E AE+IRR+ A + GSS
Sbjct: 402 ---CEGGETVPDSDELGRVIAETMGEGGREVAARAEEIRRKTEA------AVTEANGSSV 452
Query: 455 QNLTALI 461
+N+ L+
Sbjct: 453 ENVQRLV 459
>At1g06000 unknown protein
Length = 435
Score = 182 bits (463), Expect = 3e-46
Identities = 138/466 (29%), Positives = 216/466 (45%), Gaps = 62/466 (13%)
Query: 15 IPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHT------VPFPS 68
IP+ +GHM+P D+ RG VT++ TP N++ +D+ L +LH +PFPS
Sbjct: 14 IPFPQSGHMVPHLDLTHQILLRGATVTVLVTPKNSSYLDA-LRSLHSPEHFKTLILPFPS 72
Query: 69 RQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPA----DCVV-ADYCFPW 123
+P G+ES L+ + + L +P+ F+ P D ++ + + PW
Sbjct: 73 HPC-IPSGVESLQQ-LPLEAIVHMFDALSRLHDPLVDFLSRQPPSDLPDAILGSSFLSPW 130
Query: 124 IDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSSFLISGLPHGDIALNASPPEALTACVE 183
I+ + I +SF P ++ M A+ S F + TA E
Sbjct: 131 INKVADAFSIKSISFLPINAHSISVMWAQEDRSFF----------------NDLETATTE 174
Query: 184 PLLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHE 243
S+G++INSF +L+ R H+ W +GP K +RG +
Sbjct: 175 --------SYGLVINSFYDLEPEFVETVKTRFLNHHRIWTVGPLL----PFKAGVDRGGQ 222
Query: 244 KSVVSTDELLSWL-STKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEK 302
S+ ++ +WL S + +SV+YV FGS + EQ +A +E SSV F+W V
Sbjct: 223 SSIPPA-KVSAWLDSCPEDNSVVYVGFGSQIRLTAEQTAALAAALEKSSVRFIWAVRDAA 281
Query: 303 RGKEEEEEEEKEKWLPEGFEE----KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEA 358
+ + +E +P GFEE KG+++RGWAPQ +IL H +VG++LTH G S LE
Sbjct: 282 KKVNSSDNSVEEDVIPAGFEERVKEKGLVIRGWAPQTMILEHRAVGSYLTHLGWGSVLEG 341
Query: 359 VSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKA 418
+ GV ++ WP+ D F+ L+ + V VG R+ V S +K
Sbjct: 342 MVGGVMLLAWPMQADHFFNTTLIVDKLRAAVRVG----------ENRDSVPDS----DKL 387
Query: 419 VRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
R L E R + +L A A++ GGSSY+NL L+A +
Sbjct: 388 ARILAESAREDLPERVTLMKLREKAMEAIKEGGSSYKNLDELVAEM 433
>At1g51210 putative glucosyl transferase
Length = 433
Score = 175 bits (444), Expect = 4e-44
Identities = 129/403 (32%), Positives = 194/403 (48%), Gaps = 57/403 (14%)
Query: 16 PYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLH-------TVPFPS 68
PY A GH++PL D+ RG V+II TP N + P L H T+PFP
Sbjct: 25 PYPAQGHLLPLLDLTHQLCLRGLTVSIIVTPKNLPYLS---PLLSAHPSAVSVVTLPFPH 81
Query: 69 RQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCV--VADYCFPWIDD 126
+ +P G+E+ + ++ LREPI ++ HP V ++D+ W
Sbjct: 82 HPL-IPSGVENVKDLGGYGNPL-IMASLRQLREPIVNWLSSHPNPPVALISDFFLGW--- 136
Query: 127 LTTELHIPRLSFNPSPLFALCAMRAK-------RGSSSFLISGLPHGDIALNAS-----P 174
T +L IPR +F S F + + +S LP + P
Sbjct: 137 -TKDLGIPRFAFFSSGAFLASILHFVSDKPHLFESTEPVCLSDLPRSPVFKTEHLPSLIP 195
Query: 175 PEALTACVEPLLRKELK--SHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRG 232
L+ +E + + S+G + N+ L+ +Y+++ ++ ++ + +GP S V G
Sbjct: 196 QSPLSQDLESVKDSTMNFSSYGCIFNTCECLE-EDYMEYVKQKVSENRVFGVGPLSSV-G 253
Query: 233 TAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSV 292
+KE + S V LLSWL SVLY+CFGS + EQ ++A G+E S
Sbjct: 254 LSKEDSV-----SNVDAKALLSWLDGCPDDSVLYICFGSQKVLTKEQCDDLALGLEKSMT 308
Query: 293 EFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEK----GMILRGWAPQLLILGHPSVGAFLT 348
FVWVV K+ +P+GFE++ GMI+RGWAPQ+ +L H +VG FL
Sbjct: 309 RFVWVV--------------KKDPIPDGFEDRVAGRGMIVRGWAPQVAMLSHVAVGGFLI 354
Query: 349 HCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEV 391
HCG NS LEA+++G I+ WP+ DQF +L+ E GV V V
Sbjct: 355 HCGWNSVLEAMASGTMILAWPMEADQFVDARLVVEHMGVAVSV 397
>At4g15480 indole-3-acetate beta-glucosyltransferase like protein
Length = 490
Score = 151 bits (382), Expect = 7e-37
Identities = 135/480 (28%), Positives = 215/480 (44%), Gaps = 42/480 (8%)
Query: 9 PLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHTVPFPS 68
P+ + + + GH+ PL + L AS+G VT +TT + + + P S
Sbjct: 17 PIHVMLVSFQGQGHVNPLLRLGKLIASKGLLVTFVTTELWGKKMRQANKIVDGELKPVGS 76
Query: 69 RQVGLPDGIESFSSSADLQTTFKVH------QGIKLLREPIQLFME-HHPADCVVADYCF 121
+ E ++ D + F ++ GI+ + + ++ + E + P C++ +
Sbjct: 77 GSIRFEFFDEEWAEDDDRRADFSLYIAHLESVGIREVSKLVRRYEEANEPVSCLINNPFI 136
Query: 122 PWIDDLTTELHIPRLSFNPSPLFALCAM-RAKRGSSSFLISGLPHGDIALNASPP---EA 177
PW+ + E +IP A + GS SF P D+ L P +
Sbjct: 137 PWVCHVAEEFNIPCAVLWVQSCACFSAYYHYQDGSVSFPTETEPELDVKLPCVPVLKNDE 196
Query: 178 LTACVEPLLR------------KEL-KSHGVLINSFVELDGHEYVKHYERTTGGHKAWLL 224
+ + + P R K L KS VLI+SF L+ + V Y + K +
Sbjct: 197 IPSFLHPSSRFTGFRQAILGQFKNLSKSFCVLIDSFDSLE--QEVIDYMSSLCPVKT--V 252
Query: 225 GPASLVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMA 284
GP V T KS TD+ L WL ++ SSV+Y+ FG++ EQ+ E+A
Sbjct: 253 GPLFKVARTVTSDVSGDICKS---TDKCLEWLDSRPKSSVVYISFGTVAYLKQEQIEEIA 309
Query: 285 CGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRGWAPQLLILGHPSVG 344
G+ S + F+WV+ + E ++ + KGMI+ W PQ +L HPSV
Sbjct: 310 HGVLKSGLSFLWVIRPPPHDLKVETHVLPQELKESSAKGKGMIV-DWCPQEQVLSHPSVA 368
Query: 345 AFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRE 404
F+THCG NS +E++S+GVP++ P GDQ + +V GV +G G E
Sbjct: 369 CFVTHCGWNSTMESLSSGVPVVCCPQWGDQVTDAVYLIDVFKTGVR-------LGRGATE 421
Query: 405 RERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALIAHL 464
ERVV + EK + + GE+AE++R+ + A AAV PGGSS +N + L
Sbjct: 422 -ERVVPREEVAEKLLEATV--GEKAEELRKNALKWKAEAEAAVAPGGSSDKNFREFVEKL 478
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,554,202
Number of Sequences: 26719
Number of extensions: 533277
Number of successful extensions: 3382
Number of sequences better than 10.0: 133
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 2867
Number of HSP's gapped (non-prelim): 201
length of query: 481
length of database: 11,318,596
effective HSP length: 103
effective length of query: 378
effective length of database: 8,566,539
effective search space: 3238151742
effective search space used: 3238151742
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0367.1


