

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0360.7
(485 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
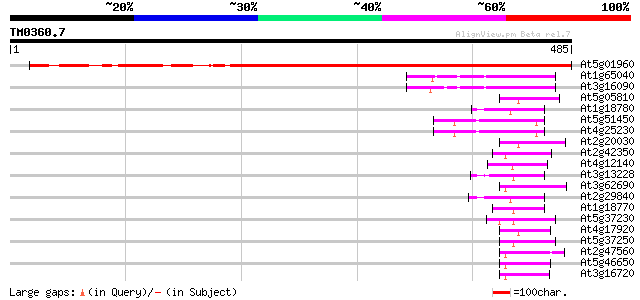
Score E
Sequences producing significant alignments: (bits) Value
At5g01960 unknown protein 536 e-152
At1g65040 66 4e-11
At3g16090 putative zinc finger protein 63 3e-10
At5g05810 putative protein 53 3e-07
At1g18780 hypothetical protein 53 4e-07
At5g51450 unknown protein 52 7e-07
At4g25230 unknown protein 52 1e-06
At2g20030 putative RING zinc finger protein 51 1e-06
At2g42350 putative RING zinc finger protein 50 3e-06
At4g12140 putative protein 50 4e-06
At3g13228 unknown protein 50 4e-06
At3g62690 RING-H2 zinc finger protein ATL5 49 6e-06
At2g29840 hypothetical protein 49 6e-06
At1g18770 hypothetical protein 49 6e-06
At5g37230 putative protein 49 8e-06
At4g17920 putative protein 48 1e-05
At5g37250 putative protein 48 1e-05
At2g47560 unknown protein (At2g47560) 48 1e-05
At5g46650 putative protein 47 2e-05
At3g16720 putative RING zinc finger protein 47 2e-05
>At5g01960 unknown protein
Length = 426
Score = 536 bits (1381), Expect = e-152
Identities = 281/470 (59%), Positives = 336/470 (70%), Gaps = 48/470 (10%)
Query: 18 SPSSSSGGFNSSDDHNNGSVSWYGMRLPSVNPLRSPLSLLLDYSGILTQTHDSGSVIVNN 77
+P +S F S N+ L + N +++P+S LL+YSG+
Sbjct: 3 TPGASHDSFRGSPRRNS--------ILSASNIIQAPISTLLEYSGLFRARPS-------- 46
Query: 78 GVPASEIRSQVQMQPFDDAVGGSAGEVAIRIIGAGEHD-QNQIGEVGFDGCDNLMGERRG 136
P+ E + V DD+ G S GEVAIRIIG E D + + G L+G
Sbjct: 47 --PSHEAETLVS----DDSSGLSNGEVAIRIIGNTEQDAETDTNALREPGHSELLGS--- 97
Query: 137 AGMSPLDDDPEGRGGAAVNETVPLVSSSASLAGSVQGDGDAAGNGAESDSRGSSSYQRYD 196
S DP G ++ P GD AAG+ A DS YQRYD
Sbjct: 98 ---SATQVDPMGGASEGASQAAP-------------GD-PAAGDAASRDS----PYQRYD 136
Query: 197 IQQVAKWIEQILPFSLLLLIVFIRQHLQGFFVTIWISAVMFKSNEIVKKQTALKGDRKVS 256
IQQ A+WIEQILPFSLLLL+VFIRQHLQGFFV IWI+AVMFKSN+I+KKQTALKG+R +S
Sbjct: 137 IQQAARWIEQILPFSLLLLVVFIRQHLQGFFVAIWIAAVMFKSNDILKKQTALKGERHIS 196
Query: 257 VLVGISFAFILQVTCIYWWYRNDDLLYPLIMLPPRPTP-FWHSVFIILVNDVLVRQAAMV 315
L+GIS AF V +YWW+R DDLLYPLIMLPP+ P FWH++FII+VND LVRQA+M+
Sbjct: 197 ALIGISVAFTAHVVGVYWWFRKDDLLYPLIMLPPKSIPPFWHAIFIIVVNDTLVRQASMI 256
Query: 316 FKCFLLIYYKNGKGHNFRRQAQMLTLVEYMLLLYRALLPTLVWYRFFLNRDYGSLFSSLT 375
FKCFLL+YYKN +G N+R+Q Q+LTLVEY +LLYR+LLPT VWYRFFLN+DYGSLFSSL
Sbjct: 257 FKCFLLMYYKNSRGRNYRKQGQLLTLVEYFMLLYRSLLPTPVWYRFFLNKDYGSLFSSLM 316
Query: 376 TGLYLTFKLTSIIEKVRGFISAFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPV 435
TGLYLTFKLTS++EKV+ F +A KALSRK+VHYG YAT EQVNAAGDLCAICQEKMH+P+
Sbjct: 317 TGLYLTFKLTSVVEKVQSFFTALKALSRKEVHYGSYATTEQVNAAGDLCAICQEKMHTPI 376
Query: 436 LLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTSLFFQLF 485
LLRCKH+FCE+CVSEWFERERTCPLCRALVKPADL++FGDGSTSLFFQ+F
Sbjct: 377 LLRCKHMFCEDCVSEWFERERTCPLCRALVKPADLKSFGDGSTSLFFQIF 426
>At1g65040
Length = 537
Score = 66.2 bits (160), Expect = 4e-11
Identities = 48/131 (36%), Positives = 68/131 (51%), Gaps = 6/131 (4%)
Query: 344 YMLLLYRALLPTLVWYRFFLN--RDYGSLFSSLTTGLYLTFKLTSIIEKVRGFISAFKAL 401
+ L L R LL ++ FFL +YG L L LY TF+ I +V ++ K
Sbjct: 214 FYLELVRDLLHLSMYLCFFLMIFMNYG-LPLHLIRELYETFRNFKI--RVTDYLRYRKIT 270
Query: 402 SRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLC 461
S + + AT E++++ C IC+E+M S L C H+F C+ W ER+ TCP C
Sbjct: 271 SNMNDRFPD-ATPEELSSNDATCIICREEMTSAKKLVCGHLFHVHCLRSWLERQNTCPTC 329
Query: 462 RALVKPADLQT 472
RALV PA+ T
Sbjct: 330 RALVVPAENAT 340
>At3g16090 putative zinc finger protein
Length = 492
Score = 63.2 bits (152), Expect = 3e-10
Identities = 45/131 (34%), Positives = 67/131 (50%), Gaps = 6/131 (4%)
Query: 344 YMLLLYRALLPTLVWYRFF--LNRDYGSLFSSLTTGLYLTFKLTSIIEKVRGFISAFKAL 401
+ L L R LL ++ FF + +YG L LY TF+ I +V ++ K
Sbjct: 214 FYLELIRDLLHLSMYICFFFVIFMNYGVPLHLLRE-LYETFRNFQI--RVSDYLRYRKIT 270
Query: 402 SRKDVHYGVYATMEQVNAAGDLCAICQEKMHSPVLLRCKHIFCEECVSEWFERERTCPLC 461
S + + AT E++ A+ C IC+E+M + L C H+F C+ W ER++TCP C
Sbjct: 271 SNMNDRFPD-ATPEELTASDATCIICREEMTNAKKLICGHLFHVHCLRSWLERQQTCPTC 329
Query: 462 RALVKPADLQT 472
RALV P + T
Sbjct: 330 RALVVPPENAT 340
>At5g05810 putative protein
Length = 407
Score = 53.1 bits (126), Expect = 3e-07
Identities = 24/56 (42%), Positives = 29/56 (50%), Gaps = 4/56 (7%)
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGD 475
CA+C + +LR CKH F ECV W + TCPLCR V P D+ GD
Sbjct: 146 CAVCLARFEPTEVLRLLPKCKHAFHVECVDTWLDAHSTCPLCRYRVDPEDILLIGD 201
>At1g18780 hypothetical protein
Length = 325
Score = 52.8 bits (125), Expect = 4e-07
Identities = 30/66 (45%), Positives = 33/66 (49%), Gaps = 8/66 (12%)
Query: 400 ALSRKDVHYGVYATMEQVNAAGDLCAICQEKM---HSPVLLRCKHIFCEECVSEWFERER 456
AL+RK Y V D+C IC E+ S V L C H F EECV EWF R
Sbjct: 258 ALNRK-----TYKKASGVVCENDVCTICLEEFDDGRSIVTLPCGHEFDEECVLEWFVRSH 312
Query: 457 TCPLCR 462
CPLCR
Sbjct: 313 VCPLCR 318
>At5g51450 unknown protein
Length = 577
Score = 52.0 bits (123), Expect = 7e-07
Identities = 32/103 (31%), Positives = 53/103 (51%), Gaps = 9/103 (8%)
Query: 367 YGSLFSSLTTGLYLTFK--LTSIIEKVRGFISAFKALSRKDVHYGVY-ATMEQVNAAGDL 423
+G F + L+L + L+SI+++++G+I AL +H + AT E++ D
Sbjct: 279 HGMAFHLVDAVLFLNIRALLSSILKRIKGYIKLRVALGA--LHAALLDATSEELRDYDDE 336
Query: 424 CAICQEKMHSPVLLRCKHIFCEECVSEWFER----ERTCPLCR 462
CAIC+E M L C H+F C+ W ++ +CP CR
Sbjct: 337 CAICREPMAKAKRLHCNHLFHLGCLRSWLDQGLNEVYSCPTCR 379
>At4g25230 unknown protein
Length = 578
Score = 51.6 bits (122), Expect = 1e-06
Identities = 32/103 (31%), Positives = 54/103 (52%), Gaps = 9/103 (8%)
Query: 367 YGSLFSSLTTGLYLTFK--LTSIIEKVRGFISAFKALSRKDVHYGVY-ATMEQVNAAGDL 423
+G F + L+L + L++I+++++G+I AL +H + AT E++ A D
Sbjct: 279 HGIAFHLVDAVLFLNIRALLSAILKRIKGYIKLRIALGA--LHAALPDATSEELRAYDDE 336
Query: 424 CAICQEKMHSPVLLRCKHIFCEECVSEWFER----ERTCPLCR 462
CAIC+E M L C H+F C+ W ++ +CP CR
Sbjct: 337 CAICREPMAKAKRLHCNHLFHLGCLRSWLDQGLNEVYSCPTCR 379
>At2g20030 putative RING zinc finger protein
Length = 390
Score = 51.2 bits (121), Expect = 1e-06
Identities = 25/62 (40%), Positives = 33/62 (52%), Gaps = 5/62 (8%)
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVK-PADLQTFGDGST 478
C++C K +LR C+H F C+ +W E+ TCPLCR V DL G+ ST
Sbjct: 124 CSVCLSKFEDVEILRLLPKCRHAFHIGCIDQWLEQHATCPLCRNRVNIEDDLSVLGNSST 183
Query: 479 SL 480
SL
Sbjct: 184 SL 185
>At2g42350 putative RING zinc finger protein
Length = 217
Score = 50.1 bits (118), Expect = 3e-06
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 4/55 (7%)
Query: 418 NAAGDLCAIC----QEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPA 468
+ AG CA+C +EK ++ +L CKH+F CV W + TCP+CR +P+
Sbjct: 94 DVAGTECAVCLSLLEEKDNARMLPNCKHVFHVSCVDTWLTTQSTCPVCRTEAEPS 148
>At4g12140 putative protein
Length = 202
Score = 49.7 bits (117), Expect = 4e-06
Identities = 19/57 (33%), Positives = 32/57 (55%), Gaps = 5/57 (8%)
Query: 414 MEQVNAAGDLCAICQEKMHS-----PVLLRCKHIFCEECVSEWFERERTCPLCRALV 465
++ N D C+IC + + S P + C H+F C+ EW +R+ TCP+CR ++
Sbjct: 143 LKSFNMETDSCSICLQSLVSSSKTGPTRMSCSHVFHSSCLVEWLKRKNTCPMCRTVL 199
>At3g13228 unknown protein
Length = 325
Score = 49.7 bits (117), Expect = 4e-06
Identities = 26/67 (38%), Positives = 38/67 (55%), Gaps = 11/67 (16%)
Query: 399 KALSRKDVHYGVYATMEQVNAAGDLCAICQEKMHSP---VLLRCKHIFCEECVSEWFERE 455
K+L+RK +Y +++N G+ C IC E+ ++ V L C H F +EC +WFE
Sbjct: 258 KSLTRK-----IY---DKINYTGERCTICLEEFNAGGILVALPCGHDFDDECAVKWFETN 309
Query: 456 RTCPLCR 462
CPLCR
Sbjct: 310 HFCPLCR 316
>At3g62690 RING-H2 zinc finger protein ATL5
Length = 257
Score = 48.9 bits (115), Expect = 6e-06
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 4/62 (6%)
Query: 424 CAIC----QEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTS 479
C++C +E VL +C H+F +C+ WF +CPLCRA V+PA T + +
Sbjct: 113 CSVCLSEFEEDDEGRVLPKCGHVFHVDCIDTWFRSRSSCPLCRAPVQPAQPVTEPEPVAA 172
Query: 480 LF 481
+F
Sbjct: 173 VF 174
>At2g29840 hypothetical protein
Length = 310
Score = 48.9 bits (115), Expect = 6e-06
Identities = 26/69 (37%), Positives = 35/69 (50%), Gaps = 8/69 (11%)
Query: 397 AFKALSRKDVHYGVYATMEQVNAAGDLCAICQEKM---HSPVLLRCKHIFCEECVSEWFE 453
A ++L+RK Y V ++C+IC E+ S V L C H F +EC +WFE
Sbjct: 239 AVESLNRK-----TYKKASDVVGENEMCSICLEEFDDGRSIVALPCGHEFDDECALKWFE 293
Query: 454 RERTCPLCR 462
CPLCR
Sbjct: 294 TNHDCPLCR 302
>At1g18770 hypothetical protein
Length = 106
Score = 48.9 bits (115), Expect = 6e-06
Identities = 21/48 (43%), Positives = 29/48 (59%), Gaps = 3/48 (6%)
Query: 418 NAAGDLCAICQEKMHSP---VLLRCKHIFCEECVSEWFERERTCPLCR 462
++ G++C IC E+ V L C H F +ECV +WFE +CPLCR
Sbjct: 53 SSTGEMCIICLEEFSEGRRVVTLPCGHDFDDECVLKWFETNHSCPLCR 100
>At5g37230 putative protein
Length = 208
Score = 48.5 bits (114), Expect = 8e-06
Identities = 24/69 (34%), Positives = 36/69 (51%), Gaps = 9/69 (13%)
Query: 413 TMEQVNAAGD---LCAICQEKMHSP------VLLRCKHIFCEECVSEWFERERTCPLCRA 463
TME N + C+IC E +L C H+F + C+ +W +R+R+CPLCR
Sbjct: 139 TMELTNLGDEEETTCSICMEDFSESHDDNIILLPDCFHLFHQSCIFKWLKRQRSCPLCRR 198
Query: 464 LVKPADLQT 472
+ DL+T
Sbjct: 199 VPYEEDLET 207
>At4g17920 putative protein
Length = 289
Score = 48.1 bits (113), Expect = 1e-05
Identities = 21/48 (43%), Positives = 28/48 (57%), Gaps = 4/48 (8%)
Query: 424 CAICQEKMHSPVLLR----CKHIFCEECVSEWFERERTCPLCRALVKP 467
CAIC + +LR C H+F +EC+ WFE RTCP+CR + P
Sbjct: 110 CAICLLEFDGDHVLRLLTTCYHVFHQECIDLWFESHRTCPVCRRDLDP 157
>At5g37250 putative protein
Length = 192
Score = 47.8 bits (112), Expect = 1e-05
Identities = 21/55 (38%), Positives = 31/55 (56%), Gaps = 6/55 (10%)
Query: 424 CAICQEKMHSP------VLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQT 472
C+IC E +L C H+F + C+ EW +R+R+CPLCR + DL+T
Sbjct: 137 CSICLEDFSESHDDNIILLPDCFHLFHQNCIFEWLKRQRSCPLCRRVPYEEDLET 191
>At2g47560 unknown protein (At2g47560)
Length = 227
Score = 47.8 bits (112), Expect = 1e-05
Identities = 23/60 (38%), Positives = 33/60 (54%), Gaps = 5/60 (8%)
Query: 424 CAIC----QEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKPADLQTFGDGSTS 479
C++C +E+ +L +C H F +C+ WF TCPLCRA V+P Q GS+S
Sbjct: 108 CSVCLSEFEEEDEGRLLPKCGHSFHVDCIDTWFRSRSTCPLCRAPVQP-PFQVIETGSSS 166
>At5g46650 putative protein
Length = 289
Score = 47.4 bits (111), Expect = 2e-05
Identities = 19/49 (38%), Positives = 29/49 (58%), Gaps = 5/49 (10%)
Query: 424 CAIC-----QEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVKP 467
CAIC +E + +L C H+F +EC+ +W E +TCP+CR + P
Sbjct: 114 CAICLLEFEEEHILLRLLTTCYHVFHQECIDQWLESNKTCPVCRRNLDP 162
>At3g16720 putative RING zinc finger protein
Length = 304
Score = 47.0 bits (110), Expect = 2e-05
Identities = 20/47 (42%), Positives = 27/47 (56%), Gaps = 4/47 (8%)
Query: 424 CAIC----QEKMHSPVLLRCKHIFCEECVSEWFERERTCPLCRALVK 466
CA+C +E VL C+H F +C+ WF TCPLCR+LV+
Sbjct: 119 CAVCLSEFEESETGRVLPNCQHTFHVDCIDMWFHSHSTCPLCRSLVE 165
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,249,568
Number of Sequences: 26719
Number of extensions: 505636
Number of successful extensions: 2043
Number of sequences better than 10.0: 353
Number of HSP's better than 10.0 without gapping: 261
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 1697
Number of HSP's gapped (non-prelim): 398
length of query: 485
length of database: 11,318,596
effective HSP length: 103
effective length of query: 382
effective length of database: 8,566,539
effective search space: 3272417898
effective search space used: 3272417898
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0360.7


