

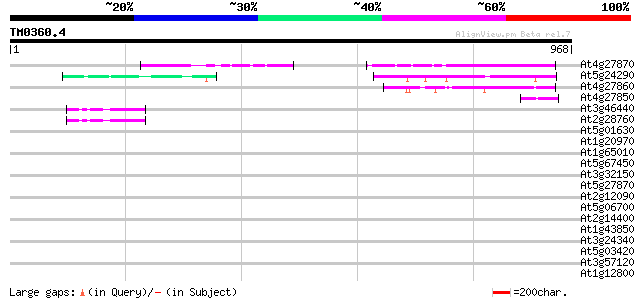
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0360.4
(968 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g27870 unknown protein 182 8e-46
At5g24290 unknown protein 160 2e-39
At4g27860 unknown protein 154 3e-37
At4g27850 putative proline-rich protein 49 2e-05
At3g46440 dTDP-glucose 4-6-dehydratases-like protein 45 3e-04
At2g28760 putative nucleotide-sugar dehydratase 43 8e-04
At5g01630 putative protein 36 0.12
At1g20970 hypothetical protein 35 0.16
At1g65010 hypothetical protein 34 0.35
At5g67450 Cys2/His2-type zinc finger protein 1 (dbj|BAA85108.1) 34 0.46
At3g32150 hypothetical protein 34 0.46
At5g27870 pectin methyl-esterase - like protein 33 0.78
At2g12090 pseudogene; similar to MURA transposase of maize Muta... 33 0.78
At5g06700 unknown protein 33 1.0
At2g14400 putative retroelement pol polyprotein 33 1.0
At1g43850 SEUSS transcriptional co-regulator 33 1.0
At3g24340 hypothetical protein 32 1.3
At5g03420 putative protein 32 1.7
At3g57120 unknown protein 32 1.7
At1g12800 unknown protein 32 1.7
>At4g27870 unknown protein
Length = 761
Score = 182 bits (462), Expect = 8e-46
Identities = 122/335 (36%), Positives = 188/335 (55%), Gaps = 22/335 (6%)
Query: 616 FLAPTTIGSLILEQSQKDVDGTSENIKNSYSSFTQSSVQSSGSSVPASVVASNKQTSGID 675
FLAP ++ EQ+Q+ +D + + + + + S S++ N T+G D
Sbjct: 408 FLAP-----MVEEQTQQKIDNDDSSTADGNHTSDKGRLSPIQPSHGMSIL--NTVTNGPD 460
Query: 676 AIFPSKP---DLAPIEEVQKDINKEISASDKKESKGGDVIVVVDREAIESTTSQTADNVP 732
+ + AP+ KD + S +D +K V+ DR I + D +
Sbjct: 461 GLKVETTIHEEGAPLLFEGKD-TPDTSTADFGLTKVTGVMDTGDRGVITGPANPEID-IS 518
Query: 733 VEGAVVPGSDTQVLIDEQPRDETGERQQWEVLKSIVYGGLVESITSLGIVSSAVSSGAAP 792
+ GS + L+ R + ++ E+LKSIVYGGL+E+ITSLG++SSA SGA+
Sbjct: 519 AGNLLEEGSLREPLM----RRVVVQGRKLEILKSIVYGGLLEAITSLGVISSAAGSGASM 574
Query: 793 LNIIALGLANLVGGLFILGHNLNDLKNNH---SGGDQQQTNAQEDQ---YHELLGRRANF 846
LNI+ LGLANL+GGL ++ HNL +L+ + + QTN +E++ Y LLGRR NF
Sbjct: 575 LNILVLGLANLLGGLILIIHNLQELREEEPIRTTTEDNQTNGREEEEGRYKRLLGRRENF 634
Query: 847 LLHAVVAVISFIIFGAVPLAIYGVLINKNYSSDVKLGIVAATSVVCIILLAIAKVYTRKP 906
LHA VA++SFII G +P +Y ++ ++ D K+ V S+ CI+LLAIAK + R P
Sbjct: 635 TLHATVAILSFIITGILPPVVYYFSFSEKHNKDYKVASVFGASLFCIVLLAIAKAHVRYP 694
Query: 907 PKYYFKTMLYYVALALGASGISYIAGILIKELLEK 941
Y K++LYY ++A+ SGISY+ G +++LLEK
Sbjct: 695 RGSYLKSILYYGSIAVSVSGISYVVGNFLEQLLEK 729
Score = 97.4 bits (241), Expect = 3e-20
Identities = 78/267 (29%), Positives = 119/267 (44%), Gaps = 44/267 (16%)
Query: 227 GSELVKEID-QDLKEFDVEVVLAKQETHDLFCPNCNSCITKRVILKKRKRNI--HSLDNK 283
G + +E+D +D++ DVE ++ KQETHDL+CPNC+SCITK+VILK+RKR I H L +
Sbjct: 183 GEVIEEEVDFEDVEYHDVENMMDKQETHDLYCPNCDSCITKKVILKRRKRKIRRHELGDS 242
Query: 284 AKRDKLDIVASSDPVNSSAQESDRGVSENVTSEIVSVETPDDNYHPEREPESDPEVFRCL 343
+ + + S+ S + +E+ VF+CL
Sbjct: 243 KRPHLTEPLFHSEDNLPSLDGGENSANESF-------------------------VFKCL 277
Query: 344 ACFSFFIPSGKGFNLFRNFGGSRKQETSQNATSITAQNLQNPSTLPGSNANWFTSLFTSN 403
+CF+ FIP G S+ Q + Q NP ++NWF+S+F N
Sbjct: 278 SCFTIFIPKGV---------SSKPIPPRQGVEGLKIQ--PNPQVEATGDSNWFSSIFGLN 326
Query: 404 KGKTATAKGNTSLEHSTTAHVEQHCSTSVTSNVPASTDIGHLEGSLADTALIKNAKPAAE 463
K ++A +G S S+ S+ V S G+L DT +P A
Sbjct: 327 KKESAIQQGGAS---SSVLEANPPPRESIVPVVNPSR--GNLSPMRKDTTGSAVVQPDAA 381
Query: 464 NGGMSSQISSTAESVKIGPGTENGIKF 490
++ + T+E V G +G KF
Sbjct: 382 TSIQVAKSNDTSEIVNNGAIVGDGQKF 408
>At5g24290 unknown protein
Length = 550
Score = 160 bits (406), Expect = 2e-39
Identities = 115/346 (33%), Positives = 175/346 (50%), Gaps = 38/346 (10%)
Query: 628 EQSQKDVDGTS--ENIKNSYSSFTQSSVQSSGSSVPASVVASNKQTSGIDAIFPSKPDL- 684
E+S D + S EN+ + ++ S V+ K+ +++ KPD+
Sbjct: 187 ERSSSDSEEKSNLENLLATQENYELYCPSCSTCITRNVVLKKRKRGKHVNSSLDLKPDIP 246
Query: 685 -------APIEEVQKDIN---KEISASDKKESKGGDVI--VVVD-----REAIESTTSQT 727
+ IEE++ + E D +E K G + +V D R +
Sbjct: 247 VVEPDEPSDIEEMESPVKVYVPETRIEDDQEDKEGTIFTCLVCDLKYFIRLGTKFLQLDY 306
Query: 728 ADNVPVEGAVVPGSDTQVLIDEQPR----DETGERQQWEVLKSIVYGGLVESITSLGIVS 783
PVE +V D + I+ GER E+LKS VYGGL E+ITSLG+VS
Sbjct: 307 IRGKPVEKSVEEYIDVRKSINTTQSPPQIQPDGERFAIELLKSTVYGGLTETITSLGVVS 366
Query: 784 SAVSSGAAPLNIIALGLANLVGGLFILGHNLNDLKNNHSGGDQQQTNAQEDQYHELLGRR 843
SA +SG++ +NI+AL +ANL GGL +L N DL+N+ ++ ++D+Y ELLGRR
Sbjct: 367 SASASGSSTMNILALAVANLAGGLIVLAQNFQDLRNS--------SDQEKDRYEELLGRR 418
Query: 844 ANFLLHAVVAVISFIIFGAVPLAIYGVLINKNYSSDVKLGIVAATSVVCIILLAIAKVYT 903
+H +VAV+S+I FG +P +Y + + KL V S+VC+ILL KVY
Sbjct: 419 TKSRIHILVAVMSYIFFGLIPPLVYAFSFYETGIKNYKLISVFLGSLVCVILLGSIKVYV 478
Query: 904 RKP------PKYYFKTMLYYVALALGASGISYIAGILIKELLEKIS 943
RKP K Y K+ YY ++ + + GISY+ G ++ E +EK+S
Sbjct: 479 RKPTNSCGSTKAYLKSAAYYTSIVVASCGISYVVGDIMGEYIEKLS 524
Score = 70.5 bits (171), Expect = 4e-12
Identities = 68/273 (24%), Positives = 110/273 (39%), Gaps = 49/273 (17%)
Query: 91 DGGAAVGLRIVAEFVEAA-KNGEAIADASVGAAESQNENSVYFDKQQDNKINGLNGFQNG 149
DG A G V+E + + + I S + NE S + N+ NG N ++
Sbjct: 71 DGSQAQGNNSVSESTSSLFSDSDPIVLESTVSETGSNEES----ETGSNEENGNNWLESS 126
Query: 150 DAATSVSLTNPGESDYDVQIENSVTQRNLCSEVNDQCMKTVDDKVPSNSEVSPLHNSRDD 209
+ D +IE + N ++D K+ +K+ E L N ++
Sbjct: 127 STNLPNVENKRQRNGEDCEIEEE--EENNERSLSDSEEKSNLEKLLGTQENYELGNEDEE 184
Query: 210 TRATTVLDFHEETNIKEGSELVKEIDQDLKEFDVEVVLAKQETHDLFCPNCNSCITKRVI 269
+ D E++N+ E +LA QE ++L+CP+C++CIT+ V+
Sbjct: 185 KNERSSSDSEEKSNL-------------------ENLLATQENYELYCPSCSTCITRNVV 225
Query: 270 LKKRKRNIHSLDNKAKRDKLDIVASSDPVNSSAQESDRGVSENVTSEIVSVETPDDNYHP 329
LKKRKR H + + + +V +P S+I +E+P Y P
Sbjct: 226 LKKRKRGKHVNSSLDLKPDIPVVEPDEP-----------------SDIEEMESPVKVYVP 268
Query: 330 EREPESDPE-----VFRCLAC-FSFFIPSGKGF 356
E E D E +F CL C +FI G F
Sbjct: 269 ETRIEDDQEDKEGTIFTCLVCDLKYFIRLGTKF 301
>At4g27860 unknown protein
Length = 611
Score = 154 bits (388), Expect = 3e-37
Identities = 112/336 (33%), Positives = 169/336 (49%), Gaps = 51/336 (15%)
Query: 646 SSFTQSSVQSSGSSVPASVVASNK--QTSGIDAIFPSKPD-----LAPI------EEVQK 692
S FT + G VP S + + Q + D F K D L PI + ++
Sbjct: 274 SRFTPRYHKEKGEVVPKSTDSKSHDDQAANTDQDFDKKTDNKRNRLTPIYPSSLEKPSKQ 333
Query: 693 DINKEISASDKKESKGGDVIVVVDREAIESTTSQTADNVPV------------------- 733
+NKE DK+ + D++ + T +Q + P+
Sbjct: 334 TVNKETQNHDKEAADP-------DQDVDKETENQKSHLTPIYPSPLEQPSKQIINKETQT 386
Query: 734 EGAVVPGSDTQVLIDEQPRDETGERQQWEVLKSIVYGGLVESITSLGIVSSAVSSGAAPL 793
E + P + +++ +PR + G + E+LKSIVYGGL ESITSL V+SA +SGA+ L
Sbjct: 387 EPMLPPNAQSEIPNSVEPR-KGGNKV--EILKSIVYGGLTESITSLCTVTSAAASGASTL 443
Query: 794 NIIALGLANLVGGLFILGHNLNDL------KNNHSGGDQQQTNAQEDQYHELLGRRANFL 847
N++ALG+ANL GL + H+L +L K ++ ++ +ED+Y E+LGRR
Sbjct: 444 NVLALGVANLSSGLLLTVHSLQELINEKPRKQTNTDDSPEEGEGEEDRYEEVLGRREYSR 503
Query: 848 LHAVVAVISFIIFGAVPLAIYGVLINKNYS--SDVKLGIVAATSVVCIILLAIAKVYTRK 905
+H V+A+ SF+IFG +P +YG K + K+ V A S++CI+LL+IAK Y K
Sbjct: 504 IHRVIAISSFVIFGLIPPLVYGFSFRKKMEKRQEYKVLAVYAVSLLCIVLLSIAKAYVSK 563
Query: 906 PPKYYFKTMLYYVALALGASGISYIAGILIKELLEK 941
+ Y KT+ Y A ASG S G L+ + LEK
Sbjct: 564 -KRDYVKTLFRYTTTATTASGFSQFVGYLVSQWLEK 598
Score = 40.0 bits (92), Expect = 0.006
Identities = 41/171 (23%), Positives = 69/171 (39%), Gaps = 23/171 (13%)
Query: 189 TVDDKVPSNSEVSPLHNSRDDTRATTVLDFHEETNIKEGSELVKEIDQDLKEFDVEVVLA 248
+VD SE + ++ + +R+ + E + +L + + D + DVE+V+
Sbjct: 40 SVDFSQEQGSESNEAIDTENGSRSVDKNQYSETEVVVRAKDL--QTEPDSLDDDVEIVIK 97
Query: 249 KQETHDLFCPNCNSCITKRVILKKRKRNIHSLDN-----------KAKRDKLDIVAS--S 295
Q + ++CP C ITK V L K H+ D+ +DK V S S
Sbjct: 98 NQHKYYIYCPCCGEDITKTVKLVKISDPKHTKDHDKAVDSDTENGSKSKDKNTKVPSWFS 157
Query: 296 DPVNSSAQESDRGVSENVTSEIVSV--------ETPDDNYHPEREPESDPE 338
D + DRG V SE++ E P + E++ S P+
Sbjct: 158 DFIQPLFSSEDRGKKGVVDSELLGTYEDLGIIGEEPSIDVSNEKDRPSFPK 208
>At4g27850 putative proline-rich protein
Length = 577
Score = 48.5 bits (114), Expect = 2e-05
Identities = 25/66 (37%), Positives = 37/66 (55%), Gaps = 2/66 (3%)
Query: 881 KLGIVAATSVVCIILLAIAKVYTRKPPKYYFKTMLYYVALALGASGISYIAGILIKELLE 940
K +V A S+VC+I L+ AK Y + K +TM Y +A GAS S+IA ++ ++ E
Sbjct: 506 KAAVVLAASLVCVISLSFAKAYAFRMQK--LRTMAEYTGMAFGASAFSFIASRIVSDIFE 563
Query: 941 KISNSE 946
K E
Sbjct: 564 KFGFHE 569
Score = 35.0 bits (79), Expect = 0.20
Identities = 20/65 (30%), Positives = 30/65 (45%), Gaps = 3/65 (4%)
Query: 220 EETNIKEGSELVKEIDQDLKEFDVEVVLAKQETHDLFCPNCNSCITKRVILKKRKRNIHS 279
EE N E E + + D + +Q+TH+L+CP C + ITK L + + S
Sbjct: 30 EENNNNESQEETRTSRSPNEALDE---IRRQQTHNLYCPKCKTNITKSAELVVKNQETDS 86
Query: 280 LDNKA 284
KA
Sbjct: 87 YKTKA 91
>At3g46440 dTDP-glucose 4-6-dehydratases-like protein
Length = 341
Score = 44.7 bits (104), Expect = 3e-04
Identities = 33/135 (24%), Positives = 58/135 (42%), Gaps = 17/135 (12%)
Query: 99 RIVAEFVEAAKNGEAIADASVGAAESQNENSVYFDKQQDNKINGLNGFQNGDAATSVSLT 158
R+V+ F+ A GEA+ G +Q + Y + ++GL GD +++
Sbjct: 213 RVVSNFIAQALRGEALTVQKPG---TQTRSFCYVS----DMVDGLMRLMEGDDTGPINIG 265
Query: 159 NPGESDYDVQIENSVTQRNLCSEVNDQCMKTVDDKVPSNSEVSPLHNSRDDTRATTVLDF 218
NPGE T L V + +++ K+ N+ P D T+A VL +
Sbjct: 266 NPGE----------FTMVELAETVKELINPSIEIKMVENTPDDPRQRKPDITKAKEVLGW 315
Query: 219 HEETNIKEGSELVKE 233
+ ++EG L++E
Sbjct: 316 EPKVKLREGLPLMEE 330
>At2g28760 putative nucleotide-sugar dehydratase
Length = 343
Score = 43.1 bits (100), Expect = 8e-04
Identities = 33/135 (24%), Positives = 56/135 (41%), Gaps = 17/135 (12%)
Query: 99 RIVAEFVEAAKNGEAIADASVGAAESQNENSVYFDKQQDNKINGLNGFQNGDAATSVSLT 158
R+V+ F+ A GEA+ G +Q + Y + + GL GD +++
Sbjct: 215 RVVSNFIAQALRGEALTVQKPG---TQTRSFCYVS----DMVEGLMRLMEGDQTGPINIG 267
Query: 159 NPGESDYDVQIENSVTQRNLCSEVNDQCMKTVDDKVPSNSEVSPLHNSRDDTRATTVLDF 218
NPGE T L V + V+ K+ N+ P D ++A VL +
Sbjct: 268 NPGE----------FTMVELAETVKELIKPDVEIKMVENTPDDPRQRKPDISKAKEVLGW 317
Query: 219 HEETNIKEGSELVKE 233
+ ++EG L++E
Sbjct: 318 EPKVKLREGLPLMEE 332
>At5g01630 putative protein
Length = 1136
Score = 35.8 bits (81), Expect = 0.12
Identities = 21/64 (32%), Positives = 33/64 (50%), Gaps = 6/64 (9%)
Query: 192 DKVPSNSEVSPLHNSRDDTRATTVLDFHEETNIKEGSELVKEIDQDLKEFDVEVVLAKQE 251
+K SNS VSPLH+S R+ + N+ G L+K+ D + E + + ++K
Sbjct: 336 NKHTSNSFVSPLHSSSKQFRSVNL------ENLASGGNLIKKFDTAVDETNCALNISKPA 389
Query: 252 THDL 255
TH L
Sbjct: 390 THGL 393
>At1g20970 hypothetical protein
Length = 1498
Score = 35.4 bits (80), Expect = 0.16
Identities = 152/796 (19%), Positives = 281/796 (35%), Gaps = 116/796 (14%)
Query: 15 DEEEVEVEDGALKTRQSLPHSKETLVTEVVANGGEHRNGVSSVVTDLDGSFVQKEKEVAE 74
D+ EV V+ + P + E V NG H + D DGS+ + E
Sbjct: 8 DQGEVSVKVDFENATEIKPEVVVSATKEDVVNGISHGGSNNGNGNDTDGSY----DFITE 63
Query: 75 IGSVGENNSHFHDDRVDGGAAVGLRIVAEFVEAAKNGEAIADASVGAAESQNENSVYFDK 134
+VG DD V+ +++V+ + D G + S+ D
Sbjct: 64 NDTVG-------DDFVE----------SDYVKPVDDANVEKDLKEGENVKVDAPSIADDD 106
Query: 135 ----QQDNKINGLNGFQNGDAATSVSLTNPGESDYDVQIENSVTQRNLCSEVNDQCMKTV 190
QD++ + ++ D + P +S+ + +E SV Q++ N +
Sbjct: 107 VLGVSQDSQTLEKSELESTDDGPEEVVEIP-KSEVEDSLEKSVDQQH---PGNGHLESGL 162
Query: 191 DDKVPSNSEVSPLHNSRDDTRATTVLDFHE-ETNIKEGSEL---------VKEIDQDLKE 240
+ KV S EV LH+S ++ T + E E I+ SE ++ ++ ++
Sbjct: 163 EGKVESKEEVEQLHDSEVGSKDLTKNNVEEPEVEIESDSETDVEGHQGDKIEAQEKSDRD 222
Query: 241 FDVEVVLAKQETHDLFCPNCNSCITKRVILKKRKRNIHSLDNKAKRDKLDIVASSDPVNS 300
DV L E + P + + + ++ + D + + ++ +N
Sbjct: 223 LDVSQDLKLNENVEKH-PVDSDEVRESELVSAKVSPTEPSDGGMDLGQPTVTDPAETING 281
Query: 301 SAQESDRGVSENVT-SEIVSVETPDDNYHPEREPESDPEVFRCLACFSFFIPSGKGFNLF 359
S +D SE VT E VSVE N HP E E + S +P
Sbjct: 282 SESVNDHVGSEPVTVLEPVSVE----NGHPPVESELER---------SSDVPFTSVAEKV 328
Query: 360 RNFGGSRKQETSQNATSITAQNLQNPSTLPGSNANWFTSLFTSNKGKTATAKGNTSLEHS 419
G ++ ++ + P+ NA SL + GK + + S S
Sbjct: 329 NASDGEVLPDSGTVDVVVSEVSSDVPAETQALNA---ISLDSQPSGKDSVVENGNSKSES 385
Query: 420 TTAHVEQHCSTSVTSNVPASTDIGHLEGSLADTALIKNAK-PAAENGGMSSQISSTAESV 478
+ ++ S + A D +GS+ ++A P + GG ISS + V
Sbjct: 386 EDSKMQ--------SEIGAVDDGSVSDGSINTHPESQDASDPTCDQGG-KQHISSEVKEV 436
Query: 479 KIGPGTE---NGIKFHTASGNEPLADQKCLDSSFQNDFSSIKSTGGVMNGVQSAGQDFID 535
P +E + + +G+E A L + Q S + +G V A + +
Sbjct: 437 LDAPASEEISDAVIVAKDNGSE-AAISDGLSCTNQQGSESDEISGLVEKLPSHALHEVVS 495
Query: 536 LSSKEQLLTGNLKTNDGETSVDIIKTTANVEVMSSMSFSD--GVVSKYESVQSVTTATSE 593
++ ++ + + G + + T ++ S + V K+ + V SE
Sbjct: 496 SANDTSVIVSDDTKSQGLSEDHGVDTNQTIQDDCSAELEEVTDVNVKHAPNEKVQGDNSE 555
Query: 594 KHFNAGETAKVSVLNPSEGIPGFLAPTTIGSLILEQSQKDVDGTSENI---------KNS 644
+ N G ++ ++ +P G L S + + S NI K +
Sbjct: 556 GNLNVGGDVCLNSAEEAKELP-------TGDLSGNASHESAETLSTNIDEPLSLLDTKTA 608
Query: 645 YSSFTQSSVQSSGSSVPASVVASNKQTSGIDAIFPSKPDLAPIEEVQKDIN--------- 695
S F +SS +G ++ + Q+ A P + E+ +++N
Sbjct: 609 VSDFAESSAGVAGEIDAVAMESEAAQSIKQCAEAHVAPSIIEDGEIDREVNCGSEVNVTK 668
Query: 696 ------------KEISASDKKESKGGDVIVVVDREAIESTTSQTADNV-PVEGAVVPGSD 742
KE+S ++ + K I + A S S+ +E VV +D
Sbjct: 669 TTPVAVREDIPPKEVSEMEESDVKERSSINTDEEVATASVASEIKTCAQDLESKVVTSTD 728
Query: 743 T-----QVLIDEQPRD 753
T + +D QP +
Sbjct: 729 TIHTGAKDCVDSQPAE 744
>At1g65010 hypothetical protein
Length = 1318
Score = 34.3 bits (77), Expect = 0.35
Identities = 22/89 (24%), Positives = 42/89 (46%), Gaps = 2/89 (2%)
Query: 504 CLDSSFQNDFSSIKSTGGVMNGVQSAGQDFIDLSSKEQLLTGNLKTNDGETSVDIIKTTA 563
C+ S + + SS + ++N ++ + +D +E L NLK +GE V ++ T
Sbjct: 520 CVKKSEEENSSSQEEVSRLVNLLKESEEDACARKEEEASLKNNLKVAEGE--VKYLQETL 577
Query: 564 NVEVMSSMSFSDGVVSKYESVQSVTTATS 592
SM + ++ K E +++VT S
Sbjct: 578 GEAKAESMKLKESLLDKEEDLKNVTAEIS 606
>At5g67450 Cys2/His2-type zinc finger protein 1 (dbj|BAA85108.1)
Length = 245
Score = 33.9 bits (76), Expect = 0.46
Identities = 26/94 (27%), Positives = 38/94 (39%), Gaps = 8/94 (8%)
Query: 319 SVETPDDNYHPEREPESDPEVFRCLACFSFFIPSGKGFNLFRNFGGSRKQETSQNATSIT 378
+V++P P R SD ++C C GK F+ ++ GG + TSIT
Sbjct: 76 AVQSPPLPPLPSRASPSDHRDYKCTVC-------GKSFSSYQALGGHKTSHRKPTNTSIT 128
Query: 379 AQNLQNPSTLPGSNANWFTSLFTSNKGKTATAKG 412
+ N Q S SN+ T N G + G
Sbjct: 129 SGN-QELSNNSHSNSGSVVINVTVNTGNGVSQSG 161
>At3g32150 hypothetical protein
Length = 241
Score = 33.9 bits (76), Expect = 0.46
Identities = 27/105 (25%), Positives = 46/105 (43%), Gaps = 2/105 (1%)
Query: 233 EIDQDLKEFDVEVVLAKQETHDLFCPNCNSCITKRVILKKRKRNIHSLDNKAKRDKLDIV 292
E++ D+KE++ ++L Q + F + + V+++KR R + + +
Sbjct: 83 EMESDIKEYESNLLLLDQTHDEDFSEEQERSVFEAVLVEKRSRLAALTSSSFNPQQFEEF 142
Query: 293 ASSDPVNSSAQESDRGVSENVTSEI--VSVETPDDNYHPEREPES 335
+ P S + G SE V EI SVETP+ P P S
Sbjct: 143 FTELPPLSESGLDWAGPSEPVEPEISVTSVETPEPVVIPPEVPPS 187
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 33.1 bits (74), Expect = 0.78
Identities = 27/89 (30%), Positives = 38/89 (42%), Gaps = 5/89 (5%)
Query: 364 GSRKQETSQNATSITAQNLQNPSTLPGSNANWFTSLFTSNKGKTATAKGNTSLEHSTTAH 423
G +K + AQ +Q + +PG + LF+ N +T G +SL +TT
Sbjct: 527 GIKKLSDEEILKFTPAQYIQGDAWIPGKGVPYILGLFSGNGSTNSTVTG-SSLSSNTT-- 583
Query: 424 VEQHCSTSVTSNVPASTDIGHLEGSLADT 452
E S S S GHL GS +DT
Sbjct: 584 -ESSDSPSTVVTPSTSPPAGHL-GSPSDT 610
>At2g12090 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 248
Score = 33.1 bits (74), Expect = 0.78
Identities = 30/129 (23%), Positives = 55/129 (42%), Gaps = 20/129 (15%)
Query: 621 TIGSLILEQSQK-----DVDGTSENIKNSYSSFTQSSVQSSGSSVPASVVASN------- 668
T+ + E+ QK +V+ +E + S ++SS Q+ GS + +N
Sbjct: 110 TLKQIHAEEIQKNMQIDEVEDENEKEEASEEEESRSSSQTLGSDSDSEEAETNEELACAN 169
Query: 669 ------KQTSGIDAIFPSKPDLAPIEEVQKDINKEISASDKKESKGGDVIVVVDREAIES 722
+Q G+ A+ + + + + KD+N E S DK + D V+V++ E
Sbjct: 170 PVEEEERQDDGLTAVIEEEEESSSTSD--KDVNVEKSVEDKGDEDERDEDVIVEKPVEER 227
Query: 723 TTSQTADNV 731
T + NV
Sbjct: 228 TIEEDIANV 236
>At5g06700 unknown protein
Length = 608
Score = 32.7 bits (73), Expect = 1.0
Identities = 49/195 (25%), Positives = 75/195 (38%), Gaps = 19/195 (9%)
Query: 386 STLPGSNANWFTSLFTSNKGKTATAKGNTSLEHSTTAHVEQHCSTSVTSNVPASTDIGHL 445
S P S++ WF+++FTS+ T T NTS S + + + +VTS P +
Sbjct: 57 SPSPNSSSPWFSNIFTSS--STTTTSDNTS--GSQFSSIFSYILPNVTSTKPTNRS-SDA 111
Query: 446 EGSLADTALIKNAKPAAENGGMSSQISSTAESV--------KIGPGTENGIKFHTASGNE 497
SL+ A ++NG + + T V I GT K +T+S
Sbjct: 112 TDSLSVNATSPPLNSNSKNGTLQTPAPETHTPVAKNTTFESPIVNGTNPDAKNNTSS--H 169
Query: 498 PLADQKCLDSSFQNDFSSIKSTGGV-MNGVQSAGQDFIDLSSKEQLLTGNLKTNDGETSV 556
PL K + N + T V N S +S L N +N S
Sbjct: 170 PLLSDKSSTTGSNNQSRTTADTETVNRNQTTSPAPSKAPVSVD---LKTNSSSNSSTASS 226
Query: 557 DIIKTTANVEVMSSM 571
K T V+++SS+
Sbjct: 227 TPKKQTKTVDLVSSV 241
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 32.7 bits (73), Expect = 1.0
Identities = 28/127 (22%), Positives = 54/127 (42%), Gaps = 17/127 (13%)
Query: 230 LVKEIDQDLKEFD-VEVVLAKQETHDLFC-------PNCNS-----CITKRVILKKRKRN 276
+VK++ ++ ++F+ + + + T D P NS CI++R I ++++
Sbjct: 812 VVKDLAKNFRKFELIRIPRGQNTTADALAALASTSDPEVNSIIPVECISERSIKEEKEAF 871
Query: 277 IHSLDNKAKRDKLDIVASSDPV----NSSAQESDRGVSENVTSEIVSVETPDDNYHPERE 332
+ + A RD+ D PV + A V EN+ E++ D E E
Sbjct: 872 VVTRSRAAARDRGDTAVELPPVKRRKSKEATVPKPAVQENIGIELIEEVISDAKIEAETE 931
Query: 333 PESDPEV 339
P D ++
Sbjct: 932 PWVDEDI 938
>At1g43850 SEUSS transcriptional co-regulator
Length = 877
Score = 32.7 bits (73), Expect = 1.0
Identities = 45/212 (21%), Positives = 80/212 (37%), Gaps = 30/212 (14%)
Query: 366 RKQETSQNATSITAQNLQNPSTLPGSNANWFTSLFTSNKGKTATA-------KGNTSLEH 418
++Q SQN S Q+ + + + G+ +N F + T+T+ + + H
Sbjct: 611 QQQTVSQNTNS--DQSSRQVALMQGNPSNGVNYAFNAASASTSTSSIAGLIHQNSMKGRH 668
Query: 419 STTAH-----------VEQHCSTSVTSNVPASTDIGHLEGSLADTALIKNAKPAAENGGM 467
A+ V+ +S + VP+S+ H + N ++NG
Sbjct: 669 QNAAYNPPNSPYGGNSVQMQSPSSSGTMVPSSSQQQHNLPTFQSPTSSSNNNNPSQNGIP 728
Query: 468 SSQISSTAESVKIGPGTENGIKFHTASGNEPLADQKCLDSSFQNDFSSIKSTGGVMNGVQ 527
S + S + E GNE + QK L+ N+ + S+GG M G
Sbjct: 729 SVNHMGSTNSPAMQQAGE-------VDGNESSSVQKILNEILMNNQAHNNSSGGSMVGHG 781
Query: 528 SAGQD---FIDLSSKEQLLTGNLKTNDGETSV 556
S G D +++S LL N+ T++
Sbjct: 782 SFGNDGKGQANVNSSGVLLMNGQVNNNNNTNI 813
>At3g24340 hypothetical protein
Length = 1132
Score = 32.3 bits (72), Expect = 1.3
Identities = 57/243 (23%), Positives = 96/243 (39%), Gaps = 37/243 (15%)
Query: 66 VQKEKEVAEIGSVGENNSHFHDDRVDGGAAVGLRIVAEFVEAAKNGEAIADASVGAAESQ 125
+ + +V +G+V N H DD G A+V V +F E DA V S
Sbjct: 132 IDDDDDVVFVGTVQRENDHVEDDDNVGSASVISPRVCDFDE--------DDAKV----SG 179
Query: 126 NENSVYFDKQQDNKINGLNGFQNGDAATSVSLTNPGESDYDVQIENSVTQRNLCSEVNDQ 185
EN + D D G +N V N G D+ ++++ + + V+D+
Sbjct: 180 KENPLSPDDDDDVVFLGTIAGEN----QHVEDVNAGSEVCDILLDDANLRGEEKTYVSDE 235
Query: 186 CMKTVDDKVPSNSEVSPLH----NSRDDT----RATTVLDFHEETNIKEGSELVKEI--D 235
V S+ E PL +SR++ R + D E+ N + S+ V E
Sbjct: 236 ---VVSLSSSSDDEEDPLEELGTDSREEVSGEDRDSGESDMDEDANDSDSSDYVGESSDS 292
Query: 236 QDLKEFDVEVVLAKQE---THDLFCPNCNSCITKRVILKKRKRNIHSLDNKAKRDKLDIV 292
D++ D + V ++ E T D C +++V K+ R K D ++++
Sbjct: 293 SDVESSDSDFVCSEDEEGGTRD--DATCEKNPSEKVYHHKKSRTFR---RKHNFDVINLL 347
Query: 293 ASS 295
A S
Sbjct: 348 AKS 350
>At5g03420 putative protein
Length = 583
Score = 32.0 bits (71), Expect = 1.7
Identities = 40/189 (21%), Positives = 77/189 (40%), Gaps = 8/189 (4%)
Query: 424 VEQHCSTSVTSNVPASTDI--GHLEGSLADTALIKNAKPAAENGGMSSQISSTAESVKIG 481
+E+ C+ V + +T+I G L D++ +K A + G +S SS A + +
Sbjct: 118 MEEDCNELVADSEDNNTNIETGGSRACLEDSST-DLSKEAEKQGSLSKDESSLAGVLSLE 176
Query: 482 PGTEN-GIKFHTASGNEPLADQKCLDSSFQNDFSSIKSTGGVMNGVQSAGQDFIDLSSKE 540
N G H+ E + ++S N+ + I+++ + + QD D SS
Sbjct: 177 NSFSNLGDSNHSGEITEKIFK---IESVELNEIADIENSSSEASVFANHSQDLYDTSSCP 233
Query: 541 QLLTGNLKTNDGETSVDIIKT-TANVEVMSSMSFSDGVVSKYESVQSVTTATSEKHFNAG 599
+ GN+ + E D+ K + + +S + + S++ V AT +
Sbjct: 234 DIEAGNVSMTEDEEVNDVDKDFSLTFDHYTSPTSNHYTSPDLNSIKHVDIATGSSYDLTS 293
Query: 600 ETAKVSVLN 608
E +V N
Sbjct: 294 ENTMTNVEN 302
>At3g57120 unknown protein
Length = 456
Score = 32.0 bits (71), Expect = 1.7
Identities = 26/93 (27%), Positives = 43/93 (45%), Gaps = 6/93 (6%)
Query: 385 PSTLPGSNANWFTSLFTSNKGKTATAKGNTSLEHSTTAHVEQHCSTSVTSNVPASTDIGH 444
PS G + + S TSN G T T T+ +S S + +S+V + T +
Sbjct: 37 PSQSQGRHPSTRRSPSTSNTGTTTT----TTTSNSNKTGASSSSSGAASSSVASRTSLAS 92
Query: 445 LEGSLADTALIKNAKP--AAENGGMSSQISSTA 475
L SL + I N AA N +++++SS++
Sbjct: 93 LRESLPENPHIYNVSEIRAATNNFLANRLSSSS 125
>At1g12800 unknown protein
Length = 767
Score = 32.0 bits (71), Expect = 1.7
Identities = 26/105 (24%), Positives = 48/105 (44%), Gaps = 9/105 (8%)
Query: 682 PDLAPIEEVQKDINKEISASDKKESKGGDVIVVVDREAIESTTSQTADNVPVEGAVVPGS 741
PD + EV IN E+S+ K E D+++V DRE + +S + V + +
Sbjct: 497 PDSSLDSEVATTINGEVSSDMKLE----DLLMVYDREKQKFLSSFVGQKIKVNVVMANRN 552
Query: 742 DTQVLIDEQPRDETGERQQWEVLKSIVYGG-----LVESITSLGI 781
+++ +PR+ E ++ L + + G ++ IT GI
Sbjct: 553 SRKLIFSMRPRENEEEVEKKRTLMAKLRVGDVVKCCIKKITYFGI 597
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,333,966
Number of Sequences: 26719
Number of extensions: 938926
Number of successful extensions: 2995
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 2944
Number of HSP's gapped (non-prelim): 120
length of query: 968
length of database: 11,318,596
effective HSP length: 109
effective length of query: 859
effective length of database: 8,406,225
effective search space: 7220947275
effective search space used: 7220947275
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0360.4


