

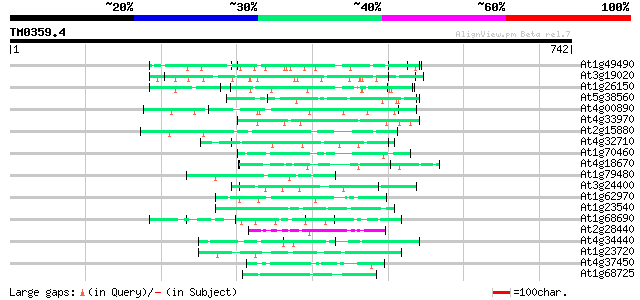
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0359.4
(742 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g49490 hypothetical protein 70 6e-12
At3g19020 hypothetical protein 67 3e-11
At1g26150 Pto kinase interactor, putative 60 3e-09
At5g38560 putative protein 58 2e-08
At4g00890 57 4e-08
At4g33970 extensin-like protein 56 8e-08
At2g15880 unknown protein 55 1e-07
At4g32710 putative protein kinase 55 2e-07
At1g70460 putative protein kinase 53 7e-07
At4g18670 extensin-like protein 52 1e-06
At1g79480 hypothetical protein 52 1e-06
At3g24400 protein kinase, putative 50 4e-06
At1g62970 unknown protein 50 5e-06
At1g23540 putative serine/threonine protein kinase 50 5e-06
At1g68690 protein kinase, putative 49 1e-05
At2g28440 En/Spm-like transposon protein 49 1e-05
At4g34440 serine/threonine protein kinase - like 47 5e-05
At1g23720 unknown protein 46 9e-05
At4g37450 arabinogalactan protein AGP18 45 1e-04
At1g68725 putative arabinogalactan protein AGP19 45 1e-04
>At1g49490 hypothetical protein
Length = 847
Score = 69.7 bits (169), Expect = 6e-12
Identities = 92/397 (23%), Positives = 146/397 (36%), Gaps = 66/397 (16%)
Query: 186 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKT-KRKPDDGDDSESEDGP-PKKKQKKK 243
P+P+ P PPK ++ + +E + ++P K++ K +P +DS + P P++ K +
Sbjct: 431 PSPK-PQPPKHESPKP---EEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEESPKPE 486
Query: 244 VRIVVKPTR----------------VEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDL 287
+ +PT+ + + RR+ PP +V + +P + S
Sbjct: 487 QPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPP 546
Query: 288 NLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPT 347
+ PI + +P P+ S P SPP S P P+ P + P
Sbjct: 547 S-----PIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASP---PPPSPPPPVHSPPPPP 598
Query: 348 SRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDP-------------SAPRAS 394
S P P P+ S P P P P P T P P +
Sbjct: 599 VFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQA 658
Query: 395 ERPAARTT-------DTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEE 447
P++ TT ++D S +PV P V S PS+ + S+ E
Sbjct: 659 PTPSSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQV--------PTPSSSES 710
Query: 448 YYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSD 507
Y SP + P P + P P S SNQS ++P P++E H T +
Sbjct: 711 YQAPNLSPVQAPTPVQAPTTSSETSQVPTP---SSESNQSPSQAPTPILE--PVHAPTPN 765
Query: 508 PNAPMINIGSPQGASEAHSSNHLASPEP---NLSIIP 541
S + S S + +PEP N S +P
Sbjct: 766 SKPVQSPTPSSEPVSSPEQSEEVEAPEPTPVNPSSVP 802
Score = 51.6 bits (122), Expect = 2e-06
Identities = 86/370 (23%), Positives = 122/370 (32%), Gaps = 73/370 (19%)
Query: 186 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESED---GPPKKKQKK 242
P P P + Q K E+ +P K P++ +D P K ++
Sbjct: 463 PQPSKPEDSPKPEQPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRS 522
Query: 243 KVRIVVKPTRVEPAAAVVRRTEPPARVTRSS--AHSSKPAL-ASDDDLNLFDALPISALL 299
V+ TRV P + PP+ + HS P + +S +++ P A
Sbjct: 523 PPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASP 582
Query: 300 QHSS---------------------NPLTPIPESQPAAQTTSPP---HSPRSSFFQPSPT 335
S +P P P P + SPP +SP F P PT
Sbjct: 583 PPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPT 642
Query: 336 E----------------APLWNLLQNPTSRSEDPTSLLTIPYD---PL-----SSEPIVQ 371
P Q PT SE S + P P+ SSEP
Sbjct: 643 HNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQV 702
Query: 372 DQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESI 431
P +++ P S AP + P TT +++S TP S SN S S
Sbjct: 703 PTPSSSESYQAPNLSPVQAPTPVQAP---TTSSETSQVPTP---------SSESNQSPSQ 750
Query: 432 RKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRS 491
+ + P+P P PE+ +E P+P P N SSV S
Sbjct: 751 APTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQ---SEEVEAPEPTPV----NPSSVPS 803
Query: 492 PHPLVETSDP 501
P +TS P
Sbjct: 804 SSPSTDTSIP 813
Score = 45.1 bits (105), Expect = 1e-04
Identities = 64/256 (25%), Positives = 85/256 (33%), Gaps = 28/256 (10%)
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDP 353
P+ SS P TP QP+ + P H P P E +N +
Sbjct: 411 PVMPKPSDSSKPETPKTPEQPSPKPQPPKHE------SPKPEEP------ENKHELPKQK 458
Query: 354 TSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPV 413
S P P S Q +PE + QP+ +P+ P + A T D A +PV
Sbjct: 459 ESPKPQPSKPEDSPKPEQPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYDA-SPV 517
Query: 414 SFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYY-----LTCPSPRRYPGPRPERLVD 468
N PP ++ + S Y + P P Y P P +
Sbjct: 518 K---NRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYS 574
Query: 469 PDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSN 528
P P+ P P S V SP P S P S P P + SP S +
Sbjct: 575 PPPPVASPPPP----SPPPPVHSPPPPPVFSPPPPVFSPP--PPSPVYSPPPPSHSPPPP 628
Query: 529 HLASPEPNLSIIPYTH 544
+ P P S P TH
Sbjct: 629 VYSPPPPTFS-PPPTH 643
Score = 45.1 bits (105), Expect = 1e-04
Identities = 54/233 (23%), Positives = 83/233 (35%), Gaps = 48/233 (20%)
Query: 302 SSNPLTPIPES-QPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIP 360
+S P PE P +S P +P++ QPSP P ++ + + E+P + +P
Sbjct: 401 TSEPKPSKPEPVMPKPSDSSKPETPKTPE-QPSPKPQPP----KHESPKPEEPENKHELP 455
Query: 361 YDPLSSEPIV---QDQPEPNQTEPQ--PRTSDPSAPRASERPAARTTDTDSSTAFTPV-S 414
S +P +D P+P Q +P+ P+ P P + +P + + T P +
Sbjct: 456 KQKESPKPQPSKPEDSPKPEQPKPEESPKPEQPQIPEPT-KPVSPPNEAQGPTPDDPYDA 514
Query: 415 FPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQ 474
P+ SPP E R V P +P
Sbjct: 515 SPVKNRRSPPPPKVEDTR----------------------------------VPPPQPPM 540
Query: 475 PDPEPAQSV-SNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHS 526
P P P + S V SP P V +S P P P+ + P HS
Sbjct: 541 PSPSPPSPIYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHS 593
Score = 42.4 bits (98), Expect = 0.001
Identities = 52/240 (21%), Positives = 77/240 (31%), Gaps = 30/240 (12%)
Query: 185 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKKQKKKV 244
PP P F PP + + + ++ P + T+ + +S+ P
Sbjct: 632 PPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSP-------- 683
Query: 245 RIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSN 304
V PT V+ + T+ P T SS+ S + S P+ A S
Sbjct: 684 --VQAPTPVQSSTPSSEPTQVP---TPSSSESYQAPNLSPVQA----PTPVQAPTTSSET 734
Query: 305 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPL 364
P P S+ + P P+P P+ Q+PT SE +S P
Sbjct: 735 SQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPV----QSPTPSSEPVSS-------PE 783
Query: 365 SSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPP 424
SE + + PEP P S + S P D D P PP
Sbjct: 784 QSEEV--EAPEPTPVNPSSVPSSSPSTDTSIPPPENNDDDDDGDFVLPPHIGFQYASPPP 841
>At3g19020 hypothetical protein
Length = 951
Score = 67.4 bits (163), Expect = 3e-11
Identities = 84/375 (22%), Positives = 128/375 (33%), Gaps = 31/375 (8%)
Query: 186 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKKQKKKVR 245
P E P P + K + + ESS++ + K +P ++S P ++ K +
Sbjct: 485 PKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEES 544
Query: 246 IVVKPTRVE-PAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDAL-----PISALL 299
+P + E P + +PP + T S KP + +A P+ + +
Sbjct: 545 PKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQEQPPKTEAPKMGSPPLESPV 604
Query: 300 QHSSNPLTPIPESQPA--------AQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSE 351
+ +PI + +P +TTSP P S P P +P P
Sbjct: 605 PNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSP-------PPPVFS 657
Query: 352 DPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFT 411
P + + P S P V P P P P P P S P + +
Sbjct: 658 PPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPP 717
Query: 412 PVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEE----YYLTCPSPRRYPGPR----P 463
PV P + SPP S I + + P P P P P
Sbjct: 718 PVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPP 777
Query: 464 ERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNA--PMINIGSPQGA 521
+ P P+ P P S S + SP P V + P T P A PM N +P +
Sbjct: 778 PPVHSPPPPVHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSS 837
Query: 522 SEAHSSNHLASPEPN 536
S + +P P+
Sbjct: 838 ESGEISTPVQAPTPD 852
Score = 55.8 bits (133), Expect = 8e-08
Identities = 85/389 (21%), Positives = 135/389 (33%), Gaps = 53/389 (13%)
Query: 186 PAPEFPSP----PKRKAQRKMVLDESSEESDVPLVKKTKRKPDD-GDDSESEDGP----- 235
P P+ PSP P + S + + P ++ K KP+ +S ++ P
Sbjct: 436 PKPQQPSPKPETPSHEPSNPKEPKPESPKQESPKTEQPKPKPESPKQESPKQEAPKPEQP 495
Query: 236 -PKKKQKKKVRIVVKPTRVE-------PAAAVVRRTEPPARVTRSSAHSSKPALASDDDL 287
PK + K+ +P + E P + +PP + T S KP +
Sbjct: 496 KPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQE-- 553
Query: 288 NLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPT 347
+ + S P P E+ P + + P P+ QP TEAP
Sbjct: 554 --------TPKPEESPKPQPPKQET-PKPEESPKPQPPKQE--QPPKTEAP--------- 593
Query: 348 SRSEDPTSLLTIPYDPLSSEPIVQDQPE-PNQTEPQPRTSDPSAPRASERPAARTTDTDS 406
+ P +P DP + PI + +P+ P+ + + +T+ P +P P +
Sbjct: 594 -KMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPP 652
Query: 407 STAFT---PVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGP-- 461
F+ P+ P + SPP + V + + P P P P
Sbjct: 653 PPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPV 712
Query: 462 --RPERLVDPDEPIQ-PDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSP 518
P + P P+Q P P P S + + SP P S P S P P + SP
Sbjct: 713 HSPPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSPPPPVHSPPPPP---VHSP 769
Query: 519 QGASEAHSSNHLASPEPNLSIIPYTHLQP 547
+ + P P S P H P
Sbjct: 770 PPPVHSPPPPVHSPPPPVHSPPPPVHSPP 798
Score = 55.5 bits (132), Expect = 1e-07
Identities = 81/383 (21%), Positives = 120/383 (31%), Gaps = 34/383 (8%)
Query: 186 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKKQKKKVR 245
P P+ P P K + + + P ++ KP+ S PK + K+
Sbjct: 408 PTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESPKQES 467
Query: 246 IVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNP 305
K + +P ++ P + KP + P + S P
Sbjct: 468 --PKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKP-----EESPKP 520
Query: 306 LTPIPESQPAAQ-----TTSPPHSPR---SSFFQPSPTEAPLWNLLQNPTSRSEDPTSLL 357
P PE P Q T P SP+ P P E+P + T + E+
Sbjct: 521 EPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPK-- 578
Query: 358 TIPYDPLSSEPIVQDQPE---PNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVS 414
P P +P + P+ P P P ++P RP + T+ + +P S
Sbjct: 579 --PQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQS 636
Query: 415 FPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPER-----LVDP 469
P++ PP S F + + + P P P P P + P
Sbjct: 637 PPVHSPPPPPPVHSPPPPVFSP--PPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSP 694
Query: 470 DEPIQPDPEPAQSV-----SNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEA 524
P+ P P S S V SP P V++ P S P I P
Sbjct: 695 PPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVQSPPPPPVFSPPPPAPIYSPPPPPVHSP 754
Query: 525 HSSNHLASPEPNLSIIPYTHLQP 547
H P P S P H P
Sbjct: 755 PPPVHSPPPPPVHSPPPPVHSPP 777
Score = 48.5 bits (114), Expect = 1e-05
Identities = 70/323 (21%), Positives = 112/323 (34%), Gaps = 37/323 (11%)
Query: 205 DESSEESDVPLVKK----TKRKPDDGDDSESEDGP---PKKKQKKKVRIVVKPTRVEPAA 257
++ S + +P+V + +K K G S P P K + + + P EP+
Sbjct: 370 NQKSAKECLPVVSRPVDCSKDKCAGGGGGGSNPSPKPTPTPKAPEPKKEINPPNLEEPSK 429
Query: 258 AVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQ 317
+ P + S KP S + N + P S Q S P P+ + Q
Sbjct: 430 PKPEESPKPQQP------SPKPETPSHEPSNPKEPKPESPK-QESPKTEQPKPKPESPKQ 482
Query: 318 TTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPN 377
+ +P+ +P P + Q P E P P + +P Q+ P+P
Sbjct: 483 ESPKQEAPKPEQPKPKPESPKQESSKQEPPKPEESPKPEPPKPEESPKPQPPKQETPKPE 542
Query: 378 QT-----------------EPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVI 420
++ +PQP + P S +P + T + P +
Sbjct: 543 ESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQEQPPKTEAPKMGSP--PL 600
Query: 421 DSPPSNTSESIRKFMEIRKEKVS-ALEEYYLTCP-SPRRYPGPRPERLVDPDEPIQPDPE 478
+SP N + R + S + EE T P SP + P P + P P+ P
Sbjct: 601 ESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPPP 660
Query: 479 PAQSVSNQSSVRSPHPLVETSDP 501
P S V SP P V + P
Sbjct: 661 PMH--SPPPPVYSPPPPVHSPPP 681
Score = 38.9 bits (89), Expect = 0.011
Identities = 40/170 (23%), Positives = 61/170 (35%), Gaps = 11/170 (6%)
Query: 250 PTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPI 309
P V V PP HS P + S + + P + P+TP+
Sbjct: 763 PPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPSPIYSPP-PPVFSPPPKPVTPL 821
Query: 310 PES-QPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEP 368
P + P A +P S P P ++ P+ + P + P SEP
Sbjct: 822 PPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDSNHSPV-FKSSPAPSPDSEP 880
Query: 369 IVQ-----DQPE---PNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAF 410
V+ +PE P Q+E P +S PS+ + + A + D D F
Sbjct: 881 EVEAPVPSSEPEVEAPKQSEATPSSSPPSSNPSPDVTAPPSEDNDDGDNF 930
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 60.5 bits (145), Expect = 3e-09
Identities = 71/255 (27%), Positives = 99/255 (37%), Gaps = 30/255 (11%)
Query: 293 LPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQ------NP 346
+P+S + S P P PA +SPP P SS P PTEAP + NP
Sbjct: 90 IPVSPPPEPSPPPPLPTEAPPPANPVSSPP--PESSPPPPPPTEAPPTTPITSPSPPTNP 147
Query: 347 TSRSEDPTSLLTI--PYDPLSSEPIVQDQPEPNQTEPQPRTSD-PSAPR------ASERP 397
E P SL P +PL +V P + P P S+ P PR ASERP
Sbjct: 148 PPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPASERP 207
Query: 398 AARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRR 457
+ +D++ + P P PP + S + PSP
Sbjct: 208 STPPSDSEHPSP-PPPGHPKRREQPPPPGSKRPT-------PSPPSPSDSKRPVHPSP-- 257
Query: 458 YPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGS 517
P P PE + P +P PDP P+ S S + + + S P S P+ P N +
Sbjct: 258 -PSP-PEETLPPPKP-SPDPLPSNSSSPPTLLPPSSVVSPPSPPRKSVSGPDNPSPNNPT 314
Query: 518 PQGASEAHSSNHLAS 532
P + + S +A+
Sbjct: 315 PVTDNSSSSGISIAA 329
Score = 58.5 bits (140), Expect = 1e-08
Identities = 78/332 (23%), Positives = 108/332 (32%), Gaps = 56/332 (16%)
Query: 185 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKK-------TKRKPDDGDDSESEDGPPK 237
PP P FP R+ E ++ P + P+ S S GPP
Sbjct: 28 PPQPSFPGDNATSPTREPTNGNPPETTNTPAQSSPPPETPLSSPPPEPSPPSPSLTGPPP 87
Query: 238 KKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISA 297
V P P + PPA S S P + A P +
Sbjct: 88 TTIP-----VSPPPEPSPPPPLPTEAPPPANPVSSPPPESSPPPPPPTE-----APPTTP 137
Query: 298 LLQHS--SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTS 355
+ S +NP P PES P+ PP +P P+ +P +L P S P
Sbjct: 138 ITSPSPPTNPPPP-PESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPPR 196
Query: 356 LLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSF 415
L P P S P P + P P P P+ E+P + + + +P
Sbjct: 197 HLPSP--PASERP---STPPSDSEHPSP--PPPGHPKRREQPPPPGSKRPTPSPPSPSDS 249
Query: 416 PINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPER------LVDP 469
V SPPS E T P P+ P P P L+ P
Sbjct: 250 KRPVHPSPPSPPEE---------------------TLPPPKPSPDPLPSNSSSPPTLLPP 288
Query: 470 DEPIQPDPEPAQSVS--NQSSVRSPHPLVETS 499
+ P P +SVS + S +P P+ + S
Sbjct: 289 SSVVSPPSPPRKSVSGPDNPSPNNPTPVTDNS 320
Score = 55.8 bits (133), Expect = 8e-08
Identities = 69/271 (25%), Positives = 88/271 (32%), Gaps = 54/271 (19%)
Query: 279 PALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAA---QTTSPPHSPRSSFFQPSPT 335
PA A ++++L +L +S PL +P QP+ TSP P +
Sbjct: 4 PAQAPREEVSLSPSL--------ASPPLMALPPPQPSFPGDNATSPTREPTNG------- 48
Query: 336 EAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASE 395
NP + P P PLSS P P P+ T P P T S P
Sbjct: 49 ---------NPPETTNTPAQSSPPPETPLSSPPPEPSPPSPSLTGPPPTTIPVSPPPEPS 99
Query: 396 RPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSP 455
P T+ P N + SPP +S A +T PSP
Sbjct: 100 PPPPLPTEAPP---------PANPVSSPPPESSPP-------PPPPTEAPPTTPITSPSP 143
Query: 456 RRYPGPRPE---RLVDPDEPIQPDPEPAQSVSNQSSVRS-PHPLVETSDP---HL----G 504
P P PE L PD P P P P + S R P P P HL
Sbjct: 144 PTNPPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPA 203
Query: 505 TSDPNAPMINIGSPQGASEAHSSNHLASPEP 535
+ P+ P + P H P P
Sbjct: 204 SERPSTPPSDSEHPSPPPPGHPKRREQPPPP 234
Score = 39.3 bits (90), Expect = 0.008
Identities = 45/194 (23%), Positives = 71/194 (36%), Gaps = 24/194 (12%)
Query: 359 IPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPIN 418
+ P + P + P P + P + P+ + P TT+T + ++ P P++
Sbjct: 12 VSLSPSLASPPLMALPPPQPSFPGDNATSPTREPTNGNPP-ETTNTPAQSS-PPPETPLS 69
Query: 419 VIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYP-GPRPERLVDPDEPIQPDP 477
PS S S LT P P P P PE P P + P
Sbjct: 70 SPPPEPSPPSPS-------------------LTGPPPTTIPVSPPPEPSPPPPLPTEAPP 110
Query: 478 EPAQSVSNQSSVRSPHPLVET-SDPHLGTSDPNAPMINIGSPQGASEAHSSNHLASPEPN 536
PA VS+ SP P T + P + P+ P P+ + + ++P P
Sbjct: 111 -PANPVSSPPPESSPPPPPPTEAPPTTPITSPSPPTNPPPPPESPPSLPAPDPPSNPLPP 169
Query: 537 LSIIPYTHLQPTSL 550
++P +H P L
Sbjct: 170 PKLVPPSHSPPRHL 183
>At5g38560 putative protein
Length = 681
Score = 57.8 bits (138), Expect = 2e-08
Identities = 65/269 (24%), Positives = 96/269 (35%), Gaps = 55/269 (20%)
Query: 287 LNLFDALPISALLQHSSNPLTPIP-ESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQN 345
++L LPI + +S+ P P ++QP + PP +P PSP ++P
Sbjct: 1 MSLVPPLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTP-----PPSPPQSP------- 48
Query: 346 PTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQ---TEPQPRTSDPSAPRASERPAARTT 402
P + + P P+ S P P P+ T P P + P P +
Sbjct: 49 -------PPVVSSSPPPPVVSSPPPSSSPPPSPPVITSPPPTVASSPPP-----PVVIAS 96
Query: 403 DTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPR 462
S+ A TP + P V PP + S S T P P+ P P
Sbjct: 97 PPPSTPATTPPAPPQTVSPPPPPDASPS--------------PPAPTTTNPPPKPSPSPP 142
Query: 463 PERLVDPDE-PIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNA--------PMI 513
E P E P P P P+ ++ P P S P +DP+ P++
Sbjct: 143 GETPSPPGETPSPPKPSPSTPTPTTTTSPPPPPATSASPPSSNPTDPSTLAPPPTPLPVV 202
Query: 514 ----NIGSPQGASEAHSSNHLASPEPNLS 538
I P G + + +N L S P S
Sbjct: 203 PREKPIAKPTGPASNNGNNTLPSSSPGKS 231
Score = 49.3 bits (116), Expect = 8e-06
Identities = 57/210 (27%), Positives = 76/210 (36%), Gaps = 25/210 (11%)
Query: 342 LLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAART 401
+L P+S S T PL ++P P P P P S P +S P +
Sbjct: 9 ILSPPSSNSS------TTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVS 62
Query: 402 TDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRR--YP 459
+ SS+ P S P VI SPP + S + I S T P+P + P
Sbjct: 63 SPPPSSS--PPPSPP--VITSPPPTVASSPPPPVVIASPPPSTPA---TTPPAPPQTVSP 115
Query: 460 GPRPERLVDPDEPIQPDPEPAQSVSNQSSVRS--------PHPLVETSDPHLGTSDPNAP 511
P P+ P P +P P S S S P P T P TS P P
Sbjct: 116 PPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPP 175
Query: 512 MINIGSPQGASEAHSSNHLASPEPNLSIIP 541
+ P +S + LA P L ++P
Sbjct: 176 ATSASPP--SSNPTDPSTLAPPPTPLPVVP 203
Score = 40.4 bits (93), Expect = 0.004
Identities = 40/165 (24%), Positives = 55/165 (33%), Gaps = 12/165 (7%)
Query: 265 PPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHS 324
PP S S P ++S + + P S+ S P P+ S P +SPP
Sbjct: 36 PPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSS----SPPPSPPVITSPPPTVASSPP-- 89
Query: 325 PRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPR 384
P P P+ T P P P ++ P + P P P P
Sbjct: 90 PPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSPPGETPSPP 149
Query: 385 TSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSE 429
PS P+ S TT T +P P P SN ++
Sbjct: 150 GETPSPPKPSPSTPTPTTTT------SPPPPPATSASPPSSNPTD 188
Score = 32.3 bits (72), Expect = 0.99
Identities = 25/92 (27%), Positives = 37/92 (40%), Gaps = 13/92 (14%)
Query: 304 NPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDP 363
+P P P + TTSPP P +S PS +P++ + PT L +P +
Sbjct: 154 SPPKPSPSTPTPTTTTSPPPPPATSASPPSSNPT-------DPSTLAPPPTPLPVVPREK 206
Query: 364 LSSEPIVQDQPEPNQTEPQPRTSDPSAPRASE 395
++P N T P S+P SE
Sbjct: 207 PIAKPTGPASNNGNNTLPS------SSPGKSE 232
>At4g00890
Length = 431
Score = 57.0 bits (136), Expect = 4e-08
Identities = 74/305 (24%), Positives = 113/305 (36%), Gaps = 45/305 (14%)
Query: 263 TEPPARVTRSSAHSSKPA--LASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTS 320
++PP R + + S P L +LP+S PL+P + +P +QT +
Sbjct: 28 SQPPRRTVQETLSQSPPQSPLFHPQSPPEPKSLPLS--------PLSPKSKEEPESQTMT 79
Query: 321 PPHSPR----SSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEP 376
P S + SP ++P P S L ++P PL++EP P
Sbjct: 80 PLMSKHVKTHRNLLIQSPPKSP-------PESTESQQQFLASVPLRPLTTEPKTPLSPSS 132
Query: 377 NQTEPQPRTSDPSAPRAS-----ERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESI 431
+ S PS P S RP+ T+ P+ P +PPS+ S +
Sbjct: 133 TLKATEESQSQPSPPLESIVETWFRPSPPTSTETGDETQLPIP-PPQEAKTPPSSPSMML 191
Query: 432 RKFMEIRKE------KVSALEEYYLTCP--SPRRYPGPRPERLVDPDE------PIQPDP 477
E + +++E L P S P P P + +D +E PI P
Sbjct: 192 NATEEFESQPKPPLLPSKSIDETRLRSPLMSQASSPPPLPSKSIDENETRSQSPPISPPK 251
Query: 478 EPAQSVSNQSSVRSPHPLV--ETSDPHLGTS--DPNAPMINIGSPQGASEAHSSNHLASP 533
Q+ S S SP PL+ + S+ H S P +P I S S + +P
Sbjct: 252 SDKQARSQTHSSPSPPPLLSPKASENHQSKSPMPPPSPTAQISLSSLKSPIPSPATITAP 311
Query: 534 EPNLS 538
P S
Sbjct: 312 PPPFS 316
Score = 43.1 bits (100), Expect = 6e-04
Identities = 77/361 (21%), Positives = 127/361 (34%), Gaps = 51/361 (14%)
Query: 177 LDDFVSRLPP-APEF--PSPPKRKAQRKMVLDESSEESD-----VPLVKKTKRKPDDGDD 228
+ + +S+ PP +P F SPP+ K+ L S+E PL+ K + +
Sbjct: 35 VQETLSQSPPQSPLFHPQSPPEPKSLPLSPLSPKSKEEPESQTMTPLMSKHVKTHRNLLI 94
Query: 229 SESEDGPPKKKQKKK---VRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDD 285
PP+ + ++ + ++P EP + + T S P L S
Sbjct: 95 QSPPKSPPESTESQQQFLASVPLRPLTTEPKTPL--SPSSTLKATEESQSQPSPPLESIV 152
Query: 286 DLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSF-------FQPSPTEAP 338
+ + P S + PIP P + +PP SP QP P P
Sbjct: 153 ETWFRPSPPTST--ETGDETQLPIP---PPQEAKTPPSSPSMMLNATEEFESQPKPPLLP 207
Query: 339 LWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQP-EPNQTEPQPRTSDPSAPRASERP 397
S+S D T L + SS P + + + N+T Q S P +P S++
Sbjct: 208 ---------SKSIDETRLRSPLMSQASSPPPLPSKSIDENETRSQ---SPPISPPKSDKQ 255
Query: 398 AARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRR 457
A T + S P ++ S +S +L PSP
Sbjct: 256 ARSQTHSSPS--------PPPLLSPKASENHQSKSPMPPPSPTAQISLSSLKSPIPSPAT 307
Query: 458 YPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSD----PNAPMI 513
P P P P P+P+ + ++SP P + ++ H P+A ++
Sbjct: 308 ITAPPPP-FSSPLSQTTPSPKPSLPQIEPNQIKSPSPKLTNTESHASPEQNLVKPDANLM 366
Query: 514 N 514
N
Sbjct: 367 N 367
>At4g33970 extensin-like protein
Length = 699
Score = 55.8 bits (133), Expect = 8e-08
Identities = 69/270 (25%), Positives = 97/270 (35%), Gaps = 34/270 (12%)
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPR----SSFFQPSPTEA-PLWNLLQNPTSRSEDPTSL 356
SS P P P +P T+P H P + +PSP P+ PT+ +P+ +
Sbjct: 416 SSTPSKPSPVHKPTPVPTTPVHKPTPVPTTPVQKPSPVPTTPVQKPSPVPTTPVHEPSPV 475
Query: 357 LTIPYD---PLSSEPIVQDQPEPNQTEPQP-----------RTSDPSAPRASERPAARTT 402
L P D P+ S P+ +P+P + PQP R S P AP S P +
Sbjct: 476 LATPVDKPSPVPSRPV--QKPQPPKESPQPDDPYDQSPVTKRRSPPPAPVNSPPPPVYSP 533
Query: 403 DTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPR 462
+P P++ PP + + V + + P P P P
Sbjct: 534 PPPPPPVHSPPP-PVHSPPPPPVYSPPPPPPPVHSPPPPVFSPPPPVYSPPPPVHSPPP- 591
Query: 463 PERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLV-----ETSDPHLGTSDPNAPMIN--- 514
P P P+ P P S V SP P V S P + + P P IN
Sbjct: 592 PVHSPPPPAPVHSPPPPVHSPPPPPPVYSPPPPVFSPPPSQSPPVVYSPPPRPPKINSPP 651
Query: 515 IGSPQGASEAHSSN---HLASPEPNLSIIP 541
+ SP A H +P + IIP
Sbjct: 652 VQSPPPAPVEKKETPPAHAPAPSDDEFIIP 681
>At2g15880 unknown protein
Length = 727
Score = 55.5 bits (132), Expect = 1e-07
Identities = 83/357 (23%), Positives = 111/357 (30%), Gaps = 56/357 (15%)
Query: 174 EIDLDDFVSRLPPAPEFPSPPKRKAQRKMVLDESSEE------------SDVPLVKKTKR 221
+I LDD + LP P+ S + +D S ++ S P+ +
Sbjct: 348 QIALDDTRNCLPDRPKQRSAKECAVVISRPVDCSKDKCAGGSSQATPSKSPSPVPTRPVH 407
Query: 222 KPDDGDDSESEDGPPKKKQKKKVRIVVKPTRVE------PAAAVVRRTEPPARVTRSSAH 275
KP +S + P + K R P + P A T PP T S H
Sbjct: 408 KPQPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPPPASSPPTSPPVHSTPSPVH 467
Query: 276 SSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPT 335
+P S + +D P+ S P P+ P SP HSP P
Sbjct: 468 KPQPPKESPQPNDPYDQSPVKF---RRSPPPPPVHSPPPP----SPIHSPPPPPVYSPPP 520
Query: 336 EAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASE 395
P+++ P S P + P P+ S P P P P P P P S
Sbjct: 521 PPPVYSPPPPPPVYSPPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSP 580
Query: 396 RPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSP 455
P S PV P + SPP P P
Sbjct: 581 PPPV------YSPPPPPVHSPPPPVHSPPPPVHS-----------------------PPP 611
Query: 456 RRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPM 512
Y P P + P P+ P P S V SP P V + P S P P+
Sbjct: 612 PVYSPPPPPPVHSPPPPVFSPPPPVH--SPPPPVYSPPPPVYSPPPPPVKSPPPPPV 666
Score = 38.5 bits (88), Expect = 0.014
Identities = 56/263 (21%), Positives = 74/263 (27%), Gaps = 17/263 (6%)
Query: 181 VSRLPPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKKQ 240
V P P PPK Q D+S + P S PP
Sbjct: 459 VHSTPSPVHKPQPPKESPQPNDPYDQSPVKFRRSPPPPPVHSPPPPSPIHSPPPPPVYSP 518
Query: 241 KKKVRIVVKP----TRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPIS 296
+ P P V PP HS P + S P+
Sbjct: 519 PPPPPVYSPPPPPPVYSPPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPV- 577
Query: 297 ALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSL 356
HS P P P P HSP P P P+++ P S P +
Sbjct: 578 ----HSPPPPVYSPPPPPVHSPPPPVHSPPPPVHSPPP---PVYSPPPPPPVHSPPPP-V 629
Query: 357 LTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASE----RPAARTTDTDSSTAFTP 412
+ P S P V P P + P P P P P + T + P
Sbjct: 630 FSPPPPVHSPPPPVYSPPPPVYSPPPPPVKSPPPPPVYSPPLLPPKMSSPPTQTPVNSPP 689
Query: 413 VSFPINVIDSPPSNTSESIRKFM 435
P +++PP + I F+
Sbjct: 690 PRTPSQTVEAPPPSEEFIIPPFI 712
>At4g32710 putative protein kinase
Length = 731
Score = 54.7 bits (130), Expect = 2e-07
Identities = 77/282 (27%), Positives = 103/282 (36%), Gaps = 48/282 (17%)
Query: 253 VEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPES 312
+ P+++ T PPA S P S D + A P+S L S+P P+P
Sbjct: 3 LSPSSSPAPATSPPAM--------SLPPADSVPDTSSPPAPPLSPLPPPLSSP-PPLPSP 53
Query: 313 QP-AAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNP----------------TSRSEDPTS 355
P +A T SPP P S P P E+P LL++P P
Sbjct: 54 PPLSAPTASPPPLPVESPPSP-PIESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPLP 112
Query: 356 LLTIPYDPLSSEPIVQDQPEPNQTEPQPRT----SDPSAPRASERPAARTTDTDS-STAF 410
L P S P + P N P PR+ S P AS P + +F
Sbjct: 113 FLPAKPSPPPSSPPSETVPPGNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPSF 172
Query: 411 TPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYP----------- 459
P PIN PS + S+ + K +S + P P++ P
Sbjct: 173 PP---PINSSPPNPSPNTPSLPETSPPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFFGPA 229
Query: 460 GPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDP 501
G P+ V P PI P EP S + S +P PLV S P
Sbjct: 230 GQLPDGTVAP--PIGPVIEPKTSPAESISPGTPQPLVPKSLP 269
Score = 47.8 bits (112), Expect = 2e-05
Identities = 56/220 (25%), Positives = 73/220 (32%), Gaps = 15/220 (6%)
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDP 353
P S+ +S P +P + T+SPP P S P + PL +P S
Sbjct: 5 PSSSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPL----PSPPPLSAPT 60
Query: 354 TSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPV 413
S +P + S PI P ++ P P PS P P F P
Sbjct: 61 ASPPPLPVESPPSPPIESPPPPLLESPPPPPLESPSPP----SPHVSAPSGSPPLPFLPA 116
Query: 414 SFPINVIDSPPSNTSESIRKF----MEIRKEKVSALEEYYLTCPS-PRRYPGPRPERLVD 468
P SPPS T + E + PS PRR GP+P
Sbjct: 117 K-PSPPPSSPPSETVPPGNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPS-FPP 174
Query: 469 PDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDP 508
P P+P P ++S PL T P T P
Sbjct: 175 PINSSPPNPSPNTPSLPETSPPPKPPLSTTPFPSSSTPPP 214
>At1g70460 putative protein kinase
Length = 710
Score = 52.8 bits (125), Expect = 7e-07
Identities = 62/235 (26%), Positives = 87/235 (36%), Gaps = 46/235 (19%)
Query: 302 SSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTE-APLWNLLQNPTSRSEDPTSLLTIP 360
S +P + P P+A + PP + P PT+ AP +P + S P +L ++P
Sbjct: 2 SDSPTSSPPA--PSADSAPPPDTSSDGSAAPPPTDSAPP----PSPPADSSPPPALPSLP 55
Query: 361 YDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVI 420
S P V P P P D + P +S P + P+ FP I
Sbjct: 56 PAVFSPPPTVSSPPPPPLDSSPPPPPDLTPPPSSPPPP-------DAPPPIPIVFP-PPI 107
Query: 421 DSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPE-- 478
DSPP ++ S P P + P P D P P PE
Sbjct: 108 DSPPPESTNSP---------------------PPPEVFEPPPPPADEDESPPAPPPPEQL 146
Query: 479 PAQSVSNQSSVRSP---HPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHL 530
P + S Q + P HP TS P P+AP + +P A +SS+ L
Sbjct: 147 PPPASSPQGGPKKPKKHHPGPATSPP-----APSAPATSPPAPPNAPPRNSSHAL 196
Score = 36.2 bits (82), Expect = 0.068
Identities = 58/257 (22%), Positives = 81/257 (30%), Gaps = 36/257 (14%)
Query: 177 LDDFVSRLPPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPP 236
+ D + PPAP S P D SS+ S P + P DS PP
Sbjct: 1 MSDSPTSSPPAPSADSAPPP--------DTSSDGSAAPPPTDSAPPPSPPADSS----PP 48
Query: 237 KKKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPIS 296
+ P V + PP + SS P + + + PI
Sbjct: 49 PALPSLPPAVFSPPPTVSSPPPPPLDSSPPPPPDLTPPPSSPPPPDAPPPIPIVFPPPID 108
Query: 297 ALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSL 356
+ S+N S P + PP P SP P L P S +
Sbjct: 109 SPPPESTN-------SPPPPEVFEPPPPPADE--DESPPAPPPPEQLPPPASSPQGG--- 156
Query: 357 LTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFP 416
P P P P P TS P+ P A R ++ S+ A P++ P
Sbjct: 157 ---PKKPKKHHPGPATSPPAPSA---PATSPPAPPNAPPRNSSHALPPKSTAAGGPLTSP 210
Query: 417 INVIDS------PPSNT 427
+ S PP+N+
Sbjct: 211 SRGVPSSGNSVPPPANS 227
>At4g18670 extensin-like protein
Length = 839
Score = 52.0 bits (123), Expect = 1e-06
Identities = 68/277 (24%), Positives = 101/277 (35%), Gaps = 58/277 (20%)
Query: 304 NPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDP 363
+P +P P SPP +P PSP+ P +P S + P S T P
Sbjct: 422 SPPSPSISPSPPITVPSPPTTPSPGGSPPSPSIVP------SPPSTTPSPGSPPTSP--- 472
Query: 364 LSSEPIVQDQPEPNQTEPQPRTSDPSAPR----ASERPAARTTDTDSSTAFTPVSFPINV 419
+ P P + T P P S PS+P P++ TT + + +P ++
Sbjct: 473 --TTPTPGGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPSP-----SI 525
Query: 420 IDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYP----GPRPERLVDPDEPIQP 475
SPP +T PSP P P P PI
Sbjct: 526 SPSPP-------------------------ITVPSPPSTPTSPGSPPSPSSPTPSSPIPS 560
Query: 476 DPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMIN----IGSPQGASEAHSSNHLA 531
P P+ + S ++ P++ S P G S P++P I SP SS +
Sbjct: 561 PPTPSTPPTPISPGQNSPPII-PSPPFTGPSPPSSPSPPLPPVIPSPPIVGPTPSSPPPS 619
Query: 532 SPEPNLSIIPYTHLQPTSLSECINIFYHEASLRLRNV 568
+P P + P+ HL LS + + H RLR++
Sbjct: 620 TPTPGTLLHPH-HLLLPQLSHLPHQYLHH---RLRHI 652
Score = 47.8 bits (112), Expect = 2e-05
Identities = 59/205 (28%), Positives = 74/205 (35%), Gaps = 51/205 (24%)
Query: 303 SNPLTPIPESQPAAQTTSPP--HSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIP 360
S+P TP P P + T+P SP SS PSP +P S S P+ +T+P
Sbjct: 483 SSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPG--------GSPPSPSISPSPPITVP 534
Query: 361 YDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVI 420
P S P P P+ + P P + PS P S P T +P +I
Sbjct: 535 SPP--STPTSPGSP-PSPSSPTPSSPIPSPPTPSTPP----------TPISPGQNSPPII 581
Query: 421 DSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPI-QPDP-E 478
SPP T PSP P P P V P PI P P
Sbjct: 582 PSPP-------------------------FTGPSPPSSPSP-PLPPVIPSPPIVGPTPSS 615
Query: 479 PAQSVSNQSSVRSPHPLVETSDPHL 503
P S ++ PH L+ HL
Sbjct: 616 PPPSTPTPGTLLHPHHLLLPQLSHL 640
Score = 37.0 bits (84), Expect = 0.040
Identities = 61/243 (25%), Positives = 80/243 (32%), Gaps = 68/243 (27%)
Query: 305 PLTPIPES--QPAAQTTSPP---------HSPRSSFFQPSPTEAPLWNLLQNPTSRSEDP 353
P P S Q AA ++ PP S R PSP P +P S S P
Sbjct: 373 PARPAQRSPGQCAAFSSLPPVDCGSFGCGRSTRPPVVVPSPPTTPSPG--GSPPSPSISP 430
Query: 354 TSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPV 413
+ +T+P P + P P + P P PS P + P + T S T TP
Sbjct: 431 SPPITVPSPPTT--------PSPGGSPPSPSIV-PSPPSTTPSPGSPPT---SPTTPTPG 478
Query: 414 SFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPI 473
SPPS+ T P+P P P P
Sbjct: 479 G-------SPPSSP-----------------------TTPTPGGSP---------PSSPT 499
Query: 474 QPDP---EPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHL 530
P P P+ + P P + S P S P+ P + GSP S S+ +
Sbjct: 500 TPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTP-TSPGSPPSPSSPTPSSPI 558
Query: 531 ASP 533
SP
Sbjct: 559 PSP 561
Score = 30.8 bits (68), Expect = 2.9
Identities = 27/106 (25%), Positives = 41/106 (38%), Gaps = 10/106 (9%)
Query: 293 LPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSED 352
LP+ +H N +P P + + PP P S P+PT ++ + P
Sbjct: 687 LPLYLTCRHRLNLASPPPPAPYYYSSPQPPPPPHYSLPPPTPT----YHYISPP----PP 738
Query: 353 PTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPA 398
PT + + P P S P ++ P P T PS S P+
Sbjct: 739 PTPIHSPP--PQSHPPCIEYSPPPPPTVHYNPPPPPSPAHYSPPPS 782
>At1g79480 hypothetical protein
Length = 356
Score = 52.0 bits (123), Expect = 1e-06
Identities = 54/220 (24%), Positives = 81/220 (36%), Gaps = 29/220 (13%)
Query: 235 PPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVT------------------RSSAHS 276
PP + P P+++ + PP +T SS++
Sbjct: 50 PPLSVPGNAPPFCINPPNTPPSSSYPGLSPPPGPITLPNPPDSSSNPNSNPNPPESSSNP 109
Query: 277 SKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHS--PRSSFFQPSP 334
+ P +S+ + N + + + SSNP P S P + P S P P+P
Sbjct: 110 NPPDSSSNPNSNPNPPVTVPNPPESSSNPNPPDSSSNPNSNPNPPESSSNPNPPVTVPNP 169
Query: 335 TEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEP----IVQDQPEPNQTEPQPRTSDPSA 390
E+ + NP S +P +TIPY P SS P IV PE T P P P +
Sbjct: 170 PES---SSNPNPPESSSNPNPPITIPYPPESSSPNPPEIVPSPPESGYT-PGPVLGPPYS 225
Query: 391 PRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSES 430
P + + SS P+ +P + PS T S
Sbjct: 226 EPGPSTPTG-SIPSPSSGFLPPIVYPPPMAPPSPSVTPTS 264
Score = 33.1 bits (74), Expect = 0.58
Identities = 49/220 (22%), Positives = 78/220 (35%), Gaps = 38/220 (17%)
Query: 327 SSFFQPSPTEAPLWNLLQNPTSRSEDP----TSLLTIPYDPLSSEPIVQDQPEPNQTEPQ 382
+S Q T+ L L +S + P T+L PY L + + P P P
Sbjct: 8 TSLLQKMMTQLNLAQPLDYSSSSNTQPYGVSTTLTLPPYVSLPPLSVPGNAP-PFCINPP 66
Query: 383 PRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKV 442
S P S P T ++ P S P +PP ++S
Sbjct: 67 NTPPSSSYPGLSPPPGPITLPNPPDSSSNPNSNP-----NPPESSSN------------- 108
Query: 443 SALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPH 502
P+P P +P+ P+ P P +S SN + S +P
Sbjct: 109 ----------PNP---PDSSSNPNSNPNPPVTV-PNPPESSSNPNPPDSSSNPNSNPNPP 154
Query: 503 LGTSDPNAPMINIGSPQGASEAHSSNHLASPEPNLSIIPY 542
+S+PN P+ P+ +S + ++P P ++ IPY
Sbjct: 155 ESSSNPNPPVTVPNPPESSSNPNPPESSSNPNPPIT-IPY 193
>At3g24400 protein kinase, putative
Length = 694
Score = 50.4 bits (119), Expect = 4e-06
Identities = 66/241 (27%), Positives = 78/241 (31%), Gaps = 40/241 (16%)
Query: 305 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPL 364
PL P QP T PP + P P L L P PT++ IP
Sbjct: 17 PLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTALPPALPPP----PPPTTVPPIPPSTP 72
Query: 365 SSEPIVQDQP-EPNQTEPQ----PRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINV 419
S P + P P+ T P P + PS P P A T S TP P +
Sbjct: 73 SPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAIT----PSPPLTPSPLPPSP 128
Query: 420 IDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEP 479
P S SI PSP P P P + P P P P P
Sbjct: 129 TTPSPPPPSPSI---------------------PSPPLTPSPPPSSPLRPSSP--PPPSP 165
Query: 480 AQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIG--SPQGASEAHSSNHLASPEPNL 537
A + S P P T P +G+ P P G SP S S+ A L
Sbjct: 166 ATPSTPPRS--PPPPSTPTPPPRVGSLSPPPPASPSGGRSPSTPSTTPGSSPPAQSSKEL 223
Query: 538 S 538
S
Sbjct: 224 S 224
Score = 49.3 bits (116), Expect = 8e-06
Identities = 54/211 (25%), Positives = 78/211 (36%), Gaps = 28/211 (13%)
Query: 294 PISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEA----PLWNLLQNPTSR 349
P L + P P+ S P A T SPP +P S PSPT P ++ P +
Sbjct: 90 PSPPLTPSPTTPSPPLTPSPPPAITPSPPLTP--SPLPPSPTTPSPPPPSPSIPSPPLTP 147
Query: 350 SEDPTSLLTI----PYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTD 405
S P+S L P P + + P P+ P PR S P + R+ T
Sbjct: 148 SPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTPPPRVGSLSPPPPASPSGGRSPSTP 207
Query: 406 SSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKE------KVSALEEYYLTCPSPRRYP 459
S+T + SPP+ +S+ + K + + AL + C RR
Sbjct: 208 STTPGS----------SPPAQSSKELSKGAMVGIAIGGGFVLLVALALIFFLCKKKRRRD 257
Query: 460 GPRPERLV--DPDEPIQPDPEPAQSVSNQSS 488
P + P P P P +S+ SS
Sbjct: 258 NEAPPAPIVPPPKSPSSAPPRPPHFMSSGSS 288
Score = 32.0 bits (71), Expect = 1.3
Identities = 47/190 (24%), Positives = 60/190 (30%), Gaps = 39/190 (20%)
Query: 363 PLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSST--AFTPVSFPINVI 420
P P QP P PQP P P + PA ++ A P P V
Sbjct: 6 PPGGTPSPPPQPLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTALPPALPPPPPPTTVP 65
Query: 421 DSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDP-EP 479
PPS S P P P P P P P P
Sbjct: 66 PIPPSTPS------------------------PPPPLTPSPLP-----------PSPTTP 90
Query: 480 AQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHLASPEPNLSI 539
+ ++ + SP PL + P + S P P SP S S + SP S
Sbjct: 91 SPPLTPSPTTPSP-PLTPSPPPAITPSPPLTPSPLPPSPTTPSPPPPSPSIPSPPLTPSP 149
Query: 540 IPYTHLQPTS 549
P + L+P+S
Sbjct: 150 PPSSPLRPSS 159
Score = 29.3 bits (64), Expect = 8.3
Identities = 35/160 (21%), Positives = 53/160 (32%), Gaps = 18/160 (11%)
Query: 182 SRLPPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKK-- 239
S LPP+P PSPP S PL + P S PP
Sbjct: 122 SPLPPSPTTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTP 181
Query: 240 -QKKKVRIVVKPTRVEPAA-------AVVRRTEPPARVTRSSAHSSKPALASDDDLNLFD 291
+V + P P+ + + PPA+ ++ + + +A L
Sbjct: 182 TPPPRVGSLSPPPPASPSGGRSPSTPSTTPGSSPPAQSSKELSKGAMVGIAIGGGFVLLV 241
Query: 292 ALPISALL------QHSSNPLTPI--PESQPAAQTTSPPH 323
AL + L + + P PI P P++ PPH
Sbjct: 242 ALALIFFLCKKKRRRDNEAPPAPIVPPPKSPSSAPPRPPH 281
>At1g62970 unknown protein
Length = 825
Score = 50.1 bits (118), Expect = 5e-06
Identities = 65/249 (26%), Positives = 90/249 (36%), Gaps = 49/249 (19%)
Query: 273 SAHSSKPALASDDDLNLFDALPISALLQHS--------SNPLTPIPESQP-AAQTTSPPH 323
S+ SKP L S LP+S LQ+S S+ P P SQP A P
Sbjct: 422 SSQKSKPLLVSQSSQRS-KPLPVSQSLQNSNPFPVSQPSSNSKPFPVSQPQPASNPFPVS 480
Query: 324 SPRS-----SFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQ 378
PR S QPS T P P S+ + I P +S+P V P ++
Sbjct: 481 QPRPNSQPFSMSQPSSTARPF------PASQPPAASKSFPISQPPTTSKPFVSQPPNTSK 534
Query: 379 TEP--QPRTSDPSAPRASERPAARTT-------DTDSSTAFTPVSFPINVIDSPPSNTSE 429
P QP T+ P + P ++T + S + PV SPP +T+
Sbjct: 535 PMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVFNSTQSFQSPPVSTTP 594
Query: 430 SIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSV 489
SA+ E T PSP P P + P P P Q+ + ++
Sbjct: 595 -------------SAVPE-ASTIPSP-----PAPAPVAQPTHVFNQTPPPEQTPKSGAAR 635
Query: 490 RSPHPLVET 498
P V +
Sbjct: 636 TDSEPEVSS 644
Score = 39.3 bits (90), Expect = 0.008
Identities = 69/305 (22%), Positives = 117/305 (37%), Gaps = 41/305 (13%)
Query: 185 PPAPEFPSPPKRKAQ--RKMVLDESSEESDVPLVKKTKR--KPDDGDDSESEDGPPKKKQ 240
PP+ P P + +Q + + +SS++S LV ++ + KP S P Q
Sbjct: 398 PPSNSKPFPMSQSSQNSKPFPVSQSSQKSKPLLVSQSSQRSKPLPVSQSLQNSNPFPVSQ 457
Query: 241 KKKVRIVVKPTRVEPAAAVVRRTEP-----------PARVTRSSAHSSKPALASDDDLNL 289
++ +PA+ ++P P+ R S PA + ++
Sbjct: 458 PSSNSKPFPVSQPQPASNPFPVSQPRPNSQPFSMSQPSSTARPFPASQPPAASKSFPISQ 517
Query: 290 --FDALPISALLQHSSNPLT---------PIPESQPAA--QTTSPPHSPR-SSFFQPSPT 335
+ P + ++S P+ P+P SQP Q+T P P SS P P
Sbjct: 518 PPTTSKPFVSQPPNTSKPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLP- 576
Query: 336 EAPLWNLLQNPTSR--SEDPTSL---LTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSA 390
P++N Q+ S S P+++ TIP P + P+ Q NQT P +T A
Sbjct: 577 --PVFNSTQSFQSPPVSTTPSAVPEASTIPSPPAPA-PVAQPTHVFNQTPPPEQTPKSGA 633
Query: 391 PRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRK---FMEIRKEKVSALEE 447
R P + T + +PI +S ++S R+ +++ A E+
Sbjct: 634 ARTDSEPEVSSFWTTCPYCYVLYEYPIIYEESVLKCQTKSCRRAYQAVKVPSPPPVAEED 693
Query: 448 YYLTC 452
Y C
Sbjct: 694 SYYCC 698
Score = 38.1 bits (87), Expect = 0.018
Identities = 78/327 (23%), Positives = 118/327 (35%), Gaps = 68/327 (20%)
Query: 229 SESEDGPPKKKQKKKVRIVVKPTRVEPAAAVVRRTE-------PPARVTRSSAHSSKPA- 280
S S +G + ++ KPT P + VR +E PAR + S+ SS+ A
Sbjct: 287 SRSFEGSSHRTPSTELTWASKPT---PVSEPVRHSELVPWQYSEPARQYQLSSRSSEAAQ 343
Query: 281 ---LASDDDLNLFDALPISALLQHS-------SNPLTPIPESQPAAQTTSPPHSPRSSFF 330
L S D + + P + H+ S P P P SQP + P S S
Sbjct: 344 LSLLPSVSDSS-HASQPTRSNQSHAVSKPQPVSKPHPPFPMSQPPPTSNPFPLSQPPSNS 402
Query: 331 QPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYD-----PL-------SSEPIVQDQPEPNQ 378
+P P N P S+S + L + PL +S P QP N
Sbjct: 403 KPFPMSQSSQNSKPFPVSQSSQKSKPLLVSQSSQRSKPLPVSQSLQNSNPFPVSQPSSNS 462
Query: 379 -----TEPQPRTSDP---SAPRASERPAARTTDTDSSTAF-------TPVSFPIN----- 418
++PQP S+P S PR + +P + + + ++ F SFPI+
Sbjct: 463 KPFPVSQPQP-ASNPFPVSQPRPNSQPFSMSQPSSTARPFPASQPPAASKSFPISQPPTT 521
Query: 419 --VIDSPPSNTSESIRKFMEIRKEKVSALEE----YYLTCPSPRRYPGPRPERLVDPDEP 472
S P NTS+ + K + + + TCPS +P P
Sbjct: 522 SKPFVSQPPNTSKPMPVSQPPTTSKPLPVSQPPPTFQSTCPS-------QPPAASSSLSP 574
Query: 473 IQPDPEPAQSVSNQSSVRSPHPLVETS 499
+ P QS + +P + E S
Sbjct: 575 LPPVFNSTQSFQSPPVSTTPSAVPEAS 601
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 50.1 bits (118), Expect = 5e-06
Identities = 59/237 (24%), Positives = 85/237 (34%), Gaps = 14/237 (5%)
Query: 273 SAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQP 332
S SS PA +D SAL S+P +P +S + P P S P
Sbjct: 7 SPSSSPPAPPADTAPPPETPSENSALPPVDSSPPSPPADSSSTPPLSEPSTPPPDSQLPP 66
Query: 333 SPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPR 392
P+ P P S S P P P S+E PE ++T P P P+
Sbjct: 67 LPSILPPLTDSPPPPSDSSPPVDSTPSPPPPTSNES--PSPPEDSETPPAP----PNESN 120
Query: 393 ASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTC 452
+ P ++ + ++ +P P N +SPP + + S L+ T
Sbjct: 121 DNNPPPSQDLQSPPPSSPSPNVGPTNP-ESPPLQSPPAPPASDPTNSPPASPLDP---TN 176
Query: 453 PSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPN 509
P P + GP +P+ P P P + V SP + P GT PN
Sbjct: 177 PPPIQPSGPATSPPANPNAPPSPFPTVPPKTPSSGPVVSP----SLTSPSKGTPTPN 229
Score = 38.5 bits (88), Expect = 0.014
Identities = 48/205 (23%), Positives = 64/205 (30%), Gaps = 61/205 (29%)
Query: 185 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKKQKKKV 244
PP PSPP + E SE P + P D +S PP
Sbjct: 86 PPVDSTPSPPPPTSNESPSPPEDSETPPAPPNESNDNNPPPSQDLQS---PP-------- 134
Query: 245 RIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSN 304
P+ P PP + S PA + D N A P+ +
Sbjct: 135 -----PSSPSPNVGPTNPESPPLQ--------SPPAPPASDPTNSPPASPL--------D 173
Query: 305 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPL 364
P P P QP+ TSPP +P + P+ T+P
Sbjct: 174 PTNP-PPIQPSGPATSPPANPNA------------------------PPSPFPTVPPKTP 208
Query: 365 SSEPIVQDQPEPNQTEPQPRTSDPS 389
SS P+V P+ T P T P+
Sbjct: 209 SSGPVV----SPSLTSPSKGTPTPN 229
Score = 32.3 bits (72), Expect = 0.99
Identities = 48/206 (23%), Positives = 73/206 (35%), Gaps = 42/206 (20%)
Query: 351 EDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPR--ASERPAARTTDTDSST 408
E P+S + P P + P + P N P +S PS P +S P + + +
Sbjct: 6 ESPSS--SPPAPPADTAP-PPETPSENSALPPVDSSPPSPPADSSSTPPLSEPSTPPPDS 62
Query: 409 AFTPVSF---PINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPER 465
P+ P+ PPS++S + + PSP P P
Sbjct: 63 QLPPLPSILPPLTDSPPPPSDSSPPVD------------------STPSP---PPPTSNE 101
Query: 466 LVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSP--QGASE 523
P E + P P N+S+ +P P + P + PN N SP Q
Sbjct: 102 SPSPPEDSETPPAPP----NESNDNNPPPSQDLQSPPPSSPSPNVGPTNPESPPLQSPPA 157
Query: 524 AHSSNHLASPEPNLSIIPYTHLQPTS 549
+S+ SP P + L PT+
Sbjct: 158 PPASDPTNSP-------PASPLDPTN 176
>At1g68690 protein kinase, putative
Length = 708
Score = 48.9 bits (115), Expect = 1e-05
Identities = 62/234 (26%), Positives = 88/234 (37%), Gaps = 31/234 (13%)
Query: 299 LQHSSNPLTPIPESQPAAQTTSPPH------SPRSSFFQPSPTEAPLWNLLQNPTSRSED 352
L ++++P TP P + P + PP+ P ++ P AP L + P S S
Sbjct: 23 LNNATSPATPPPVTSPLPPSAPPPNRAPPPPPPVTTSPPPVANGAPPPPLPKPPESSSPP 82
Query: 353 PTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSA-----PRASERPAARTTDTDSS 407
P + IP P S+ P P+P P P S P A P + PA+ S
Sbjct: 83 PQPV--IPSPPPSTSP----PPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSP 136
Query: 408 TAFTPVSFP---INVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPE 464
+ V P + I SPP S+ + E + PSP P RP
Sbjct: 137 SPPILVRSPPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQSPPSP---PSERPT 193
Query: 465 RLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSP 518
+ P P P S++ S +SP P E + P PN+P SP
Sbjct: 194 Q-------SPPPPSPPSPPSDRPS-QSPPPPPEDTKPQPPRRSPNSPPPTFSSP 239
Score = 46.2 bits (108), Expect = 7e-05
Identities = 47/205 (22%), Positives = 71/205 (33%), Gaps = 14/205 (6%)
Query: 235 PPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALP 294
PP + + P V A +PP SS+ +P + S P
Sbjct: 45 PPNRAPPPPPPVTTSPPPVANGAPPPPLPKPP----ESSSPPPQPVIPSPPPSTSPPPQP 100
Query: 295 ISALLQHSSNP----LTPIPESQPAAQTTSPPH---SPRSSFFQPSPTEAPLWNLLQNPT 347
+ S++P + P+P S P + PP SP P P+ P+ + P+
Sbjct: 101 VIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPS 160
Query: 348 SR---SEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDT 404
R S P S + P + + P P Q+ P P P + R S+ P DT
Sbjct: 161 DRPTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDT 220
Query: 405 DSSTAFTPVSFPINVIDSPPSNTSE 429
+ P SPP + E
Sbjct: 221 KPQPPRRSPNSPPPTFSSPPRSPPE 245
Score = 44.3 bits (103), Expect = 3e-04
Identities = 52/207 (25%), Positives = 70/207 (33%), Gaps = 56/207 (27%)
Query: 185 PPAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKKKQKKKV 244
PP P PSPP + + S S P P S PP
Sbjct: 82 PPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLP-------SSPPPPAS------ 128
Query: 245 RIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSN 304
V P R P+ ++ R+ PP+ S P SD + + +
Sbjct: 129 ---VPPPRPSPSPPILVRSPPPS----VRPIQSPPPPPSD---------------RPTQS 166
Query: 305 PLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPL 364
P P P S P+ + T P SP P+E P ++S P S P P
Sbjct: 167 PPPPSPPSPPSERPTQSPPSP--------PSERP---------TQSPPPPS----PPSPP 205
Query: 365 SSEPIVQDQPEPNQTEPQPRTSDPSAP 391
S P P P T+PQP P++P
Sbjct: 206 SDRPSQSPPPPPEDTKPQPPRRSPNSP 232
Score = 33.9 bits (76), Expect = 0.34
Identities = 26/92 (28%), Positives = 35/92 (37%), Gaps = 3/92 (3%)
Query: 451 TCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNA 510
+ P P R P P P P P P +SS P P++ + P TS P
Sbjct: 42 SAPPPNRAPPPPPPVTTSPPPVANGAPPPPLPKPPESSSPPPQPVIPSPPP--STSPPPQ 99
Query: 511 PMINIGSPQGASEAHSSNHL-ASPEPNLSIIP 541
P+I P + L +SP P S+ P
Sbjct: 100 PVIPSPPPSASPPPALVPPLPSSPPPPASVPP 131
Score = 29.3 bits (64), Expect = 8.3
Identities = 39/160 (24%), Positives = 58/160 (35%), Gaps = 37/160 (23%)
Query: 184 LPPAPEFP-----SPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDDGDDSESEDGPPKK 238
+PP P P PP R + +L S P V+ + P D ++ PP
Sbjct: 116 VPPLPSSPPPPASVPPPRPSPSPPIL----VRSPPPSVRPIQSPPPPPSDRPTQSPPPPS 171
Query: 239 KQKKKVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISAL 298
+PT+ P + P R T+S S P+ SD
Sbjct: 172 PPSPPSE---RPTQSPP-------SPPSERPTQSPPPPSPPSPPSD-------------- 207
Query: 299 LQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQP--SPTE 336
+ S +P P +++P S P+SP +F P SP E
Sbjct: 208 -RPSQSPPPPPEDTKPQPPRRS-PNSPPPTFSSPPRSPPE 245
>At2g28440 En/Spm-like transposon protein
Length = 268
Score = 48.5 bits (114), Expect = 1e-05
Identities = 53/185 (28%), Positives = 79/185 (42%), Gaps = 26/185 (14%)
Query: 316 AQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQPE 375
AQ SP + S +PS T++PL +P+S E+ + P P SS +D P
Sbjct: 23 AQEESPSPAAVSPGREPS-TDSPL-----SPSSSPEEDS-----PLSPSSSPE--EDSPL 69
Query: 376 PNQTEPQPRTSDPSAPRAS---ERPAARTTDTDSSTAFTPVSFPINVIDSPPSNTSESIR 432
P + P+ P AP +S + P A ++ + + P S P PPS++ E+
Sbjct: 70 PPSSSPEE--DSPLAPSSSPEVDSPLAPSSSPEVDSPQPPSSSPEADSPLPPSSSPEANS 127
Query: 433 KFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSP 492
K +L + PSP P P+PE P P P+P P + S+ S P
Sbjct: 128 PQSPASSPKPESLAD----SPSPPP-PPPQPE---SPSSPSYPEPAPVPAPSDDDSDDDP 179
Query: 493 HPLVE 497
P E
Sbjct: 180 EPETE 184
Score = 33.1 bits (74), Expect = 0.58
Identities = 40/158 (25%), Positives = 60/158 (37%), Gaps = 20/158 (12%)
Query: 186 PAPEFPSPPKRKAQRKMVLDESSEESDVPLVKKTKRKPDD---GDDSESEDGPPKKKQKK 242
P+P SP + + + SS E D PL + + D S ED P
Sbjct: 28 PSPAAVSPGREPSTDSPLSPSSSPEEDSPLSPSSSPEEDSPLPPSSSPEEDSPLAPSSSP 87
Query: 243 KVRIVVKPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHS 302
+V + P+ ++ V +PP+ SS + P S P S+ S
Sbjct: 88 EVDSPLAPS----SSPEVDSPQPPS----SSPEADSPLPPSSSPEANSPQSPASSPKPES 139
Query: 303 --SNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTEAP 338
+P P P QP + P SP S+ +P+P AP
Sbjct: 140 LADSPSPPPPPPQPES-----PSSP--SYPEPAPVPAP 170
Score = 30.8 bits (68), Expect = 2.9
Identities = 26/101 (25%), Positives = 39/101 (37%), Gaps = 10/101 (9%)
Query: 454 SPRRYPGPRPERLVDPDEPIQPD--PEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAP 511
SP P + D P+ P PE ++ SS PL +S P + + P +
Sbjct: 50 SPEEDSPLSPSSSPEEDSPLPPSSSPEEDSPLAPSSSPEVDSPLAPSSSPEVDSPQPPSS 109
Query: 512 MINIGSPQGASEAHSSNHLASPEPNLSIIPYTHLQPTSLSE 552
SP S +SPE N P + +P SL++
Sbjct: 110 SPEADSPLPPS--------SSPEANSPQSPASSPKPESLAD 142
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 46.6 bits (109), Expect = 5e-05
Identities = 47/182 (25%), Positives = 66/182 (35%), Gaps = 21/182 (11%)
Query: 363 PLSSEPIVQDQ---PEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINV 419
P+ S P + P N T P +S P+ P + + D S +F+P P
Sbjct: 5 PVDSSPAPETSNGTPPSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAPPTQ 64
Query: 420 IDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEP 479
SPP++ S S + PSP+ P P + P+ P P P
Sbjct: 65 ETSPPTSPSSS----------------PPVVANPSPQTPENPSPP-APEGSTPVTP-PAP 106
Query: 480 AQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHLASPEPNLSI 539
Q+ SNQS R P +D T+ N +S +S SP P SI
Sbjct: 107 PQTPSNQSPERPTPPSPGANDDRNRTNGGNNNRDGSTPSPPSSGNRTSGDGGSPSPPRSI 166
Query: 540 IP 541
P
Sbjct: 167 SP 168
Score = 43.9 bits (102), Expect = 3e-04
Identities = 48/184 (26%), Positives = 70/184 (37%), Gaps = 16/184 (8%)
Query: 250 PTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPI 309
P PA T PP+ T S SS P S + A P S
Sbjct: 5 PVDSSPAPETSNGT-PPSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFS------- 56
Query: 310 PESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIP---YDPLSS 366
P P Q TSPP SP SS P P +NP+ + + ++ +T P P +
Sbjct: 57 PPPAPPTQETSPPTSPSSS---PPVVANPSPQTPENPSPPAPEGSTPVTPPAPPQTPSNQ 113
Query: 367 EPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSN 426
P P P + + RT+ + R P+ ++ +S S P ++ SPP N
Sbjct: 114 SPERPTPPSPGANDDRNRTNGGNNNRDGSTPSPPSSGNRTSGDGGSPSPPRSI--SPPQN 171
Query: 427 TSES 430
+ +S
Sbjct: 172 SGDS 175
Score = 33.5 bits (75), Expect = 0.44
Identities = 41/185 (22%), Positives = 61/185 (32%), Gaps = 27/185 (14%)
Query: 223 PDDGDDSESEDGPPKKKQKKKVRIVVKP------TRVEPAAAVVRRTEPPARVTRSSAHS 276
P +G +E PP + P + P A + T PP +S S
Sbjct: 20 PSNGTSPSNESSPPTPPSSPPPSSISAPPPDISASFSPPPAPPTQETSPP-----TSPSS 74
Query: 277 SKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPHSPRSSFFQPSPTE 336
S P +A+ Q NP P PE +PP +P + +
Sbjct: 75 SPPVVANPSP-------------QTPENPSPPAPEGSTPVTPPAPPQTPSNQSPERPTPP 121
Query: 337 APLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQD--QPEPNQTEPQPRTSDPSAPRAS 394
+P N +N T+ + T P P S D P P ++ P+ S S +
Sbjct: 122 SPGANDDRNRTNGGNNNRDGST-PSPPSSGNRTSGDGGSPSPPRSISPPQNSGDSDSSSG 180
Query: 395 ERPAA 399
P A
Sbjct: 181 NHPQA 185
>At1g23720 unknown protein
Length = 895
Score = 45.8 bits (107), Expect = 9e-05
Identities = 68/280 (24%), Positives = 95/280 (33%), Gaps = 48/280 (17%)
Query: 250 PTRVEPAAAVVRRTEPPARVTRSS-----AHSSKPALASDDDLNLFDALPISALLQHSSN 304
P PA V ++ PP V S + S KP S ++ + P
Sbjct: 595 PPYYSPAPKPVYKSPPPPYVYNSPPPPYYSPSPKPTYKSPPPPYVYSSPP---------- 644
Query: 305 PLTPIPESQPAAQTTSPPH---SPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPY 361
P P +P ++ PP+ SP ++ PSP PT +S P + + P
Sbjct: 645 PPYYSPTPKPTYKSPPPPYVYSSPPPPYYSPSP----------KPTYKSPPPPYVYSSPP 694
Query: 362 DPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVID 421
P S P+P P P S P P+ + T P V
Sbjct: 695 PPYYSPA-----PKPTYKSPPPPYVYSSPPPPYYSPSPKPTYKSPP--------PPYVYS 741
Query: 422 SPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRY-PGPRPERLVDPDEPI--QPDPE 478
SPP S +E + S Y + P P Y P P+ E P + P P
Sbjct: 742 SPPPPPYYSPSPKVEYK----SPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPP 797
Query: 479 PAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSP 518
P S S + +SP P S P T +P + SP
Sbjct: 798 PYYSPSPKVEYKSPPPPYVYSSPPPPTYYSPSPKVEYKSP 837
Score = 38.1 bits (87), Expect = 0.018
Identities = 68/316 (21%), Positives = 95/316 (29%), Gaps = 40/316 (12%)
Query: 249 KPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTP 308
KPT P V + PP + S KP S P + +S P
Sbjct: 376 KPTYKSPPPPYVYSSPPPPYYSPSP----KPVYKS----------PPPPYIYNSPPPPYY 421
Query: 309 IPESQPAAQTTSPPH---SPRSSFFQPSPT----EAPLWNLLQNPTSRSEDPTSLLTIPY 361
P +P+ ++ PP+ SP ++ PSP +P + +P P+ +
Sbjct: 422 SPSPKPSYKSPPPPYVYSSPPPPYYSPSPKLTYKSSPPPYVYSSPPPPYYSPSPKVVYKS 481
Query: 362 DPLSSEPIVQDQPEPNQTEPQPRTS--DPSAPRASERPAARTTDTDSSTAFTPVSFPINV 419
P P V P P P P+ S P P P + P +V
Sbjct: 482 PP---PPYVYSSPPPPYYSPSPKPSYKSPPPPYVYNSPPPPYYSPSPKVIYKSPPHP-HV 537
Query: 420 IDSPPSNTSESIRKFMEIRKEKVSAL------EEYYLTCPSPRRYPGPRPERLVDPDEP- 472
PP S +E + + YY P P P P P P
Sbjct: 538 CVCPPPPPCYSHSPKIEYKSPPTPYVYHSPPPPPYYSPSPKPAYKSSPPPYVYSSPPPPY 597
Query: 473 IQPDPEPA-QSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHLA 531
P P+P +S SP P + P P P + P
Sbjct: 598 YSPAPKPVYKSPPPPYVYNSPPPPYYSPSPKPTYKSPPPPYVYSSPPPPYYSPTPKPTYK 657
Query: 532 SPEPNLSIIPYTHLQP 547
SP P PY + P
Sbjct: 658 SPPP-----PYVYSSP 668
Score = 38.1 bits (87), Expect = 0.018
Identities = 64/304 (21%), Positives = 96/304 (31%), Gaps = 42/304 (13%)
Query: 249 KPTRVEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTP 308
KPT P + + PP + S KP S ++ + P P
Sbjct: 201 KPTYKSPPPPYIYSSPPPPYYSPSP----KPVYKSPPPPYVYSSPP----------PPYY 246
Query: 309 IPESQPAAQTTSPPH---SPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLS 365
P +PA ++ PP+ SP ++ PSP P++ P + P PY S
Sbjct: 247 SPSPKPAYKSPPPPYVYSSPPPPYYSPSPK--PIYKSPPPPYVYNSPPP-----PYYSPS 299
Query: 366 SEPIVQDQPEPN--QTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSP 423
+P + P P P P S P P ++ ++P P P
Sbjct: 300 PKPAYKSPPPPYVYSFPPPPYYSPSPKPVYKSPPPPYVYNSPPPPYYSPSPKPAYKSPPP 359
Query: 424 PSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPA--- 480
P S + + Y + P P Y P P +P+ P P
Sbjct: 360 PYVYSSPPPPYYSPSPKPT------YKSPPPPYVYSSPPPPYYSPSPKPVYKSPPPPYIY 413
Query: 481 -------QSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHSSNHLASP 533
S S + S +SP P S P P+ + SP + SP
Sbjct: 414 NSPPPPYYSPSPKPSYKSPPPPYVYSSPPPPYYSPSPKLTYKSSPPPYVYSSPPPPYYSP 473
Query: 534 EPNL 537
P +
Sbjct: 474 SPKV 477
Score = 34.3 bits (77), Expect = 0.26
Identities = 61/276 (22%), Positives = 80/276 (28%), Gaps = 54/276 (19%)
Query: 303 SNPLTPIPESQPAAQTTSPP-----HSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLL 357
S+P P P SPP SP ++ PSP PT +S P +
Sbjct: 89 SSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSP----------KPTYKSPPPPYVY 138
Query: 358 TIPYDPLSSE-----------PIVQDQPEPNQTEPQPRTSDPSAPRA---SERPAARTTD 403
P P S P V P P P P+ S P + P +
Sbjct: 139 NSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSPPPPYYSP 198
Query: 404 TDSSTAFTPVSFPINVIDSPP----SNTSESIRKFMEIRKEKVSALEEYYLTCPSPRR-- 457
+ T +P P + SPP S + + + K S YY P P
Sbjct: 199 SPKPTYKSPP--PPYIYSSPPPPYYSPSPKPVYKSPPPPYVYSSPPPPYYSPSPKPAYKS 256
Query: 458 ------YPGPRPERLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAP 511
Y P P +PI P P + SP P + P P P
Sbjct: 257 PPPPYVYSSPPPPYYSPSPKPIYKSPPPPYVYN------SPPPPYYSPSPKPAYKSPPPP 310
Query: 512 MINIGSPQGASEAHSSNHLASPEPNLSIIPYTHLQP 547
+ P SP P PY + P
Sbjct: 311 YVYSFPPPPYYSPSPKPVYKSPPP-----PYVYNSP 341
>At4g37450 arabinogalactan protein AGP18
Length = 209
Score = 45.4 bits (106), Expect = 1e-04
Identities = 53/189 (28%), Positives = 72/189 (38%), Gaps = 40/189 (21%)
Query: 314 PAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSEPIVQDQ 373
P+A TTSP SP + SPT AP + PT+ + P P S P
Sbjct: 35 PSAPTTSPTKSPAVT----SPTTAPA----KTPTASASSPVESPKSPAPVSESSPPPTPV 86
Query: 374 PEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSP-PSNTSESIR 432
PE + P P S P + P A DS A PV+ P+ + +P PS ++ +
Sbjct: 87 PESSPPVPAPMVSSPVSSPPVPAPVA-----DSPPA--PVAAPVADVPAPAPSKHKKTTK 139
Query: 433 KFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQ-PDP-----EPAQSVSNQ 486
K +K + + P P PE L P P + P P P S +Q
Sbjct: 140 K---SKKHQAA---------------PAPAPELLGPPAPPTESPGPNSDAFSPGPSADDQ 181
Query: 487 SSVRSPHPL 495
S S L
Sbjct: 182 SGAASTRVL 190
Score = 39.7 bits (91), Expect = 0.006
Identities = 38/166 (22%), Positives = 69/166 (40%), Gaps = 13/166 (7%)
Query: 247 VVKPTR--VEPAAAVVRRTEPPARVTRSSAHSSKPALASDDDLNLFDALPISALLQHSSN 304
+ PT+ P+A T+ PA + ++A + P ++ + ++ A + SS
Sbjct: 25 ISSPTKSPTTPSAPTTSPTKSPAVTSPTTAPAKTPTASASSPV---ESPKSPAPVSESSP 81
Query: 305 PLTPIPESQP-------AAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLL 357
P TP+PES P ++ +SPP + P+P AP+ ++ S+ + T
Sbjct: 82 PPTPVPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVADVPAPAPSKHKKTTKKS 141
Query: 358 TIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPS-APRASERPAARTT 402
+ P + P P P P + S P A ++ A +T
Sbjct: 142 KKHQAAPAPAPELLGPPAPPTESPGPNSDAFSPGPSADDQSGAAST 187
Score = 29.3 bits (64), Expect = 8.3
Identities = 34/157 (21%), Positives = 59/157 (36%), Gaps = 9/157 (5%)
Query: 379 TEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSP--PSNTSESIRKFME 436
+ P + PSAP S + T ++ A TP + + ++SP P+ SES
Sbjct: 26 SSPTKSPTTPSAPTTSPTKSPAVTSPTTAPAKTPTASASSPVESPKSPAPVSESSPPPTP 85
Query: 437 IRKEKVSALEEYYLTCPSPRRYPGPRPERLVDP-DEPIQPDPEPAQSVSNQSSVRS---- 491
+ + + S P P + P P+ P PA S +++ +S
Sbjct: 86 VPESSPPVPAPMVSSPVSSPPVPAPVADSPPAPVAAPVADVPAPAPSKHKKTTKKSKKHQ 145
Query: 492 --PHPLVETSDPHLGTSDPNAPMINIGSPQGASEAHS 526
P P E P ++ P + SP +++ S
Sbjct: 146 AAPAPAPELLGPPAPPTESPGPNSDAFSPGPSADDQS 182
>At1g68725 putative arabinogalactan protein AGP19
Length = 248
Score = 45.4 bits (106), Expect = 1e-04
Identities = 42/178 (23%), Positives = 62/178 (34%), Gaps = 16/178 (8%)
Query: 308 PIPESQPAAQTTSPPHSPRSSFFQPSPTEAPLWNLLQNPTSRSEDPTSLLTIPYDPLSSE 367
P+ +QP A +PP + + P+P AP+ + P + P + P S
Sbjct: 60 PVSAAQPPASPVTPPPAVTPTS-PPAPKVAPVISPATPPPQPPQSPPASAPTVSPPPVSP 118
Query: 368 PIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDTDSSTAFTPVSFPINVIDSPPSNT 427
P P P P P + P AS PA A +P+S P +P +
Sbjct: 119 PPAPTSPPPTPASPPPAPASPPPAPASPPPA--PVSPPPVQAPSPISLPPAPAPAPTKH- 175
Query: 428 SESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPERLVDPDEPIQPDPEPAQSVSN 485
K K ++ P+P P P L DP + P P P + N
Sbjct: 176 -----------KRKHKHKRHHHAPAPAPIPPSPPSPPVLTDPQD-TAPAPSPNTNGGN 221
Score = 33.9 bits (76), Expect = 0.34
Identities = 43/194 (22%), Positives = 58/194 (29%), Gaps = 27/194 (13%)
Query: 345 NPTSRSEDPTSLLTIPYDPLSSEPIVQDQPEPNQTEPQPRTSDPSAPRASERPAARTTDT 404
N + P + T P ++ P P P T P + P A + PA T +
Sbjct: 23 NAQGPAASPVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAV-TPTS 81
Query: 405 DSSTAFTPVSFPINVIDSPPSNTSESIRKFMEIRKEKVSALEEYYLTCPSPRRYPGPRPE 464
+ PV P PP + S T P P P P
Sbjct: 82 PPAPKVAPVISPATPPPQPPQSPPASAP------------------TVSPPPVSPPPAPT 123
Query: 465 RLVDPDEPIQPDPEPAQSVSNQSSVRSPHPLVETSDPHLGTSDPNAPMINIGSPQGASEA 524
P P P P PA S + SP P + P S + P +P
Sbjct: 124 S--PPPTPASPPPAPA---SPPPAPASPPPAPVSPPPVQAPSPISLPPAPAPAPTKHKRK 178
Query: 525 HS---SNHLASPEP 535
H +H +P P
Sbjct: 179 HKHKRHHHAPAPAP 192
Score = 31.6 bits (70), Expect = 1.7
Identities = 43/196 (21%), Positives = 61/196 (30%), Gaps = 23/196 (11%)
Query: 214 PLVKKTKRKPDDGDDSESEDGPPKKKQKKKVRIVVKPTRVEPAAAVVRRTEPPAR----V 269
P+ T P + PP V P V T PPA V
Sbjct: 31 PVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAVTPTSPPAPKVAPV 90
Query: 270 TRSSAHSSKPALASDDDLNLFDALPISALLQHSSNPLTPIPESQPAAQTTSPPH--SPRS 327
+ +P + P+S +S P P P S P A + PP SP
Sbjct: 91 ISPATPPPQPPQSPPASAPTVSPPPVSPPPAPTSPP--PTPASPPPAPASPPPAPASPPP 148
Query: 328 SFFQPSPTEAPLWNLLQNPTSRSEDPT-------------SLLTIPYDPLSSEPIVQDQP 374
+ P P +AP + + P + + PT + P P P V P
Sbjct: 149 APVSPPPVQAP--SPISLPPAPAPAPTKHKRKHKHKRHHHAPAPAPIPPSPPSPPVLTDP 206
Query: 375 EPNQTEPQPRTSDPSA 390
+ P P T+ +A
Sbjct: 207 QDTAPAPSPNTNGGNA 222
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,356,381
Number of Sequences: 26719
Number of extensions: 838853
Number of successful extensions: 6144
Number of sequences better than 10.0: 360
Number of HSP's better than 10.0 without gapping: 75
Number of HSP's successfully gapped in prelim test: 292
Number of HSP's that attempted gapping in prelim test: 4500
Number of HSP's gapped (non-prelim): 1105
length of query: 742
length of database: 11,318,596
effective HSP length: 107
effective length of query: 635
effective length of database: 8,459,663
effective search space: 5371886005
effective search space used: 5371886005
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0359.4


