

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0359.12
(1065 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
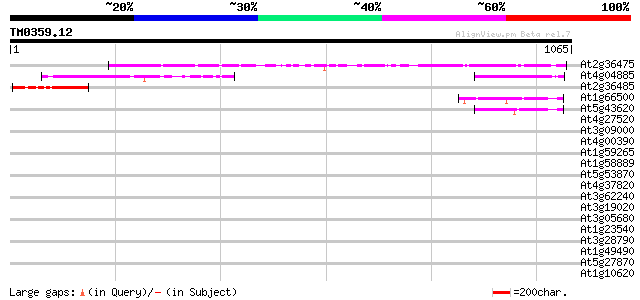
Score E
Sequences producing significant alignments: (bits) Value
At2g36475 hypothetical protein 352 5e-97
At4g04885 unknown protein 198 2e-50
At2g36485 unknown protein 113 5e-25
At1g66500 unknown protein 97 6e-20
At5g43620 S-locus protein 4-like 95 2e-19
At4g27520 early nodulin-like 2 predicted GPI-anchored protein 43 0.001
At3g09000 unknown protein 42 0.002
At4g00390 unknown protein 41 0.003
At1g59265 polyprotein, putative 41 0.004
At1g58889 polyprotein, putative 41 0.004
At5g53870 predicted GPI-anchored protein 40 0.009
At4g37820 unknown protein 40 0.009
At3g62240 putative zinc-finger protein 40 0.009
At3g19020 hypothetical protein 39 0.012
At3g05680 unknown protein 39 0.016
At1g23540 putative serine/threonine protein kinase 39 0.016
At3g28790 unknown protein 39 0.021
At1g49490 hypothetical protein 39 0.021
At5g27870 pectin methyl-esterase - like protein 38 0.035
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 38 0.035
>At2g36475 hypothetical protein
Length = 788
Score = 352 bits (904), Expect = 5e-97
Identities = 300/883 (33%), Positives = 421/883 (46%), Gaps = 133/883 (15%)
Query: 188 VHHSMRHLFGTWRGVFPSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRPPPSIHVN 247
+H +MRHLFGTW+GVF QTLQ IEKELGF +GS++ +T R++ QSQRPP SIHVN
Sbjct: 1 MHSNMRHLFGTWKGVFHPQTLQLIEKELGFNAKSDGSAAVVSTARAEPQSQRPPHSIHVN 60
Query: 248 PKYLERQRLQQSSRTKG-VDDMSGAIANSNDDPEMPDRALGVKRPRADPSNNQRAHRDAF 306
PKYLERQRLQQS RTKG V D+ N D + +R + N +R RD
Sbjct: 61 PKYLERQRLQQSGRTKGMVTDVPETAPNLTRDSDRLERVSSI--ASGGSWNIRRPQRDLL 118
Query: 307 NDSVREKSISASYGGNEYGSDLSRNLGMGTGRTGGRVAELGSDKSGYNKAAGAGVAGTIS 366
++ + EK I + G +Y SDL N G R+ + G +K Y A IS
Sbjct: 119 SEPLYEKDIESIAGEYDYASDLPHNSRSVIKNVGSRITDDGCEKQWY--GATNRDPDLIS 176
Query: 367 GQRNGLSIKNNFLNTEASMILDARNQ-PRQNINSLQRGVMSNSWKHSEEEEFMWDEMNAG 425
QR+GL K+ N + + + + P +NI GV +SWK+SEEEEFMWD M++
Sbjct: 177 DQRDGLHSKSRTSNYATARVENLESSGPSRNI-----GVPYDSWKNSEEEEFMWD-MHSR 230
Query: 426 LTGHDAASVPNNLSKDSWTSDDENLEVEDNHHQSRNPFGANVDTEMSIESQATERKQLPA 485
L+ D A++ K+ + DE+ +E +H + P + +D P
Sbjct: 231 LSETDVATIN---PKNELHAPDESERLESENHLLKRPRFSALD---------------PR 272
Query: 486 FRPHPSLSWKLQEQHSIDELNQKPGHSDGFVSTLGALPTNTNSAATRMGNRSLLPKATIG 545
F P S + EQ + GH F S TN S ATR G + P+ +
Sbjct: 273 FDPANSTNSYSSEQKDPSSI----GHW-AFSS------TNATSTATRKG---IQPQPRVA 318
Query: 546 LAGNLGQLHSVGDESPSRESSLRQQSPPPVSVHLPHLMQNLAEQDHPQTH-------KVS 598
+G L PS S +QSP +H QN+ +QD + H + S
Sbjct: 319 SSGIL----------PSSGSGSDRQSP----LHDSTSKQNVTKQDVRRAHSLPQRDPRAS 364
Query: 599 KFSGGLHSQNI-KDISPALPPNIQVGKFHRSQLRDLQSGSFSSMTFQPSHKQQLGSSHTE 657
+F QN+ +D S LP + +F + +R+L F S
Sbjct: 365 RFPA---KQNVPRDDSVRLPSS--SSQFKNTNMRELPVEIFDSK---------------- 403
Query: 658 VTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSI 717
+A P L+ T QP + A K G LSN + S D N P
Sbjct: 404 -SAAENAPGLTLA--SEATGQPNMSDLLEAVMKSGILSNNSTCGAI-KEESHDEVN-PGA 458
Query: 718 LGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPAS 777
L + + S P L P+S +A+ L R K S+ P + A+
Sbjct: 459 L----TLPAASKPKTL--PIS-LATDNLLARLKVEQSSAPLV--------------SCAA 497
Query: 778 SNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSIT 837
S +VQ S + A +P++ LL+SLV+KGLISA PS D S + +
Sbjct: 498 SLTGITSVQTSKEKSKASDPLSCLLSSLVSKGLISASKTELPSAPSITQEHSPDHSTNSS 557
Query: 838 TSSSLPGASVSGSA-VPVPSTKDEVDDGAKTPISLSESTSTEIRNFIGFEFKPDVIRELH 896
S S+ A S V PST +V G P SE++ +E ++ IG +F+ D IRELH
Sbjct: 558 MSVSVVPADAQPSVLVKGPSTAPKV-KGLAAP---SETSKSEPKDLIGLKFRADKIRELH 613
Query: 897 PSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHATREREHSGLIKASR-WYLKSSDWV 955
PSVIS LFDD PH C+ CS++LK +E+ DRH+E H ++ E SG R W+ K +W+
Sbjct: 614 PSVISSLFDDLPHLCTSCSVRLKQKEELDRHMELHDKKKLELSGTNSKCRVWFPKVDNWI 673
Query: 956 VGKVESLSENEFADSVDRYGKETDRNQEDAM-VLADENQCLCVLCGELFEEFYCQENGEW 1014
K L E E+ + + E + ED V ADE QC C+LCGE+FE+++ QE +W
Sbjct: 674 AAKAGEL-EPEYEEVL----SEPESAIEDCQAVAADETQCACILCGEVFEDYFSQEMAQW 728
Query: 1015 MFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCLSENPVSSV 1057
MFKGA Y+ N N S GPI+H CL+ + + S+
Sbjct: 729 MFKGASYLTNPPAN--------SEASGPIVHTGCLTTSSLQSL 763
>At4g04885 unknown protein
Length = 808
Score = 198 bits (503), Expect = 2e-50
Identities = 136/376 (36%), Positives = 185/376 (49%), Gaps = 60/376 (15%)
Query: 61 RLRAAANSRDLESSDFGRGDGGGGYHPRPPPFQELVAQYKSALAELTFNSKPIITNLTII 120
R +A N R+ E GGG PP E+V Y+ L ELTFNSKPIIT+LTII
Sbjct: 39 RFKALLNQREDEF--------GGGEEVLPPSMDEIVQLYEVVLGELTFNSKPIITDLTII 90
Query: 121 AGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSIVKNIGRGYVKYFAARLPEVFCKA 180
AGE + + IA +C I+E P EQKLPSLYLLDSIVKNIGR Y +YF++RLPEVFC A
Sbjct: 91 AGEQREHGEGIANAICTRILEAPVEQKLPSLYLLDSIVKNIGRDYGRYFSSRLPEVFCLA 150
Query: 181 YRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRP 240
YR P++H SMRHLFGTW VFP L+ I+ +L + A N SS + S+ +P
Sbjct: 151 YRQAHPSLHPSMRHLFGTWSSVFPPPVLRKIDMQLQLSSAANQSSVGA------SEPSQP 204
Query: 241 PPSIHVNPKYLER----------QRLQQSSRTKGVDDMSGAIANSNDDPEMPDRALGVKR 290
IHVNPKYL R + + S+R G + + G + D E P
Sbjct: 205 TRGIHVNPKYLRRLEPSAAENNLRGINSSARVYGQNSL-GGYNDFEDQLESPSSL----- 258
Query: 291 PRADPSNNQRAHRDAFNDSVREKSISASYGGNEYGSDLSRNLGMGTGRTGGRVAELGSDK 350
+ P R D N S+ + N GM GR +
Sbjct: 259 -SSTPDGFTRRSNDGANP-----------------SNQAFNYGM------GRATSRDDEH 294
Query: 351 SGYNKAAGAGVAGTISGQRNGLSIKNNFLNTEASMILDARNQPRQNINSLQRGVMSNSWK 410
+ + G +R I ++T + + N+P +++N + M W+
Sbjct: 295 MEWRRKENLGQGN--DHERPRALIDAYGVDTSKHVTI---NKPIRDMNGM-HSKMVTPWQ 348
Query: 411 HSEEEEFMWDEMNAGL 426
++EEEEF W++M+ L
Sbjct: 349 NTEEEEFDWEDMSPTL 364
Score = 118 bits (295), Expect = 2e-26
Identities = 63/174 (36%), Positives = 95/174 (54%), Gaps = 10/174 (5%)
Query: 883 IGFEFKPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHATRER--EHSG 940
+G EF D+++ + S IS L+ D P C+ C L+ K QE+ +H++WH T+ R ++
Sbjct: 625 LGLEFDADMLKIRNESAISALYGDLPRQCTTCGLRFKCQEEHSKHMDWHVTKNRMSKNHK 684
Query: 941 LIKASRWYLKSSDWVVGKVESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCG 1000
+ +W++ +S W+ G E+L + ++ ED V ADE+Q C LCG
Sbjct: 685 QNPSRKWFVSASMWLSG-AEALGAEAVPGFLPTEPTTEKKDDEDMAVPADEDQTSCALCG 743
Query: 1001 ELFEEFYCQENGEWMFKGAVYV--PNSDINDDMGIRDASTGGGPIIHARCLSEN 1052
E FE+FY E EWM+KGAVY+ P D D S GPI+HA+C E+
Sbjct: 744 EPFEDFYSDETEEWMYKGAVYMNAPEESTTD----MDKSQ-LGPIVHAKCRPES 792
>At2g36485 unknown protein
Length = 158
Score = 113 bits (283), Expect = 5e-25
Identities = 72/146 (49%), Positives = 93/146 (63%), Gaps = 9/146 (6%)
Query: 6 RRPFDRSREPG-LKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAARLRA 64
RRPFDRSR+PG +K+PR+ EE R N ++RQFL QR A + ++ +++R R
Sbjct: 5 RRPFDRSRDPGPMKKPRLSEESIRPV---NSNARQFLSQRTLGTATA-VTVPPASSRFRV 60
Query: 65 AANSRDLESSDFGRGDGGGGYHPRPP-PFQELVAQYKSALAELTFNSKPIITNLTIIAGE 123
+ R+ ESS Y P+P P ELV QYKSALAELTFNSKPIITNLTIIAGE
Sbjct: 61 SG--RETESSIVS-DPSREAYQPQPVHPHYELVNQYKSALAELTFNSKPIITNLTIIAGE 117
Query: 124 NQAAEQAIAATVCANIIEVPSEQKLP 149
N A +A+ +C NI+EV ++ P
Sbjct: 118 NVHAAKAVVTAICNNILEVNTQFSCP 143
>At1g66500 unknown protein
Length = 416
Score = 96.7 bits (239), Expect = 6e-20
Identities = 62/213 (29%), Positives = 95/213 (44%), Gaps = 31/213 (14%)
Query: 853 PVPSTKDEVD-----DGAKTPISLSESTSTEIRNFIGFEFKPDVIRELHPSVISGLFDDF 907
P+ +K+ D + K +L S S + + F+ P + H SVI L+ D
Sbjct: 194 PIVLSKELTDLLSLLNNEKEKKTLEASNSDSLPVGLSFD-NPSSLNVRHESVIKSLYSDM 252
Query: 908 PHHCSICSLKLKFQEQFDRHLEWHATREREHSGLI-------KASRWYLKSSDWVVGKV- 959
P CS C L+ K QE+ +H++WH + R K+ W +S W+
Sbjct: 253 PRQCSSCGLRFKCQEEHSKHMDWHVRKNRSVKTTTRLGQQPKKSRGWLASASLWLCAATG 312
Query: 960 -ESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKG 1018
E++ F + + K D + MV ADE+Q C LC E FEEF+ E+ +WM+K
Sbjct: 313 GETVEVASFGGEMQK-KKGKDEEPKQLMVPADEDQKNCALCVEPFEEFFSHEDDDWMYKD 371
Query: 1019 AVYVPNSDINDDMGIRDASTGGGPIIHARCLSE 1051
AVY+ T G I+H +C+ E
Sbjct: 372 AVYL---------------TKNGRIVHVKCMPE 389
>At5g43620 S-locus protein 4-like
Length = 410
Score = 94.7 bits (234), Expect = 2e-19
Identities = 57/182 (31%), Positives = 82/182 (44%), Gaps = 31/182 (17%)
Query: 883 IGFEF-KPDVIRELHPSVISGLFDDFPHHCSICSLKLKFQEQFDRHLEWHATREREHSGL 941
+G F P + H SVI L+ D P C+ C ++ K QE+ +H++WH + R
Sbjct: 220 VGLSFDNPSSLNVRHESVIKSLYSDMPRQCTSCGVRFKCQEEHSKHMDWHVRKNRSVKTT 279
Query: 942 IKASRWYLKSSDWVV------------GKVESLSENEFADSVDRYGKETDRNQEDAMVLA 989
+ + KS W+ G VE S F + E D+ Q+ MV A
Sbjct: 280 TRLGQQPKKSRGWLASASLWLCAPTGGGTVEVAS---FGGGEMQKKNEKDQVQKQHMVPA 336
Query: 990 DENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDASTGGGPIIHARCL 1049
DE+Q C LC E FEEF+ E +WM+K AVY+ T G I+H +C+
Sbjct: 337 DEDQKNCALCVEPFEEFFSHEADDWMYKDAVYL---------------TKNGRIVHVKCM 381
Query: 1050 SE 1051
E
Sbjct: 382 PE 383
>At4g27520 early nodulin-like 2 predicted GPI-anchored protein
Length = 349
Score = 42.7 bits (99), Expect = 0.001
Identities = 47/194 (24%), Positives = 76/194 (38%), Gaps = 24/194 (12%)
Query: 690 KGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRP 749
KG KL+ + I+ R+P++ P S+P + PP A + P
Sbjct: 120 KGQKLNVVVISARIPSTAQSPHAAAPG----------SSTPGSMTPPGGAHS-------P 162
Query: 750 KDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKG 809
K +S P P + PP + P +SS + A S A + +P++ +
Sbjct: 163 KSSSPVSPTTSPPGSTTPPGGAHSPKSSSAVSPATSPPGSMAPKSGSPVSPTTSPPAPPK 222
Query: 810 LISAETETTAKVPSEMLTRLEDQSDSITTSS----SLPGASVSGSAVPV---PSTKDEVD 862
S + ++A + S S +I SS S PG+ S+ PV P+ +
Sbjct: 223 STSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGSMAPKSSSPVSNSPTVSPSLA 282
Query: 863 DGAKTPISLSESTS 876
G T S S+S S
Sbjct: 283 PGGSTSSSPSDSPS 296
Score = 35.4 bits (80), Expect = 0.17
Identities = 47/193 (24%), Positives = 65/193 (33%), Gaps = 14/193 (7%)
Query: 565 SSLRQQSPPPVSVHLPHLMQNLAEQDHPQTHKVSKFSGGLHS-QNIKDISPAL-PPNIQV 622
SS PP H P ++ P GG HS ++ +SPA PP
Sbjct: 147 SSTPGSMTPPGGAHSPKSSSPVSPTTSPPGSTTPP--GGAHSPKSSSAVSPATSPPGSMA 204
Query: 623 GKFHRSQLRDLQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTT 682
K SGS S T P + S + +A P P T P++
Sbjct: 205 PK----------SGSPVSPTTSPPAPPKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSA 254
Query: 683 RFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIAS 742
+ G S+ P++N S SL G S S P+G P + S
Sbjct: 255 PMTSPPGSMAPKSSSPVSNSPTVSPSLAPGGSTSSSPSDSPSGSAMGPSGDGPSAAGDIS 314
Query: 743 PPSLGRPKDNSSA 755
P+ + SSA
Sbjct: 315 TPAGAPGQKKSSA 327
>At3g09000 unknown protein
Length = 541
Score = 42.0 bits (97), Expect = 0.002
Identities = 49/192 (25%), Positives = 78/192 (40%), Gaps = 25/192 (13%)
Query: 674 RETEQPTT-TRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAG 732
R T +P T TR T ++ P+T R SRS + ++ R + S+ +
Sbjct: 154 RSTSRPATPTRRSTTPTTS---TSRPVTTRASNSRSSTPTSRATLTAARATTSTAAPRTT 210
Query: 733 LIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSAN 792
SA ++ P+ P+ SSA K P R A P R + P S + S A +S
Sbjct: 211 TTSSGSARSATPTRSNPRP-SSASSKKPVSRPATPTRRPSTPTGPSIVSSKAPSRGTSP- 268
Query: 793 NALNPIANLLNSLVAKGLISAETETTAK-------------VPSEMLTRLEDQSDSITTS 839
+P N L+ ++G + T +++ P + T L D+ ++ S
Sbjct: 269 ---SPTVNSLSKAPSRGTSPSPTLNSSRPWKPPEMPGFSLEAPPNLRTTLADR--PVSAS 323
Query: 840 SSLPG-ASVSGS 850
PG AS GS
Sbjct: 324 RGRPGVASAPGS 335
>At4g00390 unknown protein
Length = 364
Score = 41.2 bits (95), Expect = 0.003
Identities = 40/146 (27%), Positives = 65/146 (44%), Gaps = 8/146 (5%)
Query: 733 LIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSAN 792
L PP + + + +D+SS+ + P ++ P T+A PA S SAA A S++
Sbjct: 5 LDPPTAPSSDEDDVETSEDDSSSSEEDEPIKSL--PATTAAAPAKSTAVSAATPAKSTSV 62
Query: 793 NALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAV 852
+A P + S A +E+ET + SE + S S S S + +
Sbjct: 63 SAAAPSKSTAVSAAADSDSGSESETDS--DSESTDPPKSGSGKTIASKKKDDPSSSTATL 120
Query: 853 PVPSTKDEVDDGAKTPISLSESTSTE 878
+P+ K GAK S + +TST+
Sbjct: 121 ALPAVK----SGAKRAASEAATTSTK 142
>At1g59265 polyprotein, putative
Length = 1466
Score = 40.8 bits (94), Expect = 0.004
Identities = 33/120 (27%), Positives = 54/120 (44%), Gaps = 9/120 (7%)
Query: 645 PSHKQQLGSSHTEVTAKTEKPHLSKVPLPRE-----TEQPTTTRFETAAGKGGKLSNMPI 699
P Q+ SS+ + + + P + PR+ T QPT T+ +T + + +N
Sbjct: 806 PFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNP-- 863
Query: 700 TNRLPT--SRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALP 757
TN P+ ++SL T S P+ S+ SS PP I PP L + +N++ P
Sbjct: 864 TNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAP 923
Score = 31.6 bits (70), Expect = 2.5
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query: 204 PSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRP-PPSIHVNP 248
PSQ Q++ TPA + SSS S T + S S P PPSI ++P
Sbjct: 868 PSQLAQSLS-----TPAQSSSSSPSPTTSASSSSTSPTPPSILIHP 908
>At1g58889 polyprotein, putative
Length = 1466
Score = 40.8 bits (94), Expect = 0.004
Identities = 33/120 (27%), Positives = 54/120 (44%), Gaps = 9/120 (7%)
Query: 645 PSHKQQLGSSHTEVTAKTEKPHLSKVPLPRE-----TEQPTTTRFETAAGKGGKLSNMPI 699
P Q+ SS+ + + + P + PR+ T QPT T+ +T + + +N
Sbjct: 806 PFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNP-- 863
Query: 700 TNRLPT--SRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALP 757
TN P+ ++SL T S P+ S+ SS PP I PP L + +N++ P
Sbjct: 864 TNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAP 923
Score = 31.6 bits (70), Expect = 2.5
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 6/46 (13%)
Query: 204 PSQTLQAIEKELGFTPAVNGSSSTSATLRSDSQSQRP-PPSIHVNP 248
PSQ Q++ TPA + SSS S T + S S P PPSI ++P
Sbjct: 868 PSQLAQSLS-----TPAQSSSSSPSPTTSASSSSTSPTPPSILIHP 908
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 39.7 bits (91), Expect = 0.009
Identities = 52/231 (22%), Positives = 76/231 (32%), Gaps = 10/231 (4%)
Query: 566 SLRQQSPPPVSVHLPHLMQNLAEQDHPQTHKVSKFSGGLHSQNIKD-ISPALPPNIQVGK 624
++R Q P +P + + H V+ S SQ + +SPA PP
Sbjct: 129 AVRNQPSAPAHSPVPSVSPTQPPKSHSPVSPVAPASAPSKSQPPRSSVSPAQPPKSSSPI 188
Query: 625 FHRSQLRDLQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRF 684
H L + S S T PS K SH+ + P S P + +
Sbjct: 189 SHTPALSPSHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPSHS 248
Query: 685 ETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPP 744
A + S S T PS PS S SPA P SP
Sbjct: 249 PAHAPSHSPAHAPSHSPAHSPSHSPATPKSPS-----PSSSPAQSPA--TPSPMTPQSPS 301
Query: 745 SLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNAL 795
+ P + SA P + P S P + N+ + A +++ + L
Sbjct: 302 PVSSPSPDQSAAPS--DQSTPLAPSPSETTPTADNITAPAPSPRTNSASGL 350
>At4g37820 unknown protein
Length = 532
Score = 39.7 bits (91), Expect = 0.009
Identities = 58/251 (23%), Positives = 95/251 (37%), Gaps = 29/251 (11%)
Query: 257 QQSSRTKGVDDMSGAIANSNDDPEMPDRALGVKR---PRADPSNNQRAHRDAFNDSVREK 313
++ S+ +G D+S N ++ E + V+ A+ + N+ + D+ N+ EK
Sbjct: 86 EEGSKNEGGGDVSTDKENGDEIVEREEEEKAVEENNEKEAEGTGNEEGNEDS-NNGESEK 144
Query: 314 SISASYGGNEY-----------GSDLSRNLGMGTGRTGGRVAEL-GSDKSGYNKAAGAGV 361
+ S GGNE G D S + GT E+ G KS N V
Sbjct: 145 VVDESEGGNEISNEEAREINYKGDDASSEVMHGTEEKSNEKVEVEGESKS--NSTENVSV 202
Query: 362 AGTISGQRN----GLSIKNNFLNTEASMILDARNQPRQNINSLQRGVMSNSWKHSEEEEF 417
SG +N G IK LNT + D Q ++ + G S + E E
Sbjct: 203 HEDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSELDSKTGEKGFSDSNGELPET 262
Query: 418 MWDEMNAGLTGHDAASVPNNLS------KDSWTSDDENLEVEDNHHQSR-NPFGANVDTE 470
NA T + S + S + + +DE +V+ + +S+ G N
Sbjct: 263 NLSTSNATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSEEESKVKESGKNEKDA 322
Query: 471 MSIESQATERK 481
S + ++ E K
Sbjct: 323 SSSQDESKEEK 333
>At3g62240 putative zinc-finger protein
Length = 812
Score = 39.7 bits (91), Expect = 0.009
Identities = 62/267 (23%), Positives = 98/267 (36%), Gaps = 38/267 (14%)
Query: 564 ESSLRQQSPPPVSVHLPHLMQNLAEQDHPQTHKVSKFSGGLHSQNIKDISPALPPNIQVG 623
E S ++ +PPP S +E ++ + S + D+ P+ VG
Sbjct: 342 EYSRQEPAPPPSSAP-----PGFSENNNIHVDDTDPLIQPMESLSTTDMEPSSRYLQAVG 396
Query: 624 KFHRSQLRDLQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTR 683
F R L +F ++ Q S Q + S T A L R+T + +T
Sbjct: 397 SFGGGGSR-LGESAFPPLSGQQSSGQNVESLPTNTMAAR---------LRRQTNRTSTAS 446
Query: 684 FETAAGKGGKLSNMPITNRLPTSRSLDTGNLPS-----ILGVRPSQSSGSSPAGLI---- 734
+ +G P+ NR P S+ +G S +G P Q+S SS
Sbjct: 447 AIASPSQG-----WPVINRGPGQASITSGGNHSSSGWPAIGRTPVQASSSSVQSRSHNRV 501
Query: 735 ----PPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASS----NLKSAAVQ 786
P SA+ +SS+ P + R+ QP S PP SS N K+++
Sbjct: 502 SQPRPLASAVPQAARNANRIPHSSSAPNLSDTRSLQPSH-SDFPPVSSAVVQNRKTSSTT 560
Query: 787 ASSSANNALNPIANLLNSLVAKGLISA 813
S+N P N + + + SA
Sbjct: 561 TQGSSNTQPPPDVQSANKSLIEKMRSA 587
>At3g19020 hypothetical protein
Length = 951
Score = 39.3 bits (90), Expect = 0.012
Identities = 40/147 (27%), Positives = 59/147 (39%), Gaps = 19/147 (12%)
Query: 735 PPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNA 794
PP + PP + P S PP + P + LPPA+S + +A +SS +
Sbjct: 783 PPPPVHSPPPPVHSPPPPSPIYSPPPPVFSPPPKPVTPLPPATSPMANAPTPSSSESGEI 842
Query: 795 LNPI-------------ANLLNSLVAKGLIS----AETETTAKVPSEMLTRLEDQSDSIT 837
P+ ++ +S V K + +E E A VPS +E S
Sbjct: 843 STPVQAPTPDSEDIEAPSDSNHSPVFKSSPAPSPDSEPEVEAPVPSSE-PEVEAPKQSEA 901
Query: 838 TSSSLPGASVSGSAVPVPSTKDEVDDG 864
T SS P +S V P ++D DDG
Sbjct: 902 TPSSSPPSSNPSPDVTAPPSEDN-DDG 927
Score = 34.3 bits (77), Expect = 0.39
Identities = 34/123 (27%), Positives = 49/123 (39%), Gaps = 18/123 (14%)
Query: 661 KTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGV 720
+T KP S P P + EQP T + K+ + P+ + +P D + I
Sbjct: 569 ETPKPEESPKPQPPKQEQPPKT-------EAPKMGSPPLESPVPN----DPYDASPIKKR 617
Query: 721 RPSQSSGS-----SPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPP 775
RP S S + + PPV + PP + P + P PP + PP S PP
Sbjct: 618 RPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVFSPP--PPMHSPPPPVYSPPPP 675
Query: 776 ASS 778
S
Sbjct: 676 VHS 678
>At3g05680 unknown protein
Length = 2070
Score = 38.9 bits (89), Expect = 0.016
Identities = 81/386 (20%), Positives = 137/386 (34%), Gaps = 43/386 (11%)
Query: 444 TSDDENLEVEDNHHQSRNPFGANVDTEMSIESQA---TERKQLPAFRPHPSLSWKLQEQH 500
T D N + +H + P +NVD E + R ++ R PS+S +
Sbjct: 1628 TESDANGSSQFSHMGT--PVASNVDENAQSEFSSRISVSRPEMSLIR-EPSISSDRKFVE 1684
Query: 501 SIDELNQKPGHSDGFVSTLGALP-------TNTNSAATRMGNRSLLPKATIGLAGNL-GQ 552
DE + +S G +P + NS R+G + K+ G++ G
Sbjct: 1685 QADEAKKMAPLKSAGISESGFIPAYHMPGSSGQNSIDPRVGPQGFYSKSGQQHTGHIHGG 1744
Query: 553 LHSVG---DESPSRESSLRQQSPPPVSVHLPHLMQNLAEQDHPQTHKVSKFSGGLHSQNI 609
G + + L PP VS +PH +L+ Q P ++ SGG +
Sbjct: 1745 FSGRGVYEQKVMPNQPPLPLVPPPSVSPVIPHSSDSLSNQSSPFISHGTQSSGG--PTRL 1802
Query: 610 KDISPALPPNIQVGKFHRSQLR--DLQSGSFSSMTFQPSHKQQLG------SSHTEVTAK 661
P+ P + R +QS ++ + +QQ G S + VT
Sbjct: 1803 MPPLPSAIPQYSSNPYASLPPRTSTVQSFGYNHAGVGTTEQQQSGPTIDHQSGNLSVTGM 1862
Query: 662 TEKPHLSKVP---LPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSIL 718
T P + +P R + P + G K M + +P +SL+T ++P +
Sbjct: 1863 TSYPPPNLMPSHNFSRPSSLPVPFYGNPSHQGGDKPQTMLLVPSIP--QSLNTQSIPQLP 1920
Query: 719 GVRPSQSSG--SSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQ--------PP 768
++ SQ P + PP+ I+ P G N +P + Q PP
Sbjct: 1921 SMQLSQLQRPMQPPQHVRPPIQ-ISQPSEQGVSMQNPFQIPMHQMQLMQQTQVQPYYHPP 1979
Query: 769 RTSALPPASSNLKSAAVQASSSANNA 794
+ + + AVQ A +
Sbjct: 1980 QQQEISQVQQQQQHHAVQGQQGAGTS 2005
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 38.9 bits (89), Expect = 0.016
Identities = 32/120 (26%), Positives = 46/120 (37%), Gaps = 11/120 (9%)
Query: 663 EKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSIL--GV 720
E P S P +T P T E +A +P + P S D+ + P +
Sbjct: 6 ESPSSSPPAPPADTAPPPETPSENSA--------LPPVDSSPPSPPADSSSTPPLSEPST 57
Query: 721 RPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPP-RRAAQPPRTSALPPASSN 779
P S ++PP++ PPS P +S+ P P + PP S PPA N
Sbjct: 58 PPPDSQLPPLPSILPPLTDSPPPPSDSSPPVDSTPSPPPPTSNESPSPPEDSETPPAPPN 117
Score = 34.3 bits (77), Expect = 0.39
Identities = 29/78 (37%), Positives = 37/78 (47%), Gaps = 16/78 (20%)
Query: 716 SILGVRPSQSSGSSPAGLIPPV------SAI----ASPPSLGRPKDNSS----ALPKIPP 761
S LG PS S + PA PP SA+ +SPPS P D+SS + P PP
Sbjct: 2 SDLGESPSSSPPAPPADTAPPPETPSENSALPPVDSSPPS--PPADSSSTPPLSEPSTPP 59
Query: 762 RRAAQPPRTSALPPASSN 779
+ PP S LPP + +
Sbjct: 60 PDSQLPPLPSILPPLTDS 77
Score = 30.4 bits (67), Expect = 5.6
Identities = 17/44 (38%), Positives = 23/44 (51%), Gaps = 4/44 (9%)
Query: 751 DNSSALPKIPPRRAAQPPRT----SALPPASSNLKSAAVQASSS 790
++ S+ P PP A PP T SALPP S+ S +SS+
Sbjct: 6 ESPSSSPPAPPADTAPPPETPSENSALPPVDSSPPSPPADSSST 49
>At3g28790 unknown protein
Length = 608
Score = 38.5 bits (88), Expect = 0.021
Identities = 41/156 (26%), Positives = 61/156 (38%), Gaps = 23/156 (14%)
Query: 725 SSGSSPAGLIPPVSAIASP----PSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNL 780
SSG+SP+G P + +P PS P S+ P P P +A +
Sbjct: 270 SSGASPSGSPTPTPSTPTPSTPTPSTPTP---STPTPSTPTPSTPAPSTPAAGKTSEKGS 326
Query: 781 KSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSS 840
+SA+++ S++ + + A G +S ET + D+ TSS
Sbjct: 327 ESASMKKESNSKSE--------SESAASGSVSKTKETNKGSSGDTY------KDTTGTSS 372
Query: 841 SLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTS 876
P S SGS P PST + +K S S S
Sbjct: 373 GSPSGSPSGS--PTPSTSTDGKASSKGSASASAGAS 406
Score = 33.5 bits (75), Expect = 0.66
Identities = 30/102 (29%), Positives = 40/102 (38%), Gaps = 5/102 (4%)
Query: 776 ASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDS 835
+SSN KS + S + + I S G S+ + T +V +
Sbjct: 209 SSSNTKSQGTSSKSGSESTAGSIETNTGSKTEAGSKSSSSAKTKEVSGGS----SGNTYK 264
Query: 836 ITTSSSLPGASVSGSAVPVPSTKDEVDDGAKTPISLSESTST 877
TT SS GAS SGS P PST TP + + ST
Sbjct: 265 DTTGSS-SGASPSGSPTPTPSTPTPSTPTPSTPTPSTPTPST 305
>At1g49490 hypothetical protein
Length = 847
Score = 38.5 bits (88), Expect = 0.021
Identities = 35/156 (22%), Positives = 68/156 (43%), Gaps = 13/156 (8%)
Query: 722 PSQSSGSSPAGLIPPVSA---------IASPPSLGRPKDNSS-ALPKIPPRRAAQPPRTS 771
P+ SS S + ++ PV A + P + P + S P + P +A P +
Sbjct: 669 PTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTPVQAP 728
Query: 772 ALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGL-ISAETETTAKVPS-EMLTRL 829
+S + + + +++ S + A PI +++ + + T ++ V S E +
Sbjct: 729 TTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSSPEQSEEV 788
Query: 830 EDQSDSITTSSSLPGASVS-GSAVPVPSTKDEVDDG 864
E + SS+P +S S +++P P D+ DDG
Sbjct: 789 EAPEPTPVNPSSVPSSSPSTDTSIPPPENNDDDDDG 824
Score = 33.9 bits (76), Expect = 0.51
Identities = 36/135 (26%), Positives = 52/135 (37%), Gaps = 17/135 (12%)
Query: 654 SHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKG------GKLSNMPITNRL--PT 705
S E + K E+P + P P + + P T+ + + P+ NR P
Sbjct: 466 SKPEDSPKPEQPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPYDASPVKNRRSPPP 525
Query: 706 SRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKI--PPRR 763
+ DT P +P S S P+ + P + SPP P +S P + PP
Sbjct: 526 PKVEDTRVPPP----QPPMPSPSPPSPIYSPPPPVHSPPP---PVYSSPPPPHVYSPPPP 578
Query: 764 AAQPPRTSALPPASS 778
A PP S PP S
Sbjct: 579 VASPPPPSPPPPVHS 593
Score = 31.2 bits (69), Expect = 3.3
Identities = 39/175 (22%), Positives = 55/175 (31%), Gaps = 28/175 (16%)
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPP------------- 768
P S P PP + PP + P S PP + PP
Sbjct: 582 PPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPP 641
Query: 769 -RTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLT 827
+ PP + + A SS P + S + + A T + PS T
Sbjct: 642 THNTNQPPMGAPTPTQAPTPSSETTQVPTPSSESDQSQILSP-VQAPTPVQSSTPSSEPT 700
Query: 828 RLEDQSDS------------ITTSSSLPGASVSGSAVPVPSTK-DEVDDGAKTPI 869
++ S S T P S S VP PS++ ++ A TPI
Sbjct: 701 QVPTPSSSESYQAPNLSPVQAPTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPI 755
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 37.7 bits (86), Expect = 0.035
Identities = 53/213 (24%), Positives = 81/213 (37%), Gaps = 50/213 (23%)
Query: 689 GKG-----GKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASP 743
GKG G S TN T SL + + S SP+ ++ P + SP
Sbjct: 553 GKGVPYILGLFSGNGSTNSTVTGSSLSSN----------TTESSDSPSTVVTPST---SP 599
Query: 744 PS--LGRPKDNSSAL----PKIPPRRAAQPPRTSAL------PPASSNLKSAAVQASSSA 791
P+ LG P D S++ +P + PP T ++ P + +L S + SS
Sbjct: 600 PAGHLGSPSDTPSSVVSPSTSLPAGQLGAPPATPSMVVSPSTSPPAGHLGSPSDTPSSLV 659
Query: 792 NNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSA 851
+ + +P A L S ++T ++ PS + S+ P AS S S
Sbjct: 660 SPSTSPPAGHLGS-------PSDTPSSVVTPS-------------ASPSTSPSASPSVSP 699
Query: 852 VPVPSTKDEVDDGAKTPISLSESTSTEIRNFIG 884
PS A +S S S S ++ IG
Sbjct: 700 SAFPSASPSASPSASPSVSPSASPSASPQSSIG 732
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 37.7 bits (86), Expect = 0.035
Identities = 38/148 (25%), Positives = 59/148 (39%), Gaps = 20/148 (13%)
Query: 704 PTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRR 763
P +S TG+ P ++ P P L PP S + P RP DN+ P P
Sbjct: 101 PPPQSTPTGDSPVVI---PFPKPQLPPPSLFPPPSLVNQLPD-PRPNDNNILEPINNPIS 156
Query: 764 AAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPS 823
PP T PP+ N S + P+++LL ++ +
Sbjct: 157 LPSPPSTPFSPPSQEN----------SGSQGSPPLSSLL-----PPMLPLNPNSPGNPLQ 201
Query: 824 EMLTRLEDQSDSITTSSSLPG-ASVSGS 850
+ + L +S+ + +SSS P S+SGS
Sbjct: 202 PLDSPLGGESNRVPSSSSSPSPPSLSGS 229
Score = 32.0 bits (71), Expect = 1.9
Identities = 35/126 (27%), Positives = 43/126 (33%), Gaps = 11/126 (8%)
Query: 675 ETEQPTTTRFETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLI 734
ET QP T + + S P T P N P P+ S P+
Sbjct: 39 ETTQPPATSPPSPPSPDTQTSPPPATAAQPPP------NQPPNTTPPPTPPSSPPPSITP 92
Query: 735 PPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNA 794
PP PP P +S P + P Q P S PP S L + + NN
Sbjct: 93 PPSPPQPQPPPQSTPTGDS---PVVIPFPKPQLPPPSLFPPPS--LVNQLPDPRPNDNNI 147
Query: 795 LNPIAN 800
L PI N
Sbjct: 148 LEPINN 153
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,229,456
Number of Sequences: 26719
Number of extensions: 1180242
Number of successful extensions: 4428
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 3916
Number of HSP's gapped (non-prelim): 457
length of query: 1065
length of database: 11,318,596
effective HSP length: 110
effective length of query: 955
effective length of database: 8,379,506
effective search space: 8002428230
effective search space used: 8002428230
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0359.12


