

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0358.5
(189 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
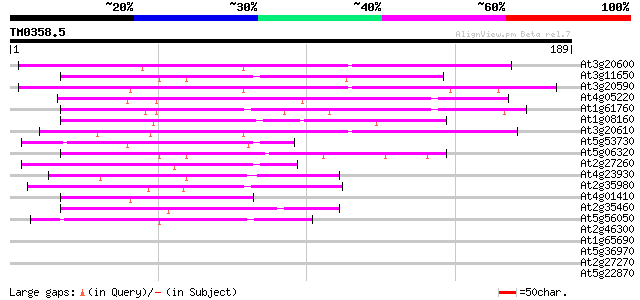
Score E
Sequences producing significant alignments: (bits) Value
At3g20600 non-race specific disease resistance protein (NDR1) 66 1e-11
At3g11650 unknown protein 57 4e-09
At3g20590 non-race specific disease resistance protein, putative 55 2e-08
At4g05220 putative protein 54 6e-08
At1g61760 hypothetical protein 50 9e-07
At1g08160 unknown protein 48 3e-06
At3g20610 non-race specific disease resistance protein, putative 45 3e-05
At5g53730 putative protein 44 4e-05
At5g06320 NDR1/HIN1-like protein 3 (NHL3) 44 4e-05
At2g27260 unknown protein 44 5e-05
At4g23930 unknown protein (At4g23930) 43 1e-04
At2g35980 similar to harpin-induced protein hin1 from tobacco 42 2e-04
At4g01410 putative hypoersensitive response protein 42 3e-04
At2g35460 similar to harpin-induced protein hin1 from tobacco 42 3e-04
At5g56050 unknown protein 41 4e-04
At2g46300 unknown protein 39 0.002
At1g65690 unknown protein 39 0.002
At5g36970 putative protein 38 0.003
At2g27270 unknown protein 38 0.003
At5g22870 putative protein 37 0.005
>At3g20600 non-race specific disease resistance protein (NDR1)
Length = 219
Score = 66.2 bits (160), Expect = 1e-11
Identities = 48/178 (26%), Positives = 83/178 (45%), Gaps = 13/178 (7%)
Query: 4 GKRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSI-----EQHPGENGSIFYSLE 58
G+ + FI GL L LWL+LR P S+ I + + +N ++ + +
Sbjct: 11 GRNCCTCCLSFIFTAGLTSLFLWLSLRADKPKCSIQNFFIPALGKDPNSRDNTTLNFMVR 70
Query: 59 IENPNKDSSIYYDDIILSF-------LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
+NPNKD IYYDD+ L+F + VG T+ F+QG K + V
Sbjct: 71 CDNPNKDKGIYYDDVHLNFSTINTTKINSSALVLVGNYTVPKFYQGHKKKAKKWGQVKPL 130
Query: 112 HGAFKPLFKAISNATAELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRKK 169
+ L + N +A ++ L T+ ++K K+K +G+ + + ++ DG ++KK
Sbjct: 131 NNQ-TVLRAVLPNGSAVFRLDLKTQVRFKIVFWKTKRYGVEVGADVEVNGDGVKAQKK 187
>At3g11650 unknown protein
Length = 240
Score = 57.4 bits (137), Expect = 4e-09
Identities = 39/140 (27%), Positives = 64/140 (44%), Gaps = 13/140 (9%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE-NGSIFYSLE----IENPNKDSSIYYDD 72
LG+ L LWL RP + + V ++ + + N ++ YSL+ I NPN+ +YYD+
Sbjct: 66 LGVAALILWLIFRPNAVKFYVADANLNRFSFDPNNNLHYSLDLNFTIRNPNQRVGVYYDE 125
Query: 73 IILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARH------GAFKPLFKAISNAT 126
+S YG Q + G + SF+QG T + + ++ GA L +
Sbjct: 126 FSVSGYYGDQ--RFGSANVSSFYQGHKNTTVILTKIEGQNLVVLGDGARTDLKDDEKSGI 183
Query: 127 AELKVALTTRFQYKTWGIKS 146
+ L ++K W IKS
Sbjct: 184 YRINAKLRLSVRFKFWFIKS 203
>At3g20590 non-race specific disease resistance protein, putative
Length = 220
Score = 55.5 bits (132), Expect = 2e-08
Identities = 53/195 (27%), Positives = 84/195 (42%), Gaps = 15/195 (7%)
Query: 4 GKRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVV-----FLSIEQHPGENGSIFYSLE 58
G++ +FI L L LWL+LR K P S+ LS +N ++ + +
Sbjct: 11 GRKCCTCFFKFIFTTRLGALILWLSLRAKKPKCSIQNFYIPALSKNLSSRDNTTLNFMVR 70
Query: 59 IENPNKDSSIYYDDIILSF-------LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
+NPNKD IYYDD+ L+F V T+ F+QG K + V
Sbjct: 71 CDNPNKDKGIYYDDVHLTFSTINTTTTNSSDLVLVANYTVPKFYQGHKKKAKKWGQVWPL 130
Query: 112 HGAFKPLFKAISNATAELKVALTTRFQYKTWGIKSK-FHGLHLQGILPIDSDG-KLSRKK 169
+ L + N +A ++ L T ++K K+K + + + + ++ DG K +K
Sbjct: 131 NNQ-TVLRAVLPNGSAVFRLDLKTHVRFKIVFWKTKWYRRIKVGADVEVNGDGVKAQKKG 189
Query: 170 KKYPLSRSSRKLARS 184
K S SS L S
Sbjct: 190 SKTKKSDSSLPLRSS 204
>At4g05220 putative protein
Length = 226
Score = 53.5 bits (127), Expect = 6e-08
Identities = 44/162 (27%), Positives = 68/162 (41%), Gaps = 12/162 (7%)
Query: 17 FLGLLVLCLWLALRPKSPSYSV---VFLSIEQHPG-ENGSIFYSLEIENPNKDSSIYYDD 72
F+G++ LWL+LRP P + + V ++Q G EN I +++ I NPN+ +Y+D
Sbjct: 56 FVGVIAFILWLSLRPHRPRFHIQDFVVQGLDQPTGVENARIAFNVTILNPNQHMGVYFDS 115
Query: 73 IILSFLYGQQEDKVGETTIGSFHQG------TGKTRSVYDTVNARHGAFKPLFKAISNAT 126
+ S Y Q + F Q TG TVN+ +A
Sbjct: 116 MEGSIYYKDQRVGLIPLLNPFFQQPTNTTIVTGTLTGASLTVNSNRWTEFSNDRAQGTVG 175
Query: 127 AELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRK 168
L + T RF+ W SK H +H + + DG + K
Sbjct: 176 FRLDIVSTIRFKLHRW--ISKHHRMHANCNIVVGRDGLILPK 215
>At1g61760 hypothetical protein
Length = 224
Score = 49.7 bits (117), Expect = 9e-07
Identities = 47/171 (27%), Positives = 72/171 (41%), Gaps = 18/171 (10%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIE--QHPG--ENGSIFYSLEIENPNKDSSIYYDDI 73
LG++ LW++L+P P + SI P E I + + NPN++ IYYD +
Sbjct: 55 LGIITFILWISLQPHRPRVHIRGFSISGLSRPDGFETSHISFKITAHNPNQNVGIYYDSM 114
Query: 74 ILSFLYGQQEDKVGETTI-GSFHQGTGKTRSVYD-------TVNARHGAFKPLFKAISNA 125
S Y +E ++G T + F+Q T S+ VN +
Sbjct: 115 EGSVYY--KEKRIGSTKLTNPFYQDPKNTSSIDGALSRPAMAVNKDRWMEMERDRNQGKI 172
Query: 126 TAELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKL--SRKKKKYPL 174
LKV RF+ TW SK H ++ + I DG L + K K+ P+
Sbjct: 173 MFRLKVRSMIRFKVYTW--HSKSHKMYASCYIEIGWDGMLLSATKDKRCPV 221
>At1g08160 unknown protein
Length = 221
Score = 48.1 bits (113), Expect = 3e-06
Identities = 40/142 (28%), Positives = 60/142 (42%), Gaps = 15/142 (10%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHP------GENGSIFYSLEIENPNKDSSIYYD 71
+GL +L +L LRPK Y+V S+++ N Y ++ NP K S+ Y
Sbjct: 51 VGLAILITYLTLRPKRLIYTVEAASVQEFAIGNNDDHINAKFSYVIKSYNPEKHVSVRYH 110
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI------SNA 125
+ +S + Q V I F Q K + +T H F A S
Sbjct: 111 SMRISTAHHNQS--VAHKNISPFKQRP-KNETRIETQLVSHNVALSKFNARDLRAEKSKG 167
Query: 126 TAELKVALTTRFQYKTWGIKSK 147
T E++V +T R YKTW +S+
Sbjct: 168 TIEMEVYITARVSYKTWIFRSR 189
>At3g20610 non-race specific disease resistance protein, putative
Length = 222
Score = 44.7 bits (104), Expect = 3e-05
Identities = 43/169 (25%), Positives = 71/169 (41%), Gaps = 9/169 (5%)
Query: 11 LMEFIGFLGLLVLCLWLA-LRPKSPSYSVVFLSIEQH-----PGENGSIFYSLEIENPNK 64
LM + LL LCLWL+ L P S+ + I +N ++ + + ++N N
Sbjct: 17 LMTLLYVSFLLALCLWLSTLVHHIPRCSIHYFYIPALNKSLISSDNTTLNFMVRLKNINA 76
Query: 65 DSSIYYDDIILSF--LYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAI 122
IYY+D+ LSF V T+ F+QG K + + + +
Sbjct: 77 KQGIYYEDLHLSFSTRINNSSLLVANYTVPRFYQGHEKKAKKWGQALPFNNQ-TVIQAVL 135
Query: 123 SNATAELKVALTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRKKKK 171
N +A +V L + +YK K+K + L L ++ DG K K+
Sbjct: 136 PNGSAIFRVDLKMQVKYKVMSWKTKRYKLKASVNLEVNEDGATKVKDKE 184
>At5g53730 putative protein
Length = 213
Score = 44.3 bits (103), Expect = 4e-05
Identities = 32/100 (32%), Positives = 50/100 (50%), Gaps = 11/100 (11%)
Query: 5 KRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSV-------VFLSIEQHPGENGSIFYSL 57
K F+ F F G L L++ +WL L P+ P +S+ + L+ N S+ +L
Sbjct: 26 KLFFTFSTFFSGLL-LIIFLVWLILHPERPEFSLTEADIYSLNLTTSSTHLLNSSVQLTL 84
Query: 58 EIENPNKDSSIYYDDIILSFLY-GQQEDKVGETTIGSFHQ 96
+NPNK IYYD +++ Y GQQ E ++ F+Q
Sbjct: 85 FSKNPNKKVGIYYDKLLVYAAYRGQQ--ITSEASLPPFYQ 122
>At5g06320 NDR1/HIN1-like protein 3 (NHL3)
Length = 231
Score = 44.3 bits (103), Expect = 4e-05
Identities = 39/146 (26%), Positives = 65/146 (43%), Gaps = 17/146 (11%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE-NGSIFYSLE----IENPNKDSSIYYDD 72
LG+ L +WL RP + + V + + + ++ Y+L+ I NPN+ +YYD+
Sbjct: 59 LGIAALIIWLIFRPNAIKFHVTDAKLTEFTLDPTNNLRYNLDLNFTIRNPNRRIGVYYDE 118
Query: 73 IILSFLYGQQEDKVGETTIGSFHQGTGKTRSV------YDTVNARHGAFKPLFKAISNA- 125
I + YG Q + I F+QG T V V G K L + +++
Sbjct: 119 IEVRGYYGDQRFGM-SNNISKFYQGHKNTTVVGTKLVGQQLVLLDGGERKDLNEDVNSQI 177
Query: 126 -TAELKVALTTRFQY---KTWGIKSK 147
+ K+ L RF++ K+W K K
Sbjct: 178 YRIDAKLRLKIRFKFGLIKSWRFKPK 203
>At2g27260 unknown protein
Length = 243
Score = 43.9 bits (102), Expect = 5e-05
Identities = 24/97 (24%), Positives = 46/97 (46%), Gaps = 6/97 (6%)
Query: 5 KRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGSIF----YSLEIE 60
+R ++ F+ LGL++ +L +RP+ P ++ LS+ N + L+
Sbjct: 60 RRLFIVFTTFLLLLGLILFIFFLIVRPQLPDVNLNSLSVSNFNVSNNQVSGKWDLQLQFR 119
Query: 61 NPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQG 97
NPN S++Y+ + + Y + + ET + F QG
Sbjct: 120 NPNSKMSLHYETALCAMYYNRV--SLSETRLQPFDQG 154
>At4g23930 unknown protein (At4g23930)
Length = 187
Score = 42.7 bits (99), Expect = 1e-04
Identities = 26/103 (25%), Positives = 54/103 (52%), Gaps = 8/103 (7%)
Query: 14 FIGFLGLLVLCLWLAL-RPKSPSYSVVFLSIEQHPGENGSIFYSLE----IENPNKDSSI 68
FI FL + L ++L + RP+ P SV + + N S+ ++ + NPN+ +
Sbjct: 17 FIVFLIIAALTVYLTVFRPRDPEISVTSVKVPSFSVANSSVSFTFSQFSAVRNPNRAAFS 76
Query: 69 YYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
+Y+++I F YG +++G T + + +G+T+ + T + +
Sbjct: 77 HYNNVIQLFYYG---NRIGYTFVPAGEIESGRTKRMLATFSVQ 116
>At2g35980 similar to harpin-induced protein hin1 from tobacco
Length = 227
Score = 42.0 bits (97), Expect = 2e-04
Identities = 29/112 (25%), Positives = 50/112 (43%), Gaps = 8/112 (7%)
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--HPGENGSIFYSL----EIE 60
F ++ I LG+ L WL +RP++ + V S+ + H + + Y+L +
Sbjct: 41 FVKVIISLIVILGVAALIFWLIVRPRAIKFHVTDASLTRFDHTSPDNILRYNLALTVPVR 100
Query: 61 NPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARH 112
NPNK +YYD I Y + + T+ F+QG T + T ++
Sbjct: 101 NPNKRIGLYYDRIEAHAYY--EGKRFSTITLTPFYQGHKNTTVLTPTFQGQN 150
>At4g01410 putative hypoersensitive response protein
Length = 227
Score = 41.6 bits (96), Expect = 3e-04
Identities = 24/70 (34%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Query: 18 LGLLVLCLWLALRPKSPSYSVV-----FLSIEQHPGENGSIFYSLEIENPNKDSSIYYDD 72
LG++ L LWL RP P +VV L+ P + S+ +S+ NPN+ SI+YD
Sbjct: 55 LGIIALILWLVYRPHKPRLTVVGAAIYDLNFTAPPLISTSVQFSVLARNPNRRVSIHYDK 114
Query: 73 IILSFLYGQQ 82
+ + Y Q
Sbjct: 115 LSMYVTYKDQ 124
>At2g35460 similar to harpin-induced protein hin1 from tobacco
Length = 238
Score = 41.6 bits (96), Expect = 3e-04
Identities = 29/100 (29%), Positives = 43/100 (43%), Gaps = 8/100 (8%)
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGS------IFYSLEIENPNKDSSIYYD 71
LG++ L LW LRP + V + + + S I + I NPN+ I+YD
Sbjct: 65 LGVVALILWFILRPNVVKFQVTEADLTRFEFDPRSHNLHYNISLNFSIRNPNQRLGIHYD 124
Query: 72 DIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNAR 111
+ + YG Q T SF+QG T V +N +
Sbjct: 125 QLEVRGYYGDQRFSAANMT--SFYQGHKNTTVVGTELNGQ 162
>At5g56050 unknown protein
Length = 283
Score = 40.8 bits (94), Expect = 4e-04
Identities = 25/100 (25%), Positives = 50/100 (50%), Gaps = 8/100 (8%)
Query: 8 YVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE-----NGSIFYSLEIENP 62
++F + I F G+ L L+LA++P++P + + + E NG + L NP
Sbjct: 108 FIFSILLIVF-GIATLILYLAVKPRTPVFDISNAKLNTILFESPVYFNGDMLLQLNFTNP 166
Query: 63 NKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTR 102
NK ++ ++++++ + + K+ + F Q GKTR
Sbjct: 167 NKKLNVRFENLMVELWFA--DTKIATQGVLPFSQRNGKTR 204
>At2g46300 unknown protein
Length = 252
Score = 38.9 bits (89), Expect = 0.002
Identities = 37/150 (24%), Positives = 59/150 (38%), Gaps = 17/150 (11%)
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIE--------QHPGENGSIFYSLEIENP 62
L F+ +G V LW PK P++S+ ++ + + +E++NP
Sbjct: 69 LFIFLLLVGTAVFYLWFD--PKLPTFSLASFRLDGFKLADDPDGASLSATAVARVEMKNP 126
Query: 63 NKDSSIYYDD--IILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARH-----GAF 115
N YY + + LS G E +GETT+ F QG + SV ++ G
Sbjct: 127 NSKLVFYYGNTAVDLSVGSGNDETGMGETTMNGFRQGPKNSTSVKVETTVKNQLVERGLA 186
Query: 116 KPLFKAISNATAELKVALTTRFQYKTWGIK 145
K L + + V T+ GIK
Sbjct: 187 KRLAAKFQSKDLVINVVAKTKVGLGVGGIK 216
>At1g65690 unknown protein
Length = 252
Score = 38.9 bits (89), Expect = 0.002
Identities = 25/103 (24%), Positives = 49/103 (47%), Gaps = 6/103 (5%)
Query: 8 YVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFL-----SIEQHPGENGSIFYSLEIENP 62
+ FL+ + +G + L+L +PK P YS+ L ++ Q + ++ +NP
Sbjct: 70 FCFLLLLVVAVGASIGILYLVFKPKLPDYSIDRLQLTRFALNQDSSLTTAFNVTITAKNP 129
Query: 63 NKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVY 105
N+ IYY+D ++ E ++ ++ F+QG T +Y
Sbjct: 130 NEKIGIYYEDGSKITVW-YMEHQLSNGSLPKFYQGHENTTVIY 171
>At5g36970 putative protein
Length = 248
Score = 38.1 bits (87), Expect = 0.003
Identities = 29/102 (28%), Positives = 50/102 (48%), Gaps = 10/102 (9%)
Query: 8 YVFLMEF--IGFLGLLVLCLWLALRPKSPSYSVVFL-----SIEQHPGENGSIFYSLEIE 60
Y L+ F I +G +V L+L RPK P Y++ L + Q + + ++ +
Sbjct: 64 YTLLVLFLLIVIVGAIVGILYLVFRPKFPDYNIDRLQLTRFQLNQDLSLSTAFNVTITAK 123
Query: 61 NPNKDSSIYYDD-IILSFLYGQQEDKVGETTIGSFHQGTGKT 101
NPN+ IYY+D +S LY + ++ ++ F+QG T
Sbjct: 124 NPNEKIGIYYEDGSKISVLY--MQTRISNGSLPKFYQGHENT 163
>At2g27270 unknown protein
Length = 231
Score = 38.1 bits (87), Expect = 0.003
Identities = 21/67 (31%), Positives = 38/67 (56%), Gaps = 6/67 (8%)
Query: 20 LLVLCL----WLALRPKSPSYSVVFLSIEQHPGENGSIFYSLEIENPNKDSSIYYDDIIL 75
LL++C+ ++A++P+ P + + L ++ P N S+ L NP+ SS+YY I L
Sbjct: 68 LLIICVFAFNYIAIQPRVPRFMLEDLYVD--PLSNTSVIVKLSSTNPSYKSSVYYRKISL 125
Query: 76 SFLYGQQ 82
L G++
Sbjct: 126 HVLVGEK 132
>At5g22870 putative protein
Length = 207
Score = 37.4 bits (85), Expect = 0.005
Identities = 27/116 (23%), Positives = 54/116 (46%), Gaps = 13/116 (11%)
Query: 8 YVFL----MEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGEN-----GSIFYSLE 58
Y+FL + F+ +G L+ WL +PK Y+V S++ N + ++++
Sbjct: 28 YIFLVILTLIFMAAVGFLIT--WLETKPKKLRYTVENASVQNFNLTNDNHMSATFQFTIQ 85
Query: 59 IENPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARHGA 114
NPN S+YY + + + ++ + T+ FHQ + + +T+ A + A
Sbjct: 86 SHNPNHRISVYYSSVEIFVKF--KDQTLAFDTVEPFHQPRMNVKQIDETLIAENVA 139
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,225,454
Number of Sequences: 26719
Number of extensions: 176068
Number of successful extensions: 475
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 434
Number of HSP's gapped (non-prelim): 44
length of query: 189
length of database: 11,318,596
effective HSP length: 94
effective length of query: 95
effective length of database: 8,807,010
effective search space: 836665950
effective search space used: 836665950
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0358.5


