

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0349b.8
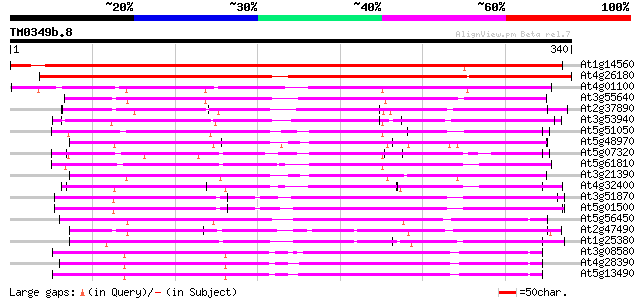
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g14560 mitochondrial carrier like protein 442 e-124
At4g26180 putative mitochondrial carrier protein 355 3e-98
At4g01100 putative carrier protein 196 1e-50
At3g55640 Ca-dependent solute carrier - like protein 167 8e-42
At2g37890 mitochondrial carrier like protein 163 1e-40
At3g53940 unknown protein 161 5e-40
At5g51050 calcium-binding transporter-like protein 154 7e-38
At5g48970 mitochondrial carrier protein-like 154 9e-38
At5g07320 peroxisomal Ca-dependent solute carrier-like protein 154 9e-38
At5g61810 peroxisomal Ca-dependent solute carrier - like protein 153 1e-37
At3g21390 unknown protein 150 1e-36
At4g32400 adenylate translocator (brittle-1) - like protein 149 2e-36
At3g51870 carrier like protein 148 4e-36
At5g01500 unknown protein 147 6e-36
At5g56450 ADP/ATP translocase-like protein 141 6e-34
At2g47490 mitochondrial carrier like protein 130 1e-30
At1g25380 unknown protein 129 2e-30
At3g08580 adenylate translocator 128 4e-30
At4g28390 ADP,ATP carrier-like protein 127 9e-30
At5g13490 adenosine nucleotide translocator 122 2e-28
>At1g14560 mitochondrial carrier like protein
Length = 331
Score = 442 bits (1136), Expect = e-124
Identities = 220/337 (65%), Positives = 262/337 (77%), Gaps = 10/337 (2%)
Query: 1 MDSSQGSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQ 60
M SSQGSTL+ V S D +PV K L+AGG AGA++KTAVAPLER+KIL Q
Sbjct: 1 MGSSQGSTLSADV--------MSLVDTLPVLAKTLIAGGAAGAIAKTAVAPLERIKILLQ 52
Query: 61 TRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNC 120
TRT F TLGV QSL K+++ +G LG YKGNGAS +RI+PYAALH+MTYE Y+ WIL
Sbjct: 53 TRTNDFKTLGVSQSLKKVLQFDGPLGFYKGNGASVIRIIPYAALHYMTYEVYRDWILEKN 112
Query: 121 PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHI 180
LGSGP +DL+AGSAAGGT+VLCTYPLDLARTKLAYQV DTR +++ G G QP +
Sbjct: 113 LPLGSGPIVDLVAGSAAGGTAVLCTYPLDLARTKLAYQVSDTRQSLRGGANGFYRQPTYS 172
Query: 181 GIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLS 240
GIK VL Y+EGG RGLYRG+GPT+ GILPYAGLKFY YE+LK HVPEEHQ S+ M L
Sbjct: 173 GIKEVLAMAYKEGGPRGLYRGIGPTLIGILPYAGLKFYIYEELKRHVPEEHQNSVRMHLP 232
Query: 241 CGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHE--DTRYRNTLDGLRKIVRNQGWRQLF 298
CGALAGLFGQT+TYPLDVV+RQMQV +LQ E + RY+NT DGL IVR QGW+QLF
Sbjct: 233 CGALAGLFGQTITYPLDVVRRQMQVENLQPMTSEGNNKRYKNTFDGLNTIVRTQGWKQLF 292
Query: 299 AGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQKSR 335
AG+SINYI+IVPS AI FT Y++MK+W+ IPP+++S+
Sbjct: 293 AGLSINYIKIVPSVAIGFTVYESMKSWMRIPPRERSK 329
>At4g26180 putative mitochondrial carrier protein
Length = 325
Score = 355 bits (910), Expect = 3e-98
Identities = 176/323 (54%), Positives = 229/323 (70%), Gaps = 11/323 (3%)
Query: 19 QKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKL 78
++ D +P++ KEL+AGGV G ++KTAVAPLER+KIL+QTR F +G+ S+NK+
Sbjct: 5 EEKNGIIDSIPLFAKELIAGGVTGGIAKTAVAPLERIKILFQTRRDEFKRIGLVGSINKI 64
Query: 79 VKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAG 138
K EG +G Y+GNGAS RIVPYAALH+M YE Y+ WI+ P GP +DL+AGS AG
Sbjct: 65 GKTEGLMGFYRGNGASVARIVPYAALHYMAYEEYRRWIIFGFPDTTRGPLLDLVAGSFAG 124
Query: 139 GTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVR-PQPAHIGIKSVLTSVYREGGARG 197
GT+VL TYPLDL RTKLAYQ +K + Q + GI + YRE GARG
Sbjct: 125 GTAVLFTYPLDLVRTKLAYQT---------QVKAIPVEQIIYRGIVDCFSRTYRESGARG 175
Query: 198 LYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALAGLFGQTLTYPLD 257
LYRGV P++ GI PYAGLKFY YE++K HVP EH++ I ++L CG++AGL GQTLTYPLD
Sbjct: 176 LYRGVAPSLYGIFPYAGLKFYFYEEMKRHVPPEHKQDISLKLVCGSVAGLLGQTLTYPLD 235
Query: 258 VVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFT 317
VV+RQMQV L +A E+TR R T+ L KI R +GW+QLF+G+SINY+++VPS AI FT
Sbjct: 236 VVRRQMQVERLYSAVKEETR-RGTMQTLFKIAREEGWKQLFSGLSINYLKVVPSVAIGFT 294
Query: 318 AYDTMKAWLGIPPQQKSRSVSAT 340
YD MK L +PP+++ + + T
Sbjct: 295 VYDIMKLHLRVPPREEPEAEAVT 317
>At4g01100 putative carrier protein
Length = 352
Score = 196 bits (499), Expect = 1e-50
Identities = 128/357 (35%), Positives = 193/357 (53%), Gaps = 50/357 (14%)
Query: 2 DSSQGSTLAGLVDNA--SIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILW 59
+S+ ST+ L + A ++ F + K L AGGVAG +S+TAVAPLER+KIL
Sbjct: 10 ESAAVSTIVNLAEEAREGVKAPSYAFKSI---CKSLFAGGVAGGVSRTAVAPLERMKILL 66
Query: 60 QTRTGGFHTL---GVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWI 116
Q + H + G Q L + + EG GL+KGNG + RIVP +A+ F +YE+ + I
Sbjct: 67 QVQNP--HNIKYSGTVQGLKHIWRTEGLRGLFKGNGTNCARIVPNSAVKFFSYEQASNGI 124
Query: 117 L--------NNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKD 168
L N L P + L AG+ AG ++ TYP+D+ R +L Q ++ +
Sbjct: 125 LYMYRQRTGNENAQLT--PLLRLGAGATAGIIAMSATYPMDMVRGRLTVQTANSPYQYR- 181
Query: 169 GIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK---- 224
GI L +V RE G R LYRG P++ G++PY GL F YE LK
Sbjct: 182 ------------GIAHALATVLREEGPRALYRGWLPSVIGVVPYVGLNFSVYESLKDWLV 229
Query: 225 ----MHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDT---- 276
+ E ++ +++ RL+CGA+AG GQT+ YPLDV++R+MQ+ ++A+ T
Sbjct: 230 KENPYGLVENNELTVVTRLTCGAIAGTVGQTIAYPLDVIRRRMQMVGWKDASAIVTGEGR 289
Query: 277 -----RYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAWLGI 328
Y +D RK VR++G+ L+ G+ N +++VPS AI+F Y+ +K LG+
Sbjct: 290 STASLEYTGMVDAFRKTVRHEGFGALYKGLVPNSVKVVPSIAIAFVTYEMVKDVLGV 346
>At3g55640 Ca-dependent solute carrier - like protein
Length = 332
Score = 167 bits (423), Expect = 8e-42
Identities = 102/308 (33%), Positives = 162/308 (52%), Gaps = 36/308 (11%)
Query: 34 ELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLG-------VCQSLNKLVKHEGFLG 86
+L+AGG+AGA SKT APL R+ IL+Q + G HT + ++++ EG
Sbjct: 37 QLLAGGLAGAFSKTCTAPLSRLTILFQVQ--GMHTNAAALRKPSILHEASRILNEEGLKA 94
Query: 87 LYKGNGASAVRIVPYAALHFMTYERYKSWIL------NNCPALGSGPFIDLLAGSAAGGT 140
+KGN + +PY++++F YE YK ++ N+ + S F+ +AG AG T
Sbjct: 95 FWKGNLVTIAHRLPYSSVNFYAYEHYKKFMYMVTGMENHKEGISSNLFVHFVAGGLAGIT 154
Query: 141 SVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYR 200
+ TYPLDL RT+LA Q + GI L S+ + G GLY+
Sbjct: 155 AASATYPLDLVRTRLAAQT---------------KVIYYSGIWHTLRSITTDEGILGLYK 199
Query: 201 GVGPTITGILPYAGLKFYTYEKLKMH--VPEEHQKSILMRLSCGALAGLFGQTLTYPLDV 258
G+G T+ G+ P + F YE L+ + H I++ L+CG+L+G+ T T+PLD+
Sbjct: 200 GLGTTLVGVGPSIAISFSVYESLRSYWRSTRPHDSPIMVSLACGSLSGIASSTATFPLDL 259
Query: 259 VKRQMQVGSLQNAAHEDTRYRNTLDG-LRKIVRNQGWRQLFAGVSINYIRIVPSAAISFT 317
V+R+ Q+ + A Y+ L G L++IV+ +G R L+ G+ Y ++VP I F
Sbjct: 260 VRRRKQLEGIGGRA---VVYKTGLLGTLKRIVQTEGARGLYRGILPEYYKVVPGVGICFM 316
Query: 318 AYDTMKAW 325
Y+T+K +
Sbjct: 317 TYETLKLY 324
>At2g37890 mitochondrial carrier like protein
Length = 337
Score = 163 bits (412), Expect = 1e-40
Identities = 102/311 (32%), Positives = 161/311 (50%), Gaps = 38/311 (12%)
Query: 33 KELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQS-------LNKLVKHEGFL 85
+ L+AGG+AGA+SKT APL R+ IL+Q + G + G S ++++ EG+
Sbjct: 43 QNLLAGGIAGAISKTCTAPLARLTILFQLQ--GMQSEGAVLSRPNLRREASRIINEEGYR 100
Query: 86 GLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGS-------GPFIDLLAGSAAG 138
+KGN + V +PY A++F YE+Y + +N P + S P + ++G AG
Sbjct: 101 AFWKGNLVTVVHRIPYTAVNFYAYEKYNLFFNSN-PVVQSFIGNTSGNPIVHFVSGGLAG 159
Query: 139 GTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGL 198
T+ TYPLDL RT+LA Q R + GI+ ++ RE G GL
Sbjct: 160 ITAATATYPLDLVRTRLAAQ---------------RNAIYYQGIEHTFRTICREEGILGL 204
Query: 199 YRGVGPTITGILPYAGLKFYTYEKLKM--HVPEEHQKSILMRLSCGALAGLFGQTLTYPL 256
Y+G+G T+ G+ P + F YE +K+ H + +++ L G LAG T TYPL
Sbjct: 205 YKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPNDSDLVVSLVSGGLAGAVSSTATYPL 264
Query: 257 DVVKRQMQVGSLQNAAHEDTRYRNTLDG-LRKIVRNQGWRQLFAGVSINYIRIVPSAAIS 315
D+V+R+MQV + A Y L G + I +++G++ ++ G+ Y ++VP I
Sbjct: 265 DLVRRRMQV---EGAGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEYYKVVPGVGIV 321
Query: 316 FTAYDTMKAWL 326
F YD ++ L
Sbjct: 322 FMTYDALRRLL 332
Score = 95.9 bits (237), Expect = 3e-20
Identities = 56/193 (29%), Positives = 96/193 (49%), Gaps = 12/193 (6%)
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGN 91
+ V+GG+AG + TA PL+ V+ + + G+ + + + EG LGLYKG
Sbjct: 149 IVHFVSGGLAGITAATATYPLDLVRTRLAAQRNAIYYQGIEHTFRTICREEGILGLYKGL 208
Query: 92 GASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLA 151
GA+ + + P A++F YE K + ++ P S + L++G AG S TYPLDL
Sbjct: 209 GATLLGVGPSLAINFAAYESMKLFWHSHRPN-DSDLVVSLVSGGLAGAVSSTATYPLDLV 267
Query: 152 RTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILP 211
R ++ + G R + + G+ +++ G +G+YRG+ P ++P
Sbjct: 268 RRRMQVEG-----------AGGRARVYNTGLFGTFKHIFKSEGFKGIYRGILPEYYKVVP 316
Query: 212 YAGLKFYTYEKLK 224
G+ F TY+ L+
Sbjct: 317 GVGIVFMTYDALR 329
Score = 82.8 bits (203), Expect = 2e-16
Identities = 66/228 (28%), Positives = 102/228 (43%), Gaps = 26/228 (11%)
Query: 121 PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHI 180
P G F +LLAG AG S CT P LAR + +Q+ +G +G RP
Sbjct: 35 PQAKLGTFQNLLAGGIAGAISKTCTAP--LARLTILFQL---QGMQSEGAVLSRP----- 84
Query: 181 GIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPE---------EH 231
++ + + E G R ++G T+ +PY + FY YEK +
Sbjct: 85 NLRREASRIINEEGYRAFWKGNLVTVVHRIPYTAVNFYAYEKYNLFFNSNPVVQSFIGNT 144
Query: 232 QKSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRN 291
+ ++ G LAG+ T TYPLD+V+ ++ + +NA + Y+ R I R
Sbjct: 145 SGNPIVHFVSGGLAGITAATATYPLDLVRTRL--AAQRNAIY----YQGIEHTFRTICRE 198
Query: 292 QGWRQLFAGVSINYIRIVPSAAISFTAYDTMKA-WLGIPPQQKSRSVS 338
+G L+ G+ + + PS AI+F AY++MK W P VS
Sbjct: 199 EGILGLYKGLGATLLGVGPSLAINFAAYESMKLFWHSHRPNDSDLVVS 246
>At3g53940 unknown protein
Length = 365
Score = 161 bits (407), Expect = 5e-40
Identities = 103/316 (32%), Positives = 161/316 (50%), Gaps = 38/316 (12%)
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQ-----TRTGGFHTLGVCQSLNKLVKHEGFLG 86
V+ L+AGG+AGA SKT APL R+ IL+Q + + + +++VK EGF
Sbjct: 70 VERLLAGGIAGAFSKTCTAPLARLTILFQIQGMQSEAAILSSPNIWHEASRIVKEEGFRA 129
Query: 87 LYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPAL-------GSGPFIDLLAGSAAGG 139
+KGN + +PY A++F YE YK+++ +N P L G + ++G AG
Sbjct: 130 FWKGNLVTVAHRLPYGAVNFYAYEEYKTFLHSN-PVLQSYKGNAGVDISVHFVSGGLAGL 188
Query: 140 TSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLY 199
T+ TYPLDL RT+L+ Q R + G+ ++ RE G GLY
Sbjct: 189 TAASATYPLDLVRTRLSAQ---------------RNSIYYQGVGHAFRTICREEGILGLY 233
Query: 200 RGVGPTITGILPYAGLKFYTYEKLK----MHVPEEHQKSILMRLSCGALAGLFGQTLTYP 255
+G+G T+ G+ P + F YE K H P + + ++ L CG+L+G+ T T+P
Sbjct: 234 KGLGATLLGVGPSLAISFAAYETFKTFWLSHRPND--SNAVVSLGCGSLSGIVSSTATFP 291
Query: 256 LDVVKRQMQVGSLQNAAHEDTRYRNTLDG-LRKIVRNQGWRQLFAGVSINYIRIVPSAAI 314
LD+V+R+MQ L+ A Y L G + I + +G R L+ G+ Y ++VP I
Sbjct: 292 LDLVRRRMQ---LEGAGGRARVYTTGLFGTFKHIFKTEGMRGLYRGIIPEYYKVVPGVGI 348
Query: 315 SFTAYDTMKAWLGIPP 330
+F ++ +K L P
Sbjct: 349 AFMTFEELKKLLSTVP 364
Score = 99.4 bits (246), Expect = 3e-21
Identities = 62/198 (31%), Positives = 100/198 (50%), Gaps = 13/198 (6%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLG 86
GV + V V+GG+AG + +A PL+ V+ + + GV + + + EG LG
Sbjct: 173 GVDISV-HFVSGGLAGLTAASATYPLDLVRTRLSAQRNSIYYQGVGHAFRTICREEGILG 231
Query: 87 LYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTY 146
LYKG GA+ + + P A+ F YE +K++ L++ P S + L GS +G S T+
Sbjct: 232 LYKGLGATLLGVGPSLAISFAAYETFKTFWLSHRPN-DSNAVVSLGCGSLSGIVSSTATF 290
Query: 147 PLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTI 206
PLDL R ++ + G R + G+ +++ G RGLYRG+ P
Sbjct: 291 PLDLVRRRMQLEG-----------AGGRARVYTTGLFGTFKHIFKTEGMRGLYRGIIPEY 339
Query: 207 TGILPYAGLKFYTYEKLK 224
++P G+ F T+E+LK
Sbjct: 340 YKVVPGVGIAFMTFEELK 357
Score = 42.0 bits (97), Expect = 5e-04
Identities = 28/97 (28%), Positives = 46/97 (46%), Gaps = 1/97 (1%)
Query: 238 RLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQL 297
RL G +AG F +T T PL + Q+ +Q+ A N +IV+ +G+R
Sbjct: 72 RLLAGGIAGAFSKTCTAPLARLTILFQIQGMQSEA-AILSSPNIWHEASRIVKEEGFRAF 130
Query: 298 FAGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQKS 334
+ G + +P A++F AY+ K +L P +S
Sbjct: 131 WKGNLVTVAHRLPYGAVNFYAYEEYKTFLHSNPVLQS 167
>At5g51050 calcium-binding transporter-like protein
Length = 487
Score = 154 bits (389), Expect = 7e-38
Identities = 94/308 (30%), Positives = 163/308 (52%), Gaps = 36/308 (11%)
Query: 26 DGVPVYVKE---LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHE 82
+G+ ++K +AGG+AGA S+TA APL+R+K+L Q + + +++ + K
Sbjct: 200 EGISKHIKRSNYFIAGGIAGAASRTATAPLDRLKVLLQIQKTDAR---IREAIKLIWKQG 256
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNC--PALGSGPFIDLLAGSAAGGT 140
G G ++GNG + V++ P +A+ F YE +K+ I N G + L AG AG
Sbjct: 257 GVRGFFRGNGLNIVKVAPESAIKFYAYELFKNAIGENMGEDKADIGTTVRLFAGGMAGAV 316
Query: 141 SVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYR 200
+ YPLDL +T+L T + G+ R + ++ + G R Y+
Sbjct: 317 AQASIYPLDLVKTRLQTY------TSQAGVAVPR-------LGTLTKDILVHEGPRAFYK 363
Query: 201 GVGPTITGILPYAGLKFYTYEKLK----MHVPEEHQKSILMRLSCGALAGLFGQTLTYPL 256
G+ P++ GI+PYAG+ YE LK ++ ++ + L++L CG ++G G T YPL
Sbjct: 364 GLFPSLLGIIPYAGIDLAAYETLKDLSRTYILQDAEPGPLVQLGCGTISGALGATCVYPL 423
Query: 257 DVVKRQMQVGSLQNAAHEDTRYRNTLDGL-RKIVRNQGWRQLFAGVSINYIRIVPSAAIS 315
VV+ +MQ R R ++ G+ R+ + +G+R L+ G+ N +++VP+A+I+
Sbjct: 424 QVVRTRMQA----------ERARTSMSGVFRRTISEEGYRALYKGLLPNLLKVVPAASIT 473
Query: 316 FTAYDTMK 323
+ Y+ MK
Sbjct: 474 YMVYEAMK 481
Score = 44.3 bits (103), Expect = 1e-04
Identities = 27/86 (31%), Positives = 43/86 (49%), Gaps = 9/86 (10%)
Query: 242 GALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGV 301
G +AG +T T PLD +K +Q+ D R R + ++ I + G R F G
Sbjct: 215 GGIAGAASRTATAPLDRLKVLLQIQKT------DARIR---EAIKLIWKQGGVRGFFRGN 265
Query: 302 SINYIRIVPSAAISFTAYDTMKAWLG 327
+N +++ P +AI F AY+ K +G
Sbjct: 266 GLNIVKVAPESAIKFYAYELFKNAIG 291
>At5g48970 mitochondrial carrier protein-like
Length = 339
Score = 154 bits (388), Expect = 9e-38
Identities = 94/326 (28%), Positives = 163/326 (49%), Gaps = 51/326 (15%)
Query: 37 AGGVAGALSKTAVAPLERVKILWQTR--------------TGGFHTLGVCQSLNKLVKHE 82
AG ++G +S++ +PL+ +KI +Q + +G G+ Q+ + + E
Sbjct: 24 AGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQATKDIFREE 83
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPA---LGSGPFIDLLAGSAAGG 139
GF G ++GN + + ++PY ++ F + KS+ + + P++ ++G+ AG
Sbjct: 84 GFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFVSGALAGC 143
Query: 140 TSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLY 199
+ L +YP DL RT LA Q G V P ++S + + G RGLY
Sbjct: 144 AATLGSYPFDLLRTILASQ----------GEPKVYPT-----MRSAFVDIIQSRGIRGLY 188
Query: 200 RGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLS--------------CGALA 245
G+ PT+ I+PYAGL+F TY+ K + + ++ + ++ CG A
Sbjct: 189 NGLTPTLVEIVPYAGLQFGTYDMFKRWMMDWNRYKLSSKIPINVDTNLSSFQLFICGLGA 248
Query: 246 GLFGQTLTYPLDVVKRQMQVGSLQN-----AAHEDTRYRNTLDGLRKIVRNQGWRQLFAG 300
G + + +PLDVVK++ Q+ LQ A E YRN LDGLR+I+ ++GW L+ G
Sbjct: 249 GTSAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYRNMLDGLRQIMISEGWHGLYKG 308
Query: 301 VSINYIRIVPSAAISFTAYDTMKAWL 326
+ + ++ P+ A++F AY+ WL
Sbjct: 309 IVPSTVKAAPAGAVTFVAYEFTSDWL 334
Score = 83.6 bits (205), Expect = 1e-16
Identities = 57/206 (27%), Positives = 95/206 (45%), Gaps = 17/206 (8%)
Query: 129 IDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTR--GTIKDGIKGVRPQPAHIGIKSVL 186
ID AG+ +GG S T PLD+ + + Q+ T G ++ + G + G+
Sbjct: 20 IDASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGA---SKYTGMVQAT 76
Query: 187 TSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHV-----PEEH-QKSILMRLS 240
++RE G RG +RG P + ++PY ++F KLK E+H S +
Sbjct: 77 KDIFREEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSFASGSTKTEDHIHLSPYLSFV 136
Query: 241 CGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAG 300
GALAG +YP D+++ ++ + E Y I++++G R L+ G
Sbjct: 137 SGALAGCAATLGSYPFDLLR------TILASQGEPKVYPTMRSAFVDIIQSRGIRGLYNG 190
Query: 301 VSINYIRIVPSAAISFTAYDTMKAWL 326
++ + IVP A + F YD K W+
Sbjct: 191 LTPTLVEIVPYAGLQFGTYDMFKRWM 216
Score = 43.1 bits (100), Expect = 2e-04
Identities = 26/101 (25%), Positives = 50/101 (48%), Gaps = 8/101 (7%)
Query: 233 KSILMRLSCGALAGLFGQTLTYPLDVVKRQMQV--------GSLQNAAHEDTRYRNTLDG 284
K L+ S GA++G +++T PLDV+K + QV G ++ ++Y +
Sbjct: 16 KRALIDASAGAISGGVSRSVTSPLDVIKIRFQVQLEPTTSWGLVRGNLSGASKYTGMVQA 75
Query: 285 LRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAW 325
+ I R +G+R + G + ++P +I FT +K++
Sbjct: 76 TKDIFREEGFRGFWRGNVPALLMVMPYTSIQFTVLHKLKSF 116
>At5g07320 peroxisomal Ca-dependent solute carrier-like protein
Length = 479
Score = 154 bits (388), Expect = 9e-38
Identities = 92/305 (30%), Positives = 165/305 (53%), Gaps = 35/305 (11%)
Query: 26 DGVPVYVKE---LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHE 82
DG+ +VK L+AGG+AGA+S+TA APL+R+K++ Q + GV ++ K+ + +
Sbjct: 197 DGISKHVKRSRLLLAGGLAGAVSRTATAPLDRLKVVLQVQRA---HAGVLPTIKKIWRED 253
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSV 142
+G ++GNG + +++ P +A+ F YE K I +G+ L+AG AG +
Sbjct: 254 KLMGFFRGNGLNVMKVAPESAIKFCAYEMLKPMIGGEDGDIGTSG--RLMAGGMAGALAQ 311
Query: 143 LCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGV 202
YP+DL +T+L + + +G K + + + ++ G R Y+G+
Sbjct: 312 TAIYPMDLVKTRL-------QTCVSEGGKAPK-------LWKLTKDIWVREGPRAFYKGL 357
Query: 203 GPTITGILPYAGLKFYTYEKLK----MHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDV 258
P++ GI+PYAG+ YE LK ++ ++ + L++LSCG +G G + YPL V
Sbjct: 358 FPSLLGIVPYAGIDLAAYETLKDLSRTYILQDTEPGPLIQLSCGMTSGALGASCVYPLQV 417
Query: 259 VKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTA 318
V+ +MQ S + ++ + NT+ G +G R + G+ N +++VP+A+I++
Sbjct: 418 VRTRMQADSSKTTMKQE--FMNTMKG-------EGLRGFYRGLLPNLLKVVPAASITYIV 468
Query: 319 YDTMK 323
Y+ MK
Sbjct: 469 YEAMK 473
Score = 83.6 bits (205), Expect = 1e-16
Identities = 63/201 (31%), Positives = 93/201 (45%), Gaps = 32/201 (15%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVK----HEGFLGLYKG 90
L+AGG+AGAL++TA+ P++ VK QT G L KL K EG YKG
Sbjct: 300 LMAGGMAGALAQTAIYPMDLVKTRLQTCVSEG---GKAPKLWKLTKDIWVREGPRAFYKG 356
Query: 91 NGASAVRIVPYAALHFMTYERYK----SWILNNCPALGSGPFIDLLAGSAAGGTSVLCTY 146
S + IVPYA + YE K ++IL + GP I L G +G C Y
Sbjct: 357 LFPSLLGIVPYAGIDLAAYETLKDLSRTYILQDTEP---GPLIQLSCGMTSGALGASCVY 413
Query: 147 PLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTI 206
PL + RT++ Q ++ T+K + + G RG YRG+ P +
Sbjct: 414 PLQVVRTRM--QADSSKTTMKQEFM----------------NTMKGEGLRGFYRGLLPNL 455
Query: 207 TGILPYAGLKFYTYEKLKMHV 227
++P A + + YE +K ++
Sbjct: 456 LKVVPAASITYIVYEAMKKNM 476
Score = 46.6 bits (109), Expect = 2e-05
Identities = 28/89 (31%), Positives = 42/89 (46%), Gaps = 9/89 (10%)
Query: 239 LSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLF 298
L G LAG +T T PLD +K +QV + L ++KI R F
Sbjct: 209 LLAGGLAGAVSRTATAPLDRLKVVLQV---------QRAHAGVLPTIKKIWREDKLMGFF 259
Query: 299 AGVSINYIRIVPSAAISFTAYDTMKAWLG 327
G +N +++ P +AI F AY+ +K +G
Sbjct: 260 RGNGLNVMKVAPESAIKFCAYEMLKPMIG 288
>At5g61810 peroxisomal Ca-dependent solute carrier - like protein
Length = 478
Score = 153 bits (386), Expect = 1e-37
Identities = 95/311 (30%), Positives = 161/311 (51%), Gaps = 37/311 (11%)
Query: 26 DGVPVYV---KELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHE 82
DG+ + K L+AGG+AGA+S+TA APL+R+K+ Q + LGV ++ K+ + +
Sbjct: 196 DGISAHAQRSKLLLAGGIAGAVSRTATAPLDRLKVALQVQRTN---LGVVPTIKKIWRED 252
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSV 142
LG ++GNG + ++ P +A+ F YE K I +G+ LLAG AG +
Sbjct: 253 KLLGFFRGNGLNVAKVAPESAIKFAAYEMLKPIIGGADGDIGTSG--RLLAGGLAGAVAQ 310
Query: 143 LCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGV 202
YP+DL +T+L V + GT K + + ++ + G R YRG+
Sbjct: 311 TAIYPMDLVKTRLQTFVSEV-GTPK--------------LWKLTKDIWIQEGPRAFYRGL 355
Query: 203 GPTITGILPYAGLKFYTYEKLK-----MHVPEEHQKSILMRLSCGALAGLFGQTLTYPLD 257
P++ GI+PYAG+ YE LK + + + L++L CG +G G + YPL
Sbjct: 356 CPSLIGIIPYAGIDLAAYETLKDLSRAHFLHDTAEPGPLIQLGCGMTSGALGASCVYPLQ 415
Query: 258 VVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFT 317
V++ +MQ S + + ++ K +R +G + + G+ N+ +++PSA+IS+
Sbjct: 416 VIRTRMQADSSKTSMGQE---------FLKTLRGEGLKGFYRGIFPNFFKVIPSASISYL 466
Query: 318 AYDTMKAWLGI 328
Y+ MK L +
Sbjct: 467 VYEAMKKNLAL 477
>At3g21390 unknown protein
Length = 335
Score = 150 bits (378), Expect = 1e-36
Identities = 98/325 (30%), Positives = 159/325 (48%), Gaps = 52/325 (16%)
Query: 37 AGGVAGALSKTAVAPLERVKILWQTRTGGFHTL------------GVCQSLNKLVKHEGF 84
AGGVAGA+S+ +PL+ +KI +Q + T G+ ++ + + EG
Sbjct: 21 AGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTKDIFREEGL 80
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSG---PFIDLLAGSAAGGTS 141
G ++GN + + +VPY ++ F + KS+ + A P++ ++G+ AG +
Sbjct: 81 SGFWRGNVPALLMVVPYTSIQFAVLHKVKSFAAGSSKAENHAQLSPYLSYISGALAGCAA 140
Query: 142 VLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRG 201
+ +YP DL RT LA Q G V P ++S S+ + G +GLY G
Sbjct: 141 TVGSYPFDLLRTVLASQ----------GEPKVYPN-----MRSAFLSIVQTRGIKGLYAG 185
Query: 202 VGPTITGILPYAGLKFYTYEKLKMHV----------------PEEHQKSILMRLSCGALA 245
+ PT+ I+PYAGL+F TY+ K P + S + L CG +
Sbjct: 186 LSPTLIEIIPYAGLQFGTYDTFKRWSMVYNKRYRSSSSSSTNPSDSLSSFQLFL-CGLAS 244
Query: 246 GLFGQTLTYPLDVVKRQMQVGSLQN-----AAHEDTRYRNTLDGLRKIVRNQGWRQLFAG 300
G + + +PLDVVK++ QV LQ A E Y+N DGL +I+R++GW L+ G
Sbjct: 245 GTVSKLVCHPLDVVKKRFQVEGLQRHPKYGARVELNAYKNMFDGLGQILRSEGWHGLYKG 304
Query: 301 VSINYIRIVPSAAISFTAYDTMKAW 325
+ + I+ P+ A++F AY+ W
Sbjct: 305 IVPSTIKAAPAGAVTFVAYELASDW 329
Score = 38.9 bits (89), Expect = 0.004
Identities = 26/95 (27%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 36 VAGGVAGALSKTAVAPLERVKILWQTRTGGFHTL-----------GVCQSLNKLVKHEGF 84
+ G +G +SK PL+ VK +Q H + L ++++ EG+
Sbjct: 239 LCGLASGTVSKLVCHPLDVVKKRFQVEGLQRHPKYGARVELNAYKNMFDGLGQILRSEGW 298
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWILNN 119
GLYKG S ++ P A+ F+ YE W N
Sbjct: 299 HGLYKGIVPSTIKAAPAGAVTFVAYELASDWFEAN 333
Score = 37.0 bits (84), Expect = 0.015
Identities = 27/99 (27%), Positives = 48/99 (48%), Gaps = 6/99 (6%)
Query: 233 KSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNA--AHEDTRYRNTLDGL----R 286
K ++ S G +AG + +T PLDV+K + QV A A +D++ + +GL +
Sbjct: 13 KRAVIDASAGGVAGAISRMVTSPLDVIKIRFQVQLEPTATWALKDSQLKPKYNGLFRTTK 72
Query: 287 KIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAW 325
I R +G + G + +VP +I F +K++
Sbjct: 73 DIFREEGLSGFWRGNVPALLMVVPYTSIQFAVLHKVKSF 111
>At4g32400 adenylate translocator (brittle-1) - like protein
Length = 392
Score = 149 bits (377), Expect = 2e-36
Identities = 97/299 (32%), Positives = 155/299 (51%), Gaps = 34/299 (11%)
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGN 91
++ L++G VAGA+S+T VAPLE ++ +GG + V + ++KHEG+ GL++GN
Sbjct: 111 LRRLLSGAVAGAVSRTVVAPLETIRTHLMVGSGGNSSTEV---FSDIMKHEGWTGLFRGN 167
Query: 92 GASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI----DLLAGSAAGGTSVLCTYP 147
+ +R+ P A+ +E + P G I LLAG+ AG + L TYP
Sbjct: 168 LVNVIRVAPARAVELFVFETVNKKL---SPPHGQESKIPIPASLLAGACAGVSQTLLTYP 224
Query: 148 LDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTIT 207
L+L +T+L Q RG K GI + RE G LYRG+ P++
Sbjct: 225 LELVKTRLTIQ----RGVYK-------------GIFDAFLKIIREEGPTELYRGLAPSLI 267
Query: 208 GILPYAGLKFYTYEKL-KMHVPEEHQKSI--LMRLSCGALAGLFGQTLTYPLDVVKRQMQ 264
G++PYA ++ Y+ L K + Q+ I + L G+LAG T T+PL+V ++ MQ
Sbjct: 268 GVVPYAATNYFAYDSLRKAYRSFSKQEKIGNIETLLIGSLAGALSSTATFPLEVARKHMQ 327
Query: 265 VGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMK 323
VG++ Y+N L L I+ ++G + G+ + +++VP+A ISF Y+ K
Sbjct: 328 VGAVSGR----VVYKNMLHALVTILEHEGILGWYKGLGPSCLKLVPAAGISFMCYEACK 382
Score = 84.3 bits (207), Expect = 8e-17
Identities = 56/200 (28%), Positives = 91/200 (45%), Gaps = 15/200 (7%)
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGAS 94
L+AG AG PLE VK + G + G+ + K+++ EG LY+G S
Sbjct: 208 LLAGACAGVSQTLLTYPLELVKTRLTIQRGVYK--GIFDAFLKIIREEGPTELYRGLAPS 265
Query: 95 AVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTK 154
+ +VPYAA ++ Y+ + + G LL GS AG S T+PL++AR
Sbjct: 266 LIGVVPYAATNYFAYDSLRKAYRSFSKQEKIGNIETLLIGSLAGALSSTATFPLEVARKH 325
Query: 155 LAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAG 214
+ + R K+ + L ++ G G Y+G+GP+ ++P AG
Sbjct: 326 MQVGAVSGRVVYKNMLHA-------------LVTILEHEGILGWYKGLGPSCLKLVPAAG 372
Query: 215 LKFYTYEKLKMHVPEEHQKS 234
+ F YE K + E +Q++
Sbjct: 373 ISFMCYEACKKILIENNQEA 392
Score = 60.1 bits (144), Expect = 2e-09
Identities = 34/90 (37%), Positives = 51/90 (55%), Gaps = 2/90 (2%)
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTR--TGGFHTLGVCQSLNKLVKHEGFLGLYK 89
++ L+ G +AGALS TA PLE + Q +G + +L +++HEG LG YK
Sbjct: 299 IETLLIGSLAGALSSTATFPLEVARKHMQVGAVSGRVVYKNMLHALVTILEHEGILGWYK 358
Query: 90 GNGASAVRIVPYAALHFMTYERYKSWILNN 119
G G S +++VP A + FM YE K ++ N
Sbjct: 359 GLGPSCLKLVPAAGISFMCYEACKKILIEN 388
Score = 56.6 bits (135), Expect = 2e-08
Identities = 31/100 (31%), Positives = 53/100 (53%), Gaps = 9/100 (9%)
Query: 236 LMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWR 295
L RL GA+AG +T+ PL+ ++ + VGS N++ E I++++GW
Sbjct: 111 LRRLLSGAVAGAVSRTVVAPLETIRTHLMVGSGGNSSTEV---------FSDIMKHEGWT 161
Query: 296 QLFAGVSINYIRIVPSAAISFTAYDTMKAWLGIPPQQKSR 335
LF G +N IR+ P+ A+ ++T+ L P Q+S+
Sbjct: 162 GLFRGNLVNVIRVAPARAVELFVFETVNKKLSPPHGQESK 201
>At3g51870 carrier like protein
Length = 381
Score = 148 bits (374), Expect = 4e-36
Identities = 93/311 (29%), Positives = 148/311 (46%), Gaps = 34/311 (10%)
Query: 28 VPVYVKELVAGGVAGALSKTAVAPLERVKILWQT------RTGGFHTLGVCQSLNKLVKH 81
VP AG +AGA +KT APL+R+K+L QT + +G +++ + K
Sbjct: 84 VPKDAAIFAAGALAGAAAKTVTAPLDRIKLLMQTHGIRLGQQSAKKAIGFIEAITLIAKE 143
Query: 82 EGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTS 141
EG G +KGN +R++PY+A+ + YE YK+ L L AG+ AG TS
Sbjct: 144 EGVKGYWKGNLPQVIRVLPYSAVQLLAYESYKNLFKGKDDQLSV--IGRLAAGACAGMTS 201
Query: 142 VLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRG 201
L TYPLD+ R +LA + P + + V S+ R+ G Y G
Sbjct: 202 TLLTYPLDVLRLRLAVE------------------PRYRTMSQVALSMLRDEGIASFYYG 243
Query: 202 VGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVKR 261
+GP++ GI PY + F ++ +K +PEE++K L L+ YPLD V+R
Sbjct: 244 LGPSLVGIAPYIAVNFCIFDLVKKSLPEEYRKKAQSSLLTAVLSAGIATLTCYPLDTVRR 303
Query: 262 QMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDT 321
QMQ+ T Y++ + I+ G L+ G N ++ +P+++I T +D
Sbjct: 304 QMQM--------RGTPYKSIPEAFAGIIDRDGLIGLYRGFLPNALKTLPNSSIRLTTFDM 355
Query: 322 MKAWLGIPPQQ 332
+K + +Q
Sbjct: 356 VKRLIATSEKQ 366
Score = 85.5 bits (210), Expect = 4e-17
Identities = 59/205 (28%), Positives = 101/205 (48%), Gaps = 19/205 (9%)
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYRE 192
AG+ AG + T PLD R KL Q R G + IG +T + +E
Sbjct: 93 AGALAGAAAKTVTAPLD--RIKLLMQTHGIR-------LGQQSAKKAIGFIEAITLIAKE 143
Query: 193 GGARGLYRGVGPTITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQT 251
G +G ++G P + +LPY+ ++ YE K + ++ Q S++ RL+ GA AG+
Sbjct: 144 EGVKGYWKGNLPQVIRVLPYSAVQLLAYESYKNLFKGKDDQLSVIGRLAAGACAGMTSTL 203
Query: 252 LTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPS 311
LTYPLDV++ ++ V + RYR ++R++G + G+ + + I P
Sbjct: 204 LTYPLDVLRLRLAV---------EPRYRTMSQVALSMLRDEGIASFYYGLGPSLVGIAPY 254
Query: 312 AAISFTAYDTMKAWLGIPPQQKSRS 336
A++F +D +K L ++K++S
Sbjct: 255 IAVNFCIFDLVKKSLPEEYRKKAQS 279
>At5g01500 unknown protein
Length = 415
Score = 147 bits (372), Expect = 6e-36
Identities = 93/314 (29%), Positives = 148/314 (46%), Gaps = 34/314 (10%)
Query: 28 VPVYVKELVAGGVAGALSKTAVAPLERVKILWQT------RTGGFHTLGVCQSLNKLVKH 81
VP AG AGA +K+ APL+R+K+L QT + +G +++ + K
Sbjct: 112 VPKDAALFFAGAFAGAAAKSVTAPLDRIKLLMQTHGVRAGQQSAKKAIGFIEAITLIGKE 171
Query: 82 EGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTS 141
EG G +KGN +RIVPY+A+ YE YK L L AG+ AG TS
Sbjct: 172 EGIKGYWKGNLPQVIRIVPYSAVQLFAYETYKKLFRGKDGQLSV--LGRLGAGACAGMTS 229
Query: 142 VLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRG 201
L TYPLD+ R +LA + P + + V ++ RE G Y G
Sbjct: 230 TLITYPLDVLRLRLAVE------------------PGYRTMSQVALNMLREEGVASFYNG 271
Query: 202 VGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVKR 261
+GP++ I PY + F ++ +K +PE++Q+ L +A YPLD ++R
Sbjct: 272 LGPSLLSIAPYIAINFCVFDLVKKSLPEKYQQKTQSSLLTAVVAAAIATGTCYPLDTIRR 331
Query: 262 QMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDT 321
QMQ+ + T Y++ LD I+ +G L+ G N ++ +P+++I T +D
Sbjct: 332 QMQL--------KGTPYKSVLDAFSGIIAREGVVGLYRGFVPNALKSMPNSSIKLTTFDI 383
Query: 322 MKAWLGIPPQQKSR 335
+K + ++ R
Sbjct: 384 VKKLIAASEKEIQR 397
Score = 85.1 bits (209), Expect = 5e-17
Identities = 61/205 (29%), Positives = 99/205 (47%), Gaps = 19/205 (9%)
Query: 133 AGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYRE 192
AG+ AG + T PLD R KL Q R G + IG +T + +E
Sbjct: 121 AGAFAGAAAKSVTAPLD--RIKLLMQTHGVRA-------GQQSAKKAIGFIEAITLIGKE 171
Query: 193 GGARGLYRGVGPTITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQT 251
G +G ++G P + I+PY+ ++ + YE K + ++ Q S+L RL GA AG+
Sbjct: 172 EGIKGYWKGNLPQVIRIVPYSAVQLFAYETYKKLFRGKDGQLSVLGRLGAGACAGMTSTL 231
Query: 252 LTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPS 311
+TYPLDV++ ++ V + YR ++R +G + G+ + + I P
Sbjct: 232 ITYPLDVLRLRLAV---------EPGYRTMSQVALNMLREEGVASFYNGLGPSLLSIAPY 282
Query: 312 AAISFTAYDTMKAWLGIPPQQKSRS 336
AI+F +D +K L QQK++S
Sbjct: 283 IAINFCVFDLVKKSLPEKYQQKTQS 307
>At5g56450 ADP/ATP translocase-like protein
Length = 330
Score = 141 bits (355), Expect = 6e-34
Identities = 95/313 (30%), Positives = 158/313 (50%), Gaps = 34/313 (10%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLG------------VCQSLNKL 78
+ K+L+AG V G + T VAP+ER K+L QT+ +G + + +
Sbjct: 30 FQKDLLAGAVMGGVVHTIVAPIERAKLLLQTQESNIAIVGDEGHAGKRRFKGMFDFIFRT 89
Query: 79 VKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPA---LGSGPFIDLLAGS 135
V+ EG L L++GNG+S +R P AL+F + Y+S + N+ + SG + +AGS
Sbjct: 90 VREEGVLSLWRGNGSSVLRYYPSVALNFSLKDLYRSILRNSSSQENHIFSGALANFMAGS 149
Query: 136 AAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGA 195
AAG T+++ YPLD+A T+LA + G GI L++++++ G
Sbjct: 150 AAGCTALIVVYPLDIAHTRLAADI------------GKPEARQFRGIHHFLSTIHKKDGV 197
Query: 196 RGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILM--RLSCGALAGLFGQTLT 253
RG+YRG+ ++ G++ + GL F ++ +K E+ + + + R +
Sbjct: 198 RGIYRGLPASLHGVIIHRGLYFGGFDTVKEIFSEDTKPELALWKRWGLAQAVTTSAGLAS 257
Query: 254 YPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAA 313
YPLD V+R++ + + E YR+TLD +KI R++G + G N R SAA
Sbjct: 258 YPLDTVRRRI----MMQSGMEHPMYRSTLDCWKKIYRSEGLASFYRGALSNMFRSTGSAA 313
Query: 314 ISFTAYDTMKAWL 326
I YD +K +L
Sbjct: 314 I-LVFYDEVKRFL 325
>At2g47490 mitochondrial carrier like protein
Length = 312
Score = 130 bits (327), Expect = 1e-30
Identities = 91/313 (29%), Positives = 159/313 (50%), Gaps = 36/313 (11%)
Query: 37 AGGVAGALSKTAVAPLERVKILWQTRTGGFHTLG--------VCQSLNKLVKHEGFLGLY 88
AG AG ++ T V PL+ +K +Q G LG + SL ++ K EG GLY
Sbjct: 19 AGAAAGVVAATFVCPLDVIKTRFQVH--GLPKLGDANIKGSLIVGSLEQIFKREGMRGLY 76
Query: 89 KGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPL 148
+G + + ++ A++F Y++ KS++ +N L G ++LA S AG + + T PL
Sbjct: 77 RGLSPTVMALLSNWAIYFTMYDQLKSFLCSNDHKLSVG--ANVLAASGAGAATTIATNPL 134
Query: 149 DLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITG 208
+ +T+L Q + G+ P + S L + E G RGLY G+ P + G
Sbjct: 135 WVVKTRLQTQGMRV---------GIVPYKSTF---SALRRIAYEEGIRGLYSGLVPALAG 182
Query: 209 ILPYAGLKFYTYEKLKMHVPEEHQKSILMRLS-----CGALAGLFGQTLTYPLDVVKRQM 263
I + ++F TYE +K+++ ++ KS+ + ++A +F TLTYP +VV+ ++
Sbjct: 183 I-SHVAIQFPTYEMIKVYLAKKGDKSVDNLNARDVAVASSIAKIFASTLTYPHEVVRARL 241
Query: 264 QVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMK 323
Q + H + RY D ++K+ G+ + G + N +R P+A I+FT+++ +
Sbjct: 242 Q----EQGHHSEKRYSGVRDCIKKVFEKDGFPGFYRGCATNLLRTTPAAVITFTSFEMVH 297
Query: 324 AWL--GIPPQQKS 334
+L IP +Q S
Sbjct: 298 RFLVTHIPSEQSS 310
Score = 75.1 bits (183), Expect = 5e-14
Identities = 57/210 (27%), Positives = 97/210 (46%), Gaps = 14/210 (6%)
Query: 118 NNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQP 177
N+ P + AG+AAG + PLD+ +T+ + G IKG
Sbjct: 4 NSHPPNSKNVLCNAAAGAAAGVVAATFVCPLDVIKTRFQVHGLPKLGDA--NIKGSL--- 58
Query: 178 AHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHV-PEEHQKSIL 236
I L +++ G RGLYRG+ PT+ +L + F Y++LK + +H+ S+
Sbjct: 59 ----IVGSLEQIFKREGMRGLYRGLSPTVMALLSNWAIYFTMYDQLKSFLCSNDHKLSVG 114
Query: 237 MRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQ 296
+ + AG T PL VVK ++Q ++ Y++T LR+I +G R
Sbjct: 115 ANVLAASGAGAATTIATNPLWVVKTRLQTQGMRVGI---VPYKSTFSALRRIAYEEGIRG 171
Query: 297 LFAGVSINYIRIVPSAAISFTAYDTMKAWL 326
L++G+ + + + AI F Y+ +K +L
Sbjct: 172 LYSGL-VPALAGISHVAIQFPTYEMIKVYL 200
>At1g25380 unknown protein
Length = 363
Score = 129 bits (325), Expect = 2e-30
Identities = 89/315 (28%), Positives = 163/315 (51%), Gaps = 35/315 (11%)
Query: 37 AGGVAGALSKTAVAPLERVKI------LWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKG 90
AG AGA++ T V PL+ +K L + G + SL ++K EG+ G+Y+G
Sbjct: 23 AGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQRGGVIITSLKNIIKEEGYRGMYRG 82
Query: 91 NGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDL 150
+ + ++P A++F Y + K + ++ L G +++A + AG + + T PL +
Sbjct: 83 LSPTIIALLPNWAVYFSVYGKLKDVLQSSDGKLSIGS--NMIAAAGAGAATSIATNPLWV 140
Query: 151 ARTKLAYQVIDTRGTIKDGIKGVRPQPA-HIGIKSVLTSVYREGGARGLYRGVGPTITGI 209
+T+L Q G+RP + + S + + E G RGLY G+ P++ G+
Sbjct: 141 VKTRLMTQ-------------GIRPGVVPYKSVMSAFSRICHEEGVRGLYSGILPSLAGV 187
Query: 210 LPYAGLKFYTYEKLKMHVPEEHQKSILMRLSCG------ALAGLFGQTLTYPLDVVKRQM 263
+ ++F YEK+K ++ + S+ LS G ++A + LTYP +V++ ++
Sbjct: 188 -SHVAIQFPAYEKIKQYMAKMDNTSV-ENLSPGNVAIASSIAKVIASILTYPHEVIRAKL 245
Query: 264 QV-GSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTM 322
Q G ++NA +T+Y +D + K+ R++G L+ G + N +R PSA I+FT Y+ M
Sbjct: 246 QEQGQIRNA---ETKYSGVIDCITKVFRSEGIPGLYRGCATNLLRTTPSAVITFTTYEMM 302
Query: 323 -KAWLGIPPQQKSRS 336
+ + + P + +RS
Sbjct: 303 LRFFRQVVPPETNRS 317
Score = 55.1 bits (131), Expect = 5e-08
Identities = 27/91 (29%), Positives = 47/91 (50%)
Query: 233 KSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQ 292
+ + GA AG T PLDV+K ++QV L A R + L+ I++ +
Sbjct: 15 REVAANAGAGATAGAIAATFVCPLDVIKTRLQVLGLPEAPASGQRGGVIITSLKNIIKEE 74
Query: 293 GWRQLFAGVSINYIRIVPSAAISFTAYDTMK 323
G+R ++ G+S I ++P+ A+ F+ Y +K
Sbjct: 75 GYRGMYRGLSPTIIALLPNWAVYFSVYGKLK 105
>At3g08580 adenylate translocator
Length = 381
Score = 128 bits (322), Expect = 4e-30
Identities = 95/307 (30%), Positives = 151/307 (48%), Gaps = 24/307 (7%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLV 79
G + + + GGV+ A+SKTA AP+ERVK+L Q + G+ + +
Sbjct: 76 GFTNFALDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGIGDCFGRTI 135
Query: 80 KHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI--DLLAGSAA 137
K EGF L++GN A+ +R P AL+F + +K G + +L +G AA
Sbjct: 136 KDEGFGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAA 195
Query: 138 GGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARG 197
G +S+L Y LD ART+LA D + K G G R G+ V + G G
Sbjct: 196 GASSLLFVYSLDYARTRLAN---DAKAAKKGG--GGR---QFDGLVDVYRKTLKTDGIAG 247
Query: 198 LYRGVGPTITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQTLTYPL 256
LYRG + GI+ Y GL F Y+ +K + + + Q S + G + +YP+
Sbjct: 248 LYRGFNISCVGIIVYRGLYFGLYDSVKPVLLTGDLQDSFFASFALGWVITNGAGLASYPI 307
Query: 257 DVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISF 316
D V+R+M + S E +Y+++LD ++I++N+G + LF G N +R V A +
Sbjct: 308 DTVRRRMMMTS-----GEAVKYKSSLDAFKQILKNEGAKSLFKGAGANILRAVAGAGV-L 361
Query: 317 TAYDTMK 323
+ YD ++
Sbjct: 362 SGYDKLQ 368
>At4g28390 ADP,ATP carrier-like protein
Length = 379
Score = 127 bits (319), Expect = 9e-30
Identities = 96/303 (31%), Positives = 146/303 (47%), Gaps = 25/303 (8%)
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLVKHEG 83
++ + + GGV+ A+SKTA AP+ERVK+L Q + G+ + VK EG
Sbjct: 79 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGISDCFARTVKDEG 138
Query: 84 FLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI--DLLAGSAAGGTS 141
L L++GN A+ +R P AL+F + +K G + +L +G AAG +S
Sbjct: 139 MLALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKEKDGYWKWFAGNLASGGAAGASS 198
Query: 142 VLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRG 201
+L Y LD ART+LA D + K G Q G+ V G GLYRG
Sbjct: 199 LLFVYSLDYARTRLAN---DAKAAKKGG------QRQFNGMVDVYKKTIASDGIVGLYRG 249
Query: 202 VGPTITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVK 260
+ GI+ Y GL F Y+ LK + + + Q S L G + +YP+D V+
Sbjct: 250 FNISCVGIVVYRGLYFGLYDSLKPVVLVDGLQDSFLASFLLGWGITIGAGLASYPIDTVR 309
Query: 261 RQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYD 320
R+M + S E +Y+++L +IV+N+G + LF G N +R V A + YD
Sbjct: 310 RRMMMTS-----GEAVKYKSSLQAFSQIVKNEGAKSLFKGAGANILRAVAGAGV-LAGYD 363
Query: 321 TMK 323
++
Sbjct: 364 KLQ 366
>At5g13490 adenosine nucleotide translocator
Length = 385
Score = 122 bits (307), Expect = 2e-28
Identities = 91/307 (29%), Positives = 146/307 (46%), Gaps = 24/307 (7%)
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLV 79
G + + + GGV+ A+SKTA AP+ERVK+L Q + G+ + +
Sbjct: 80 GFTNFAIDFMMGGVSAAVSKTAAAPIERVKLLIQNQDEMLKAGRLTEPYKGIRDCFGRTI 139
Query: 80 KHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI--DLLAGSAA 137
+ EG L++GN A+ +R P AL+F + +K G + +L +G AA
Sbjct: 140 RDEGIGSLWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 199
Query: 138 GGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARG 197
G +S+L Y LD ART+LA D++ K G + G+ V + G G
Sbjct: 200 GASSLLFVYSLDYARTRLAN---DSKSAKKGG-----GERQFNGLVDVYKKTLKSDGIAG 251
Query: 198 LYRGVGPTITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQTLTYPL 256
LYRG + GI+ Y GL F Y+ +K + + + Q S + G L +YP+
Sbjct: 252 LYRGFNISCAGIIVYRGLYFGLYDSVKPVLLTGDLQDSFFASFALGWLITNGAGLASYPI 311
Query: 257 DVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISF 316
D V+R+M + S E +Y+++ D +IV+ +G + LF G N +R V A +
Sbjct: 312 DTVRRRMMMTS-----GEAVKYKSSFDAFSQIVKKEGAKSLFKGAGANILRAVAGAGV-L 365
Query: 317 TAYDTMK 323
YD ++
Sbjct: 366 AGYDKLQ 372
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,346,136
Number of Sequences: 26719
Number of extensions: 303124
Number of successful extensions: 1194
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 59
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 604
Number of HSP's gapped (non-prelim): 167
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0349b.8


