

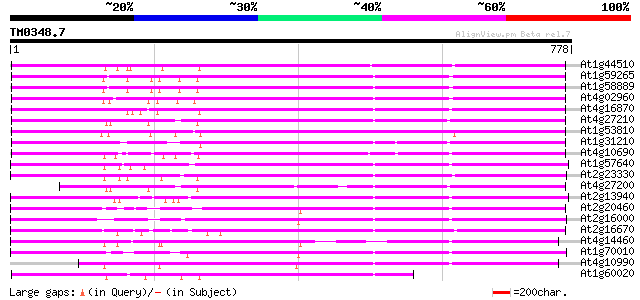
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.7
(778 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g44510 polyprotein, putative 678 0.0
At1g59265 polyprotein, putative 671 0.0
At1g58889 polyprotein, putative 669 0.0
At4g02960 putative polyprotein of LTR transposon 667 0.0
At4g16870 retrotransposon like protein 656 0.0
At4g27210 putative protein 634 0.0
At1g53810 617 e-177
At1g31210 putative reverse transcriptase 608 e-174
At4g10690 retrotransposon like protein 607 e-173
At1g57640 540 e-153
At2g23330 putative retroelement pol polyprotein 527 e-149
At4g27200 putative protein 525 e-149
At2g13940 putative retroelement pol polyprotein 523 e-148
At2g20460 putative retroelement pol polyprotein 520 e-147
At2g16000 putative retroelement pol polyprotein 512 e-145
At2g16670 putative retroelement pol polyprotein 504 e-143
At4g14460 retrovirus-related like polyprotein 488 e-138
At1g70010 hypothetical protein 486 e-137
At4g10990 putative retrotransposon polyprotein 466 e-131
At1g60020 hypothetical protein 451 e-127
>At1g44510 polyprotein, putative
Length = 1459
Score = 678 bits (1749), Expect = 0.0
Identities = 369/826 (44%), Positives = 497/826 (59%), Gaps = 62/826 (7%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRHIVETGL LL+ AS+P +W +AF TA YLINRM T L SP+ KL+G+ P++
Sbjct: 632 ERKHRHIVETGLTLLTQASVPREYWTYAFATAVYLINRMPTPVLCLQSPFQKLFGSSPNY 691
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLFH 121
+ L++FG CFP+LRPY NKL SK CVFLGYS + Y CLD + R+Y S+ V+F
Sbjct: 692 QRLRVFGCLCFPWLRPYTRNKLEERSKRCVFLGYSLTQTAYLCLDVDNNRLYTSRHVMFD 751
Query: 122 EHRFPYTTLF----PSEPFSPPTSSAEYFP-----------LSTVPIISRSMPQP----- 161
E +P+ S +PP SS+ P L + P S P P
Sbjct: 752 ESTYPFAASIREQSQSSLVTPPESSSSSSPANSGFPCSVLRLQSPPASSPETPSPPQQQN 811
Query: 162 ----------SPAPI------------STELANPGPLSPQSEASDLQSQPSPIPTGSGLA 199
SP P S ++N P +P + ++Q +P G
Sbjct: 812 DSPVSPRQTGSPTPSHHSQVRDSTLSPSPSVSNSEPTAPHENGPEPEAQSNPNSPFIGPL 871
Query: 200 STSQPAEHASS--------ESAHQEMATSSGVHAASSASTVAVPVNAHPMQTRSKSGIIK 251
P + SS +S + + AA+S S P N H M+TRSK+ I K
Sbjct: 872 PNPNPETNPSSSIEQRPVDKSTTTALPPNQTTIAATSNSRSQPPKNNHQMKTRSKNNITK 931
Query: 252 PRLNPTLLLT----HM-EPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAV 306
P+ +L + H+ EP TV QA++D KW AM +E++A N TW LVP + V
Sbjct: 932 PKTKTSLTVALTQPHLSEPNTVTQALKDKKWRFAMSDEFDAQQRNHTWDLVPPNPTQHLV 991
Query: 307 GCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRG 366
GC+W++++K P+G I+KYKARLVAKG++Q G DY+ETFSPV+K T+R++L +A+ +
Sbjct: 992 GCRWVFKLKYLPNGLIDKYKARLVAKGFNQQYGVDYAETFSPVIKATTIRVVLDVAVKKN 1051
Query: 367 WPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCKLHKALYGLKQAPRAWFHRLK 425
WPL+Q+DVNNAFL G L EEVYM QPPGF KD+ + VC+L KA+YGLKQAPRAW+ LK
Sbjct: 1052 WPLKQLDVNNAFLQGTLTEEVYMAQPPGFVDKDRPSHVCRLRKAIYGLKQAPRAWYMELK 1111
Query: 426 EVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALK 485
+ LL GF S D SLF Y+ +Y+L+YVDDII+TG + + + L F++K
Sbjct: 1112 QHLLNIGFVNSLADTSLFIYSHGTTLLYLLVYVDDIIVTGSDHKSVSAVLSSLAERFSIK 1171
Query: 486 QLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGT 545
L YFLG++ T N L L Q KY+ DLL K NM DA P++TP+ KLT HGGT
Sbjct: 1172 DPTDLHYFLGIEATR-TNTGLHLMQRKYMTDLLAKHNMLDAKPVATPLPTSPKLTLHGGT 1230
Query: 546 SLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITH 605
L+D +EYRSVVG+LQY TRP+I++AVN++ QF+ P +HW+A KR+LRYL GT TH
Sbjct: 1231 KLNDASEYRSVVGSLQYLAFTRPDIAFAVNRLSQFMHQPTSDHWQAAKRVLRYLAGTTTH 1290
Query: 606 GVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSST 665
G+ L S P+ L AF DADW D D ST+ ++LG N ISW++KKQ V+RSST
Sbjct: 1291 GIFLNSSS---PIHLHAFSDADWAGDSADYVSTNAYVIYLGRNPISWSSKKQRGVSRSST 1347
Query: 666 EAEYRSLANTTAELLWVESLLTELKIAFT-VPTVLCDNMSTVLLTHNPILHTRTKHMEMD 724
E+EYR++AN +E+ W+ SLLTEL I PT+ CDN+ + NP+ H+R KH+ +D
Sbjct: 1348 ESEYRAVANAASEIRWLCSLLTELHIRLPHGPTIFCDNIGATYICANPVFHSRMKHIALD 1407
Query: 725 LFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
FVR +Q+++L V HV + Q AD TK+LS FL R K+ V
Sbjct: 1408 YHFVRGMIQSRALRVSHVSTNDQLADALTKSLSRPHFLSARSKIGV 1453
>At1g59265 polyprotein, putative
Length = 1466
Score = 671 bits (1730), Expect = 0.0
Identities = 380/840 (45%), Positives = 502/840 (59%), Gaps = 77/840 (9%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRHIVETGL LLSHAS+P +W +AF A YLINR+ T LQ SP+ KL+G P++
Sbjct: 626 ERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNY 685
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLFH 121
L++FG AC+P+LRPYN +KL S++CVFLGYS + Y CL Q+ R+Y+S+ V F
Sbjct: 686 DKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFD 745
Query: 122 EHRFPYT----TLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPS----------PAPIS 167
E+ FP++ TL P + SS + P +T+P + +P PS P+ S
Sbjct: 746 ENCFPFSNYLATLSPVQE-QRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHAATPPSSPS 804
Query: 168 TELANPGPLSPQSEASDLQSQPS-PIPTG---SGLASTSQPAE-----HASSESAHQEMA 218
N S ++S S PS P PT +G T+QP + H+S ++
Sbjct: 805 APFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPT 864
Query: 219 TSSGVHAASSASTVA------------------------------------------VPV 236
S A S ST A P+
Sbjct: 865 NESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPL 924
Query: 237 NAHPMQTRSKSGIIKPRLNPTL---LLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGT 293
N H M TR+K+GIIKP +L L EP T QA++D++W AM E NA + N T
Sbjct: 925 NTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHT 984
Query: 294 WSLVPLP-SNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKP 352
W LVP P S+ VGC+WI+ K N DGS+N+YKARLVAKGY+Q G DY+ETFSPV+K
Sbjct: 985 WDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKS 1044
Query: 353 ITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCKLHKALY 411
++R++L +A+ R WP++Q+DVNNAFL G L ++VYM+QPPGF KD+ VCKL KALY
Sbjct: 1045 TSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALY 1104
Query: 412 GLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLI 471
GLKQAPRAW+ L+ LL GF S D SLF +YML+YVDDI++TG+ L+
Sbjct: 1105 GLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLL 1164
Query: 472 QQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPIST 531
L F++K +L YFLG++ + G L L+Q +YI DLL + NM A P++T
Sbjct: 1165 HNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTG-LHLSQRRYILDLLARTNMITAKPVTT 1223
Query: 532 PMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKA 591
PM KL+ + GT L DPTEYR +VG+LQY TRP+ISYAVN++ QF+ P EEH +A
Sbjct: 1224 PMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQA 1283
Query: 592 VKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLIS 651
+KRILRYL GT HG+ L+ L L A+ DADW D DD ST+G V+LG + IS
Sbjct: 1284 LKRILRYLAGTPNHGIFLK---KGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPIS 1340
Query: 652 WTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTH 710
W++KKQ V RSSTEAEYRS+ANT++E+ W+ SLLTEL I T P V+ CDN+ L
Sbjct: 1341 WSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCA 1400
Query: 711 NPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
NP+ H+R KH+ +D F+R +VQ+ +L V HV + Q AD TK LS T F K+ V
Sbjct: 1401 NPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
>At1g58889 polyprotein, putative
Length = 1466
Score = 669 bits (1726), Expect = 0.0
Identities = 379/840 (45%), Positives = 501/840 (59%), Gaps = 77/840 (9%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRHIVETGL LLSHAS+P +W +AF A YLINR+ T LQ SP+ KL+G P++
Sbjct: 626 ERKHRHIVETGLTLLSHASIPKTYWPYAFAVAVYLINRLPTPLLQLESPFQKLFGTSPNY 685
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLFH 121
L++FG AC+P+LRPYN +KL S++CVFLGYS + Y CL Q+ R+Y+S+ V F
Sbjct: 686 DKLRVFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFD 745
Query: 122 EHRFPYT----TLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPS----------PAPIS 167
E+ FP++ TL P + SS + P +T+P + +P PS P+ S
Sbjct: 746 ENCFPFSNYLATLSPVQE-QRRESSCVWSPHTTLPTRTPVLPAPSCSDPHHAATPPSSPS 804
Query: 168 TELANPGPLSPQSEASDLQSQPS-PIPTG---SGLASTSQPAE-----HASSESAHQEMA 218
N S ++S S PS P PT +G T+QP + H+S ++
Sbjct: 805 APFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQNNPT 864
Query: 219 TSSGVHAASSASTVA------------------------------------------VPV 236
S A S ST A P+
Sbjct: 865 NESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNNQAPL 924
Query: 237 NAHPMQTRSKSGIIKPRLNPTL---LLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGT 293
N H M TR+K+GIIKP +L L EP T QA++D++W AM E NA + N T
Sbjct: 925 NTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHT 984
Query: 294 WSLVPLP-SNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKP 352
W LVP P S+ VGC+WI+ K N DGS+N+YKAR VAKGY+Q G DY+ETFSPV+K
Sbjct: 985 WDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAETFSPVIKS 1044
Query: 353 ITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCKLHKALY 411
++R++L +A+ R WP++Q+DVNNAFL G L ++VYM+QPPGF KD+ VCKL KALY
Sbjct: 1045 TSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALY 1104
Query: 412 GLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLI 471
GLKQAPRAW+ L+ LL GF S D SLF +YML+YVDDI++TG+ L+
Sbjct: 1105 GLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLL 1164
Query: 472 QQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPIST 531
L F++K +L YFLG++ + G L L+Q +YI DLL + NM A P++T
Sbjct: 1165 HNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTG-LHLSQRRYILDLLARTNMITAKPVTT 1223
Query: 532 PMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKA 591
PM KL+ + GT L DPTEYR +VG+LQY TRP+ISYAVN++ QF+ P EEH +A
Sbjct: 1224 PMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQA 1283
Query: 592 VKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLIS 651
+KRILRYL GT HG+ L+ L L A+ DADW D DD ST+G V+LG + IS
Sbjct: 1284 LKRILRYLAGTPNHGIFLK---KGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPIS 1340
Query: 652 WTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTH 710
W++KKQ V RSSTEAEYRS+ANT++E+ W+ SLLTEL I T P V+ CDN+ L
Sbjct: 1341 WSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCA 1400
Query: 711 NPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
NP+ H+R KH+ +D F+R +VQ+ +L V HV + Q AD TK LS T F K+ V
Sbjct: 1401 NPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 667 bits (1721), Expect = 0.0
Identities = 378/847 (44%), Positives = 497/847 (58%), Gaps = 85/847 (10%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRHIVE GL LLSHAS+P +W +AF A YLINR+ T LQ SP+ KL+G P++
Sbjct: 605 ERKHRHIVEMGLTLLSHASVPKTYWPYAFSVAVYLINRLPTPLLQLQSPFQKLFGQPPNY 664
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLFH 121
+ LK+FG AC+P+LRPYN +KL SK+C F+GYS + Y CL +GR+Y S+ V F
Sbjct: 665 EKLKVFGCACYPWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFD 724
Query: 122 EHRFPYT-----------------------TLFPSEPF----------------SPPTSS 142
E FP++ T P+ P PP+S
Sbjct: 725 ERCFPFSTTNFGVSTSQEQRSDSAPNWPSHTTLPTTPLVLPAPPCLGPHLDTSPRPPSSP 784
Query: 143 AEYFPLSTVPIISRSMPQPS-PAPISTELANPGPLSPQSEASDLQSQPS---------PI 192
+ PL T + S ++P S +P S+E P PQ A Q+Q S P
Sbjct: 785 S---PLCTTQVSSSNLPSSSISSPSSSEPTAPSHNGPQPTAQPHQTQNSNSNSPILNNPN 841
Query: 193 PTGSGLASTSQ----PAEHASSESAHQEMATSSGVHAASSAST----------------- 231
P S +Q P SS + S ++ SS+ST
Sbjct: 842 PNSPSPNSPNQNSPLPQSPISSPHIPTPSTSISEPNSPSSSSTSTPPLPPVLPAPPIIQV 901
Query: 232 -VAVPVNAHPMQTRSKSGIIKPRLN---PTLLLTHMEPTTVKQAMRDDKWLQAMKEEYNA 287
PVN H M TR+K GI KP T L + EP T QAM+DD+W QAM E NA
Sbjct: 902 NAQAPVNTHSMATRAKDGIRKPNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINA 961
Query: 288 LMSNGTWSLVPLPS-NRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETF 346
+ N TW LVP P + VGC+WI+ K N DGS+N+YKARLVAKGY+Q G DY+ETF
Sbjct: 962 QIGNHTWDLVPPPPPSVTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETF 1021
Query: 347 SPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCK 405
SPV+K ++R++L +A+ R WP++Q+DVNNAFL G L +EVYM+QPPGF KD+ VC+
Sbjct: 1022 SPVIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCR 1081
Query: 406 LHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTG 465
L KA+YGLKQAPRAW+ L+ LL GF S D SLF IYML+YVDDI++TG
Sbjct: 1082 LRKAIYGLKQAPRAWYVELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITG 1141
Query: 466 DSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMAD 525
+ VL++ L F++K+ L YFLG++ + G L L+Q +Y DLL + NM
Sbjct: 1142 NDTVLLKHTLDALSQRFSVKEHEDLHYFLGIEAKRVPQG-LHLSQRRYTLDLLARTNMLT 1200
Query: 526 AAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPH 585
A P++TPM KLT H GT L DPTEYR +VG+LQY TRP++SYAVN++ Q++ P
Sbjct: 1201 AKPVATPMATSPKLTLHSGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPT 1260
Query: 586 EEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFL 645
++HW A+KR+LRYL GT HG+ L+ L L A+ DADW D DD ST+G V+L
Sbjct: 1261 DDHWNALKRVLRYLAGTPDHGIFLK---KGNTLSLHAYSDADWAGDTDDYVSTNGYIVYL 1317
Query: 646 GPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMS 704
G + ISW++KKQ V RSSTEAEYRS+ANT++EL W+ SLLTEL I + P V+ CDN+
Sbjct: 1318 GHHPISWSSKKQKGVVRSSTEAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVG 1377
Query: 705 TVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLM 764
L NP+ H+R KH+ +D F+R +VQ+ +L V HV + Q AD TK LS F
Sbjct: 1378 ATYLCANPVFHSRMKHIALDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNF 1437
Query: 765 RDKLNVV 771
K+ V+
Sbjct: 1438 SRKIGVI 1444
>At4g16870 retrotransposon like protein
Length = 1474
Score = 656 bits (1692), Expect = 0.0
Identities = 372/849 (43%), Positives = 489/849 (56%), Gaps = 88/849 (10%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRHIVETGL LL+ AS+P +W +AF A YLINRM T L SP+ KL+G+ P++
Sbjct: 627 ERKHRHIVETGLTLLTQASVPREYWPYAFAAAVYLINRMPTPVLSMESPFQKLFGSKPNY 686
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLFH 121
+ L++FG CFP+LRPY NKL S+ CVFLGYS + Y C D + R+Y S+ V+F
Sbjct: 687 ERLRVFGCLCFPWLRPYTHNKLEERSRRCVFLGYSLTQTAYLCFDVEHKRLYTSRHVVFD 746
Query: 122 EHRFPYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPS------------PAPISTE 169
E FP++ L T PL T PI+S S PS P++T
Sbjct: 747 EASFPFSNLTSQNSLPTVTFEQSSSPLVT-PILSSSSVLPSCLSSPCTVLHQQQPPVTTP 805
Query: 170 --------LANPGPLSPQ-------------------SEASDLQSQPSPIPTGSGLASTS 202
+P PLSP S +S L S+P+ P +G +
Sbjct: 806 NSPHSSQPTTSPAPLSPHRSTTMDFQVPQVRSSSPLLSSSSSLNSEPTA-PNENGPEPEA 864
Query: 203 Q---------------------------------PAEHASSESAHQEMATSSGVHAASSA 229
Q PA H T + + ++
Sbjct: 865 QSPPIGPLSNPTHEAFIGPLPNPNRNPTNEIEPTPAPHPKPVKPTTTTTTPNRTTVSDAS 924
Query: 230 STVAVPV-NAHPMQTRSKSGIIKPRLNPTLLLT-----HMEPTTVKQAMRDDKWLQAMKE 283
P N H M+TR+K+ I KP +L T EPT V QA++D KW AM +
Sbjct: 925 HQPTAPQQNQHNMKTRAKNNIKKPNTKFSLTATLPNRSPSEPTNVTQALKDKKWRFAMSD 984
Query: 284 EYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYS 343
E++A N TW LVP S + VGCKW++++K P+G+I+KYKARLVAKG++Q G DY+
Sbjct: 985 EFDAQQRNHTWDLVPHES-QLLVGCKWVFKLKYLPNGAIDKYKARLVAKGFNQQYGVDYA 1043
Query: 344 ETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TL 402
ETFSPV+K T+RL+L +A+ + W ++Q+DVNNAFL G L EEVYM QPPGF KD+ T
Sbjct: 1044 ETFSPVIKSTTIRLVLDVAVKKDWEIKQLDVNNAFLQGTLTEEVYMAQPPGFIDKDRPTH 1103
Query: 403 VCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDII 462
VC+L KA+YGLKQAPRAW+ LK+ L GF S D SLF Y +Y+L+YVDDII
Sbjct: 1104 VCRLRKAIYGLKQAPRAWYMELKQHLFNIGFVNSLSDASLFIYCHGTTFVYVLVYVDDII 1163
Query: 463 LTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVN 522
+TG I + L F++K L YFLG++ T G L L Q KYI DLL K N
Sbjct: 1164 VTGSDKSSIDAVLTSLAERFSIKDPTDLHYFLGIEATRTKQG-LHLMQRKYIKDLLAKHN 1222
Query: 523 MADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLS 582
MADA P+ TP+ KLT HGGT L+D +EYRSVVG+LQY TRP+I+YAVN++ Q +
Sbjct: 1223 MADAKPVLTPLPTSPKLTLHGGTKLNDASEYRSVVGSLQYLAFTRPDIAYAVNRLSQLMP 1282
Query: 583 DPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSC 642
P E+HW+A KR+LRYL GT THG+ L T PL L AF DADW D DD ST+
Sbjct: 1283 QPTEDHWQAAKRVLRYLAGTSTHGIFL---DTTSPLNLHAFSDADWAGDSDDYVSTNAYV 1339
Query: 643 VFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTV-PTVLCD 701
++LG N ISW++KKQ VARSSTE+EYR++AN +E+ W+ SLL++L I + P++ CD
Sbjct: 1340 IYLGKNPISWSSKKQRGVARSSTESEYRAVANAASEVKWLCSLLSKLHIRLPIRPSIFCD 1399
Query: 702 NMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRF 761
N+ L NP+ H+R KH+ +D FVR +Q+ +L V HV + Q AD TK LS F
Sbjct: 1400 NIGATYLCANPVFHSRMKHIAIDYHFVRNMIQSGALRVSHVSTRDQLADALTKPLSRAHF 1459
Query: 762 LLMRDKLNV 770
R K+ V
Sbjct: 1460 QSARFKIGV 1468
>At4g27210 putative protein
Length = 1318
Score = 634 bits (1635), Expect = 0.0
Identities = 350/796 (43%), Positives = 473/796 (58%), Gaps = 38/796 (4%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTL-QGASPYFKLYGNHPD 61
ERKHRH+VE GL++L + +P FW AF TA +LIN + T+ L + SPY KLY PD
Sbjct: 432 ERKHRHLVELGLSMLFQSHVPHKFWVEAFFTANFLINLLPTSALKESISPYEKLYDKKPD 491
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCL-DQSGRIYVSKDVLF 120
+ SL+ FGSACFP LR Y NK + S +CVFLGY+ +KGY+CL +GR+Y+S+ V+F
Sbjct: 492 YTSLRSFGSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIF 551
Query: 121 HEHRFPYTTLFPS---EPFSP-----------PTSSAEYFPLSTVPIISRSMPQPSPAPI 166
E +P++ + +P +P P S P S P+ + + P P
Sbjct: 552 DESVYPFSHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFTSADFPPLPQRK 611
Query: 167 STELANPGPLSPQSEASDLQSQPSP-------IPTGSGLASTSQPAEHASSESAHQEMAT 219
+ L P+S S AS++ +Q SP S S + A S+S
Sbjct: 612 TPLLPTLVPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQA 671
Query: 220 SSGVHAASSASTVAVPVNAHPMQTRSKSGIIKPRLNPTLL---LTHMEPTTVKQAMRDDK 276
S VH +++ N HPM TR+K GI KP L +++ EP TV A++
Sbjct: 672 SVNVHQTHAST------NVHPMVTRAKVGISKPNPRYVFLSHKVSYPEPKTVTAALKHPG 725
Query: 277 WLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQ 336
W AM EE TWSLVP S+ +G KW++R K + DG++NK KAR+VAKG+ Q
Sbjct: 726 WTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQ 785
Query: 337 VQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFE 396
+G DY ET+SPVV+ TVRL+L LA + W ++Q+DV NAFL+G L+E VYMTQP GF
Sbjct: 786 EEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAGFV 845
Query: 397 HKDK-TLVCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYML 455
K VC LHK++YGLKQ+PRAWF + LL+FGF SK DPSLF Y I +L
Sbjct: 846 DPSKPDHVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCSKSDPSLFIYAHNNNLILLL 905
Query: 456 IYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYIN 515
+YVDD+++TG+S + L A L+ F + +GQL YFLG+QV NG L ++Q KY
Sbjct: 906 LYVDDMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFLGIQVQRQQNG-LFMSQQKYAE 964
Query: 516 DLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVN 575
DLL +M P+ TP+ H DPT +RS+ G LQY T+TRP+I +AVN
Sbjct: 965 DLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVN 1024
Query: 576 KVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDR 635
VCQ + P + +KRILRY+KGTIT G+ S P L A+ D+DWG+ R
Sbjct: 1025 FVCQKMHQPTISDFHLLKRILRYIKGTITMGI---SYSRDSPTLLQAYSDSDWGNCKQTR 1081
Query: 636 RSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAF-T 694
RS G C F+G NL+SW++KK V+RSSTEAEY+SL++ +E+LW+ +LL EL+I
Sbjct: 1082 RSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPD 1141
Query: 695 VPTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTK 754
P + CDN+S V LT NP H RTKH ++D FVRE+V K+LVV+H+P Q ADIFTK
Sbjct: 1142 TPELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTK 1201
Query: 755 ALSPTRFLLMRDKLNV 770
+L F+ +R KL V
Sbjct: 1202 SLPYEAFIHLRGKLGV 1217
>At1g53810
Length = 1522
Score = 617 bits (1591), Expect = e-177
Identities = 343/810 (42%), Positives = 479/810 (58%), Gaps = 44/810 (5%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQG-ASPYFKLYGNHPD 61
ERKHRHIVE GL+++ + +PL +W +F TA ++IN + T++L SPY KLYG P+
Sbjct: 621 ERKHRHIVELGLSMIFQSKLPLKYWLESFFTANFVINLLPTSSLDNNESPYQKLYGKAPE 680
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCL-DQSGRIYVSKDVLF 120
+ +L++FG AC+P LR Y S K S +CVFLGY+ +KGY+CL +GRIY+S+ V+F
Sbjct: 681 YSALRVFGCACYPTLRDYASTKFDPRSLKCVFLGYNEKYKGYRCLYPPTGRIYISRHVVF 740
Query: 121 HEHRFP----YTTLFPSEP-------------FSPPTSSAEYFPLSTVPIISRSMPQPSP 163
E+ P Y+ L P + +P +P+S++P + +P
Sbjct: 741 DENTHPFESIYSHLHPQDKTPLLEAWFKSFHHVTPTQPDQSRYPVSSIPQPETTDLSAAP 800
Query: 164 APISTELANPGPLSPQSEASDLQSQPSPIP---TGSGLASTSQPAEHASSESAHQEMATS 220
A ++ E A P S+ ++ S S P TG AS +++S+H A S
Sbjct: 801 ASVAAETAGPNASDDTSQDNETISVVSGSPERTTGLDSASIGDSYHSPTADSSHPSPARS 860
Query: 221 SGVHAASS-------ASTVAVPV-NAHPMQTRSKSGIIKPRLNPTLLLTHM----EPTTV 268
S + A V PV N H M TR K GI KP +LLTH EP TV
Sbjct: 861 SPASSPQGSPIQMAPAQQVQAPVTNEHAMVTRGKEGISKPNKR-YVLLTHKVSIPEPKTV 919
Query: 269 KQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKAR 328
+A++ W AM+EE TW+LVP N +G W++R K + DGS++K KAR
Sbjct: 920 TEALKHPGWNNAMQEEMGNCKETETWTLVPYSPNMNVLGSMWVFRTKLHADGSLDKLKAR 979
Query: 329 LVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVY 388
LVAKG+ Q +G DY ET+SPVV+ TVRLIL +A W L+Q+DV NAFL+G L E VY
Sbjct: 980 LVAKGFKQEEGIDYLETYSPVVRTPTVRLILHVATVLKWELKQMDVKNAFLHGDLTETVY 1039
Query: 389 MTQPPGFEHKDK-TLVCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNS 447
M QP GF K K VC LHK+LYGLKQ+PRAWF R LL+FGF S DPSLF Y+S
Sbjct: 1040 MRQPAGFVDKSKPDHVCLLHKSLYGLKQSPRAWFDRFSNFLLEFGFICSLFDPSLFVYSS 1099
Query: 448 PLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLL 507
I +L+YVDD+++TG++ + L A L+ F +K +GQ+ YFLG+Q+ +G L
Sbjct: 1100 NNDVILLLLYVDDMVITGNNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQI-QTYDGGLF 1158
Query: 508 LNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITR 567
++Q KY DLL +MA+ +P+ TP+ + DPT +RS+ G LQY T+TR
Sbjct: 1159 MSQQKYAEDLLITASMANCSPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGKLQYLTLTR 1218
Query: 568 PEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMT------QPLPLL 621
P+I +AVN VCQ + P + +KRILRY+KGT++ G+ S + L
Sbjct: 1219 PDIQFAVNFVCQKMHQPSVSDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSAYESDYDLS 1278
Query: 622 AFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLW 681
A+ D+D+ + + RRS G C F+G N+ISW++KKQ V+RSSTEAEYRSL+ T +E+ W
Sbjct: 1279 AYSDSDYANCKETRRSVGGYCTFMGQNIISWSSKKQPTVSRSSTEAEYRSLSETASEIKW 1338
Query: 682 VESLLTELKIAF-TVPTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQ 740
+ S+L E+ ++ P + CDN+S V LT NP H RTKH ++D ++RE+V K+LVV+
Sbjct: 1339 MSSILREIGVSLPDTPELFCDNLSAVYLTANPAFHARTKHFDVDHHYIRERVALKTLVVK 1398
Query: 741 HVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
H+P Q ADIFTK+L F +R KL V
Sbjct: 1399 HIPGHLQLADIFTKSLPFEAFTRLRFKLGV 1428
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 608 bits (1569), Expect = e-174
Identities = 330/777 (42%), Positives = 466/777 (59%), Gaps = 41/777 (5%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRH+VE GL++L H+ P FW +F TA Y+INR+ ++ L+ SPY L+G PD+
Sbjct: 617 ERKHRHLVELGLSMLFHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALFGEKPDY 676
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCL-DQSGRIYVSKDVLFH 121
SL++FGSAC+P LRP NK S +CVFLGY+S +KGY+C +G++Y+S++V+F+
Sbjct: 677 SSLRVFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYISRNVIFN 736
Query: 122 EHRFPYTTLFPS--EPFSPPTSSA-EYFPLSTVPIISRSMPQPSPAPISTELANPGPLSP 178
E P+ + S +S P A ++ +S + + PA + P L+
Sbjct: 737 ESELPFKEKYQSLVPQYSTPLLQAWQHNKISEISV---------PAAPVQLFSKPIDLNT 787
Query: 179 QSEASDLQSQPSPIPTGSGLASTSQPAEHASSESAHQEMATSSGVHAASSASTVAVPVNA 238
+ + + P PT + S + A +A+QE +N+
Sbjct: 788 YAGSQVTEQLTDPEPTSNNEGSDEEVNPVAEEIAANQEQV-----------------INS 830
Query: 239 HPMQTRSKSGIIKPRLNPTLLLTHM---EPTTVKQAMRDDKWLQAMKEEYNALMSNGTWS 295
H M TRSK+GI KP L+ + M EP T+ AM+ W +A+ EE N + TWS
Sbjct: 831 HAMTTRSKAGIQKPNTRYALITSRMNTAEPKTLASAMKHPGWNEAVHEEINRVHMLHTWS 890
Query: 296 LVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITV 355
LVP + + KW+++ K +PDGSI+K KARLVAKG+ Q +G DY ETFSPVV+ T+
Sbjct: 891 LVPPTDDMNILSSKWVFKTKLHPDGSIDKLKARLVAKGFDQEEGVDYLETFSPVVRTATI 950
Query: 356 RLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCKLHKALYGLK 414
RL+L ++ S+GWP++Q+DV+NAFL+G L+E V+M QP GF K T VC+L KA+YGLK
Sbjct: 951 RLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVFMYQPSGFIDPQKPTHVCRLTKAIYGLK 1010
Query: 415 QAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQL 474
QAPRAWF LL +GF SK DPSLF + +Y+L+YVDDI+LTG L++ L
Sbjct: 1011 QAPRAWFDTFSNFLLDYGFVCSKSDPSLFVCHQDGKILYLLLYVDDILLTGSDQSLLEDL 1070
Query: 475 TAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQ 534
L + F++K LG YFLG+Q+ ANG L L+QT Y D+L + M+D P+ TP+
Sbjct: 1071 LQALKNRFSMKDLGPPRYFLGIQIEDYANG-LFLHQTAYATDILQQAGMSDCNPMPTPLP 1129
Query: 535 FGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKR 594
+L +PT +RS+ G LQY TITRP+I +AVN +CQ + P + +KR
Sbjct: 1130 --QQLDNLNSELFAEPTYFRSLAGKLQYLTITRPDIQFAVNFICQRMHSPTTSDFGLLKR 1187
Query: 595 ILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTA 654
ILRY+KGTI G+ P L L A+ D+D + RRST+G C+ LG NLISW+A
Sbjct: 1188 ILRYIKGTIGMGL---PIKRNSTLTLSAYSDSDHAGCKNTRRSTTGFCILLGSNLISWSA 1244
Query: 655 KKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPT-VLCDNMSTVLLTHNPI 713
K+Q V+ SSTEAEYR+L E+ W+ LL +L I +PT V CDN+S V L+ NP
Sbjct: 1245 KRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGIPQYLPTQVYCDNLSAVYLSANPA 1304
Query: 714 LHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
LH R+KH + D ++RE+V + QH+ + Q AD+FTK+L F+ +R KL V
Sbjct: 1305 LHNRSKHFDTDYHYIREQVALGLIETQHISATFQLADVFTKSLPRRAFVDLRSKLGV 1361
>At4g10690 retrotransposon like protein
Length = 1515
Score = 607 bits (1564), Expect = e-173
Identities = 350/821 (42%), Positives = 483/821 (58%), Gaps = 68/821 (8%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQ-GASPYFKLYGNHPD 61
ER+HR++ E GL+L+ H+ +P W AF T+ +L N + ++TL SPY L+G P
Sbjct: 618 ERRHRYLTELGLSLMFHSKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPV 677
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLDQ-SGRIYVSKDVLF 120
+ +L++FGSAC+P+LRPY NK S CVFLGY++ +KGY+CL +G++Y+ + VLF
Sbjct: 678 YTALRVFGSACYPYLRPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLF 737
Query: 121 HEHRFPYTTL-------------------FPSEPFSPPTSSAEY----FPLSTVPIISRS 157
E +FPY+ + F S S T S FP +TV S S
Sbjct: 738 DERKFPYSDIYSQFQTISGSPLFTAWQKGFSSTALSRETPSTNVEDIIFPSATV---SSS 794
Query: 158 MPQPSPAPISTELANPGPLSPQSEASDLQSQPSPIPTGSGLASTSQPAEHASSE---SAH 214
+P AP E A P + A D+ PSPI + S +QP E S + S
Sbjct: 795 VPTGC-APNIAETAT-APDVDVAAAHDMVVPPSPITSTS---LPTQPEESTSDQNHYSTD 849
Query: 215 QEMATSSGVHAASS-----------------ASTVAVPVNAHPMQTRSKSGIIKPRLNPT 257
E A SS + S +ST A P +HPM TR+KSGI KP NP
Sbjct: 850 SETAISSAMTPQSINVSLFEDSDFPPLQSVISSTTAAPETSHPMITRAKSGITKP--NPK 907
Query: 258 LLL-----THMEPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIY 312
L + EP +VK+A++D+ W AM EE + TW LVP + +GCKW++
Sbjct: 908 YALFSVKSNYPEPKSVKEALKDEGWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVF 967
Query: 313 RVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQI 372
+ K N DGS+++ KARLVA+GY Q +G DY ET+SPVV+ TVR IL +A W L+Q+
Sbjct: 968 KTKLNSDGSLDRLKARLVARGYEQEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQL 1027
Query: 373 DVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCKLHKALYGLKQAPRAWFHRLKEVLLQF 431
DV NAFL+ L+E V+MTQPPGFE + VCKL KA+Y LKQAPRAWF + LL++
Sbjct: 1028 DVKNAFLHDELKETVFMTQPPGFEDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKY 1087
Query: 432 GFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLD 491
GF S DPSLF Y +++L+YVDD+ILTG++ VL+QQL L + F +K +G L
Sbjct: 1088 GFICSFSDPSLFVYLKGRDVMFLLLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALH 1147
Query: 492 YFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPT 551
YFLG+Q H N L L+Q KY +DLL M+D + + TP+Q L + +PT
Sbjct: 1148 YFLGIQ-AHYHNDGLFLSQEKYTSDLLVNAGMSDCSSMPTPLQL--DLLQGNNKPFPEPT 1204
Query: 552 EYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQP 611
+R + G LQY T+TRP+I +AVN VCQ + P + +KRIL YLKGT+T G+ L
Sbjct: 1205 YFRRLAGKLQYLTLTRPDIQFAVNFVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINL-- 1262
Query: 612 CSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRS 671
S L + D+DW D RRST G C FLG N+ISW+AK+ V++SSTEAEYR+
Sbjct: 1263 -SSNTDSVLRCYSDSDWAGCKDTRRSTGGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRT 1321
Query: 672 LANTTAELLWVESLLTELKI-AFTVPTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVRE 730
L+ +E+ W+ LL E+ + +P + CDN+S V L+ NP LH+R+KH ++D ++VRE
Sbjct: 1322 LSFAASEVSWIGFLLQEIGLPQQQIPEMYCDNLSAVYLSANPALHSRSKHFQVDYYYVRE 1381
Query: 731 KVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNVV 771
+V +L V+H+P+ Q ADIFTK+L F +R KL VV
Sbjct: 1382 RVALGALTVKHIPASQQLADIFTKSLPQAPFCDLRFKLGVV 1422
>At1g57640
Length = 1444
Score = 540 bits (1390), Expect = e-153
Identities = 306/805 (38%), Positives = 452/805 (56%), Gaps = 44/805 (5%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKHRHI+ AL + +P+ FW L+A YLINR + LQG SPY LY P
Sbjct: 649 VERKHRHILNIARALRFQSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPK 708
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
+ L++FGS C+ + + +K + S+ CVF+GY KG++ D + + +VS+DV+F
Sbjct: 709 YSHLRVFGSLCYAHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIF 768
Query: 121 HEHRFPYTTL-----------------FPSEPFSPPTSSAEYFPLSTV-------PIISR 156
E FPY+ + F E P T +TV PII
Sbjct: 769 QETEFPYSKMSCNEEDERVLVDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATGPIIPE 828
Query: 157 SMPQPSPAP---ISTELANPGPLSPQSEASDL---QSQPSPIPTGSGLASTSQPAEHASS 210
+ Q S +P +S +P S + +DL + P+PI T +P + +
Sbjct: 829 -INQESSSPSEFVSLSSLDPFLASSTVQTADLPLSSTTPAPIQLRRSSRQTQKPMKLKNF 887
Query: 211 ESAHQEMATSSGVHAASSASTVAVPVNAHPMQTRSKSGIIKPRLNPTLLLTHMEPTTVKQ 270
+ + + S ++SS + V+ H + K+ + + MEPTT +
Sbjct: 888 VTNTVSVESISPEASSSSLYPIEKYVDCHRFTSSHKAFL-------AAVTAGMEPTTYNE 940
Query: 271 AMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLV 330
AM D W +AM E +L N T+S+V LP ++A+G KW+Y++K DG+I +YKARLV
Sbjct: 941 AMVDKAWREAMSAEIESLRVNQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLV 1000
Query: 331 AKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMT 390
G Q +G DY ETF+PV K TVRL L +A +R W + Q+DV+NAFL+G L+EEVYM
Sbjct: 1001 VLGNCQKEGVDYDETFAPVAKMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMK 1060
Query: 391 QPPGFEHKDKTLVCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLG 450
P GF+ D + VC+LHK+LYGLKQAPR WF +L L Q+GF S D SLF+YN+
Sbjct: 1061 LPQGFQCDDPSKVCRLHKSLYGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGI 1120
Query: 451 CIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQ 510
+++L+YVDD+I++G + Q + L S F +K LG L YFLG++V+ A G L+Q
Sbjct: 1121 FVHVLVYVDDLIISGSCPDAVAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQG-FYLSQ 1179
Query: 511 TKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEI 570
KY+ D+++++ + A P + P++ KL+ L D + YR +VG L Y +TRPE+
Sbjct: 1180 RKYVLDIISEMGLLGARPSAFPLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPEL 1239
Query: 571 SYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGS 630
SY+V+ + QF+ +P ++HW A R++RYLK G+LL S T L + +CD+D+ +
Sbjct: 1240 SYSVHTLAQFMQNPRQDHWNAAIRVVRYLKSNPGQGILL---SSTSTLQINGWCDSDYAA 1296
Query: 631 DPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELK 690
P RRS +G V LG ISW KKQ V+RSS EAEYR++A T EL+W++ +L +L
Sbjct: 1297 CPLTRRSLTGYFVQLGDTPISWKTKKQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLG 1356
Query: 691 IAFT-VPTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRA 749
++ + D+ S + L+ NP+ H RTKH+E+D F+R+ + + VPS Q A
Sbjct: 1357 VSHVQAMRIFSDSKSAIALSVNPVQHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLA 1416
Query: 750 DIFTKALSPTRFLLMRDKLNVVDKH 774
DI TKAL KL ++D H
Sbjct: 1417 DILTKALGEKEVRYFLRKLGILDVH 1441
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 527 bits (1357), Expect = e-149
Identities = 305/840 (36%), Positives = 460/840 (54%), Gaps = 76/840 (9%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKHRHI+ A L ++P+ FW + LTAT+LINR + L+G +PY L+G P
Sbjct: 659 VERKHRHILNVARACLFQGNLPVKFWGESILTATHLINRTPSAVLKGKTPYELLFGERPS 718
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
+ L+ FG C+ +RP N +K + S++CVF+GY K ++ D ++G+I+ S+DV F
Sbjct: 719 YDMLRSFGCLCYAHIRPRNKDKFTSRSRKCVFIGYPHGKKAWRVYDLETGKIFASRDVRF 778
Query: 121 HEHRFPYTTL----FPSEPFSPPTSSAEYF-PLSTV-------------PIISRSMPQP- 161
HE +PY T P P +PP + ++F P+ST P S S+ QP
Sbjct: 779 HEDIYPYATATQSNVPLPPPTPPMVNDDWFLPISTQVDSTNVDSSSSSSPAQSGSIDQPP 838
Query: 162 ---------SPAPISTELANPGPLSPQSEASDLQSQPSPIPTGSGLASTSQPAEHAS--- 209
S P+ E+ + P S S + D + + L ST + + +S
Sbjct: 839 RSIDQSPSTSTNPVPEEIGSIVPSSSPSRSIDRSTSDLSASDTTELLSTGESSTPSSPGL 898
Query: 210 ---------------------SESAHQEMATSSGVHAASSASTVAVPVNAHPMQTRSKSG 248
+ + ++ S +H+ S +P + + S SG
Sbjct: 899 PELLGKGCREKKKSVLLKDFVTNTTSKKKTASHNIHSPSQVLPSGLPTS---LSADSVSG 955
Query: 249 IIKPRLNPTL---------------LLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGT 293
L+ L +L EP K A+ +W +AM +E +AL +N T
Sbjct: 956 KTLYPLSDFLTNSGYSANHIAFMAAILDSNEPKHFKDAILIKEWCEAMSKEIDALEANHT 1015
Query: 294 WSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPI 353
W + LP +KA+ KW+Y++K N DG++ ++KARLV G Q +G D+ ETF+PV K
Sbjct: 1016 WDITDLPHGKKAISSKWVYKLKYNSDGTLERHKARLVVMGNHQKEGVDFKETFAPVAKLT 1075
Query: 354 TVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDKTLVCKLHKALYGL 413
TVR IL++A ++ W + Q+DV+NAFL+G LEEEVYM PPGF+ D + VC+L K+LYGL
Sbjct: 1076 TVRTILAVAAAKDWEVHQMDVHNAFLHGDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGL 1135
Query: 414 KQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQ 473
KQAPR WF +L L GF S D SLF+ + I++L+YVDD+I+ G+++ I +
Sbjct: 1136 KQAPRCWFSKLSTALRNIGFTQSYEDYSLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDR 1195
Query: 474 LTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPM 533
++LH F +K LG+L YFLG++V+ +G L+Q KY D++ + + P + P+
Sbjct: 1196 FKSQLHKCFHMKDLGKLKYFLGLEVSRGPDG-FCLSQRKYALDIVKETGLLGCKPSAVPI 1254
Query: 534 QFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVK 593
KL G +P +YR +VG Y TITRP++SYAV+ + QF+ P HW+A
Sbjct: 1255 ALNHKLASITGPVFTNPEQYRRLVGRFIYLTITRPDLSYAVHILSQFMQAPLVAHWEAAL 1314
Query: 594 RILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWT 653
R++RYLKG+ G+ L+ S L + A+CD+D+ + P RRS S V+LG + ISW
Sbjct: 1315 RLVRYLKGSPAQGIFLRSDS---SLIINAYCDSDYNACPLTRRSLSAYVVYLGDSPISWK 1371
Query: 654 AKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNP 712
KKQ V+ SS EAEYR++A T EL W+++LL +L + + P L CD+ + + + NP
Sbjct: 1372 TKKQDTVSYSSAEAEYRAMAYTLKELKWLKALLKDLGVHHSSPMKLHCDSEAAIHIAANP 1431
Query: 713 ILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNVVD 772
+ H RTKH+E D VR+ V K + +H+ +E Q AD+ TK+L F + L V D
Sbjct: 1432 VFHERTKHIESDCHKVRDAVLDKLITTEHIYTEDQVADLLTKSLPRPTFERLLSTLGVTD 1491
>At4g27200 putative protein
Length = 819
Score = 525 bits (1353), Expect = e-149
Identities = 303/729 (41%), Positives = 413/729 (56%), Gaps = 52/729 (7%)
Query: 69 GSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCL-DQSGRIYVSKDVLFHEHRFPY 127
GSACFP LR Y NK + S +CVFLGY+ +KGY+CL +GR+Y+S+ V+F E +P+
Sbjct: 15 GSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPF 74
Query: 128 TTLFPS---EPFSP-----------PTSSAEYFPLSTVPIISRSMPQPSPAPISTELANP 173
+ + +P +P P S P S P+ + + P P + L
Sbjct: 75 SHTYKHLHPQPRTPLLAAWLRSSDSPAPSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTL 134
Query: 174 GPLSPQSEASDLQSQPSP-------IPTGSGLASTSQPAEHASSESAHQEMATSSGVHAA 226
P+S S AS++ +Q SP S S + A S+S S VH
Sbjct: 135 VPISSVSHASNITTQQSPDFDSERTTDFDSASIGDSSHSSQAGSDSEETIQQASVNVHQT 194
Query: 227 SSASTVAVPVNAHPMQTRSKSGIIKPRLNPTLL---LTHMEPTTVKQAMRDDKWLQAMKE 283
+++ N HPM TR+K GI KP L +++ EP TV A++ W AM E
Sbjct: 195 PAST------NVHPMVTRAKVGISKPNPRYVFLSHKVSYPEPKTVTAALKHPGWTGAMTE 248
Query: 284 EYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYS 343
E TWSLVP S+ +G KW++R K + DG++NK KAR+VAKG+ Q +G DY
Sbjct: 249 EIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQEEGIDYL 308
Query: 344 ETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDKTLV 403
ET+SPVV+ TVRL+L LA + W ++Q+DV NAFL+G L+E VYMTQP ++ V
Sbjct: 309 ETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAA----NRDHV 364
Query: 404 CKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIIL 463
C LHK++YGLKQ+PRAWF + LL+FGF K DPSLF Y I +L+
Sbjct: 365 CLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCRKSDPSLFIYAHNNNLILLLL------- 417
Query: 464 TGDSMVLIQQLTAKLHSIFALKQLGQLDY-FLGVQVTHLANGSLLLNQTKYINDLLTKVN 522
+ L A L+ F + +GQ FLG+QV NG L ++Q KY DLL +
Sbjct: 418 ----SQTLTSLLAALNKEFRMTDMGQHSLTFLGIQVQRQQNG-LFMSQQKYAEDLLIAAS 472
Query: 523 MADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLS 582
M P+ TP+ H DPT +RS+ G LQY T+TRP+I +AVN VCQ +
Sbjct: 473 MEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQKMH 532
Query: 583 DPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSC 642
P + +KRILRY+KGTIT G+ S P L A+ D+DWG+ RRS G C
Sbjct: 533 QPTISDFHLLKRILRYIKGTITMGI---SYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLC 589
Query: 643 VFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAF-TVPTVLCD 701
F+G NL+SW++KK V+RSSTEAEY+SL++ +E+LW+ +LL EL+I P + CD
Sbjct: 590 TFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDTPELFCD 649
Query: 702 NMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRF 761
N+S V LT NP H RTKH ++D FVRE+V K+LVV+H+P Q ADIFTK+L F
Sbjct: 650 NLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKSLPYEAF 709
Query: 762 LLMRDKLNV 770
+ +R KL V
Sbjct: 710 IHLRGKLGV 718
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 523 bits (1348), Expect = e-148
Identities = 312/822 (37%), Positives = 444/822 (53%), Gaps = 58/822 (7%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKHRHI+ ALL AS+P+ FW + LTA YLINR ++ L G +PY L+G+ P
Sbjct: 686 VERKHRHILNVARALLFQASLPIKFWGESILTAAYLINRTPSSILSGRTPYEVLHGSKPV 745
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
+ L++FGSAC+ + +K S+ C+F+GY KG+K D + VS+DV+F
Sbjct: 746 YSQLRVFGSACYVHRVTRDKDKFGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVIF 805
Query: 121 HEHRFPYTTLFPS--EPFSPPTSSAE----YFPLST----------------------VP 152
E FPY + S S PT S + PL
Sbjct: 806 REEVFPYAGVNSSTLASTSLPTVSEDDDWAIPPLEVRGSIDSVETERVVCTTDEVVLDTS 865
Query: 153 IISRSMPQPSPAPISTELANPGPLSPQSEASDLQSQPSPIPTGSGLASTSQPAEHASSES 212
+ +P P T ++P +SP S + + + P +P S + S P + S +
Sbjct: 866 VSDSEIPNQEFVPDDTPPSSPLSVSP-SGSPNTPTTPIVVPVASPI-PVSPPKQRKSKRA 923
Query: 213 AH-----------QEMATSSGVHA----ASSASTV----AVPVNAHPMQTRSKSGIIKPR 253
H M T S +HA S +STV P+ + S R
Sbjct: 924 THPPPKLNDYVLYNAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSS---SHR 980
Query: 254 LNPTLLLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYR 313
+ ++EP K+A++ W AM E +AL N TW +V LP + A+G +W+++
Sbjct: 981 AYLAAITDNVEPKHFKEAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFK 1040
Query: 314 VKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQID 373
K N DG++ +YKARLV +G QV+G DY ETF+PVV+ TVR +L + W + Q+D
Sbjct: 1041 TKYNSDGTVERYKARLVVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMD 1100
Query: 374 VNNAFLNGVLEEEVYMTQPPGFEHKDKTLVCKLHKALYGLKQAPRAWFHRLKEVLLQFGF 433
V+NAFL+G LEEEVYM PPGF H VC+L K+LYGLKQAPR WF +L + LL+FGF
Sbjct: 1101 VHNAFLHGDLEEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGF 1160
Query: 434 KASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYF 493
S D SLF+Y + +LIYVDD+++ G+ ++Q+ L F++K LG+L YF
Sbjct: 1161 VQSYEDYSLFSYTRNNIELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYF 1220
Query: 494 LGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEY 553
LG++V+ G + L+Q KY D++ + P TP++ L G L DP Y
Sbjct: 1221 LGIEVSRGPEG-IFLSQRKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPY 1279
Query: 554 RSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCS 613
R +VG L Y TRPE+SY+V+ + QF+ +P E H+ A R++RYLKG+ G+LL +
Sbjct: 1280 RRLVGRLLYLLHTRPELSYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILL---N 1336
Query: 614 MTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLA 673
L L +CD+DW S P RRS S V LG + ISW KKQ V+ SS EAEYR+++
Sbjct: 1337 ADPDLTLEVYCDSDWQSCPLTRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMS 1396
Query: 674 NTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKV 732
E+ W+ LL EL I + P L CD+ + + + NP+ H RTKH+E D VR+ V
Sbjct: 1397 YALKEIKWLRKLLKELGIEQSTPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAV 1456
Query: 733 QAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNVVDKH 774
+ + QHV + Q AD+FTKAL +FL + KL V + H
Sbjct: 1457 RDGIITTQHVRTTEQLADVFTKALGRNQFLYLMSKLGVQNLH 1498
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 520 bits (1339), Expect = e-147
Identities = 298/775 (38%), Positives = 431/775 (55%), Gaps = 53/775 (6%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKH+HI+ AL+ ++M L +W LTA +LINR + L +P+ L G PD
Sbjct: 726 VERKHQHILNVARALMFQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLPD 785
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
+ LK FG C+ +K S+ CVFLGY KGYK LD +S +++S++V F
Sbjct: 786 YSQLKTFGCLCYSSTSSKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEF 845
Query: 121 HEHRFPYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPSPAPISTELANPGPLSPQS 180
HE FP S S T+S + P+ P S I++ L +P +SP +
Sbjct: 846 HEELFPLA----SSQQSATTASDVFTPMD---------PLSSGNSITSHLPSP-QISPST 891
Query: 181 EASDLQSQPSPIPTGSGLASTSQPAEHASSESAHQEMATSSGVHAASSASTVAVPVNAHP 240
+ S + P A + H H SS+ + + +H
Sbjct: 892 QISKRRITKFP----------------AHLQDYHCYFVNKDDSHPISSSLSYSQISPSHM 935
Query: 241 MQTRSKSGIIKPRLNPTLLLTHMEPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLP 300
+ + S I P+ + +A +W A+ +E A+ TW + LP
Sbjct: 936 LYINNISKIPIPQ-------------SYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLP 982
Query: 301 SNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILS 360
+KAVGCKW++ VK + DGS+ ++KAR+VAKGY+Q +G DY+ETFSPV K TV+L+L
Sbjct: 983 PGKKAVGCKWVFTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLK 1042
Query: 361 LAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDKT-----LVCKLHKALYGLKQ 415
++ S+ W L Q+D++NAFLNG LEE +YM P G+ T +VC+L K++YGLKQ
Sbjct: 1043 VSASKKWYLNQLDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQ 1102
Query: 416 APRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLT 475
A R WF + LL GF+ D +LF I +L+YVDDI++ + Q LT
Sbjct: 1103 ASRQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLT 1162
Query: 476 AKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQF 535
L + F L++LG L YFLG++V + G + L+Q KY +LLT +M D P S PM
Sbjct: 1163 EALKASFKLRELGPLKYFLGLEVARTSEG-ISLSQRKYALELLTSADMLDCKPSSIPMTP 1221
Query: 536 GAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRI 595
+L+K+ G L D YR +VG L Y TITRP+I++AVNK+CQF S P H AV ++
Sbjct: 1222 NIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKV 1281
Query: 596 LRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAK 655
L+Y+KGT+ G+ S L L + DADWG+ PD RRST+G +F+G +LISW +K
Sbjct: 1282 LQYIKGTVGQGLFY---SAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSK 1338
Query: 656 KQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVLCDNMSTVLLTHNPILH 715
KQ V+RSS EAEYR+LA + E+ W+ +LL L++ VP + D+ + V + NP+ H
Sbjct: 1339 KQPTVSRSSAEAEYRALALASCEMAWLSTLLLALRVHSGVPILYSDSTAAVYIATNPVFH 1398
Query: 716 TRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNV 770
RTKH+E+D VREK+ L + HV ++ Q ADI TK L P +F + K+++
Sbjct: 1399 ERTKHIEIDCHTVREKLDNGQLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSI 1453
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 512 bits (1318), Expect = e-145
Identities = 290/779 (37%), Positives = 431/779 (55%), Gaps = 53/779 (6%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKH+HI+ AL+ + +PL W LTA +LINR + L +PY L G P
Sbjct: 715 VERKHQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPV 774
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
++ L+ FG C+ P +K S+ C+FLGY S +KGYK +D +S +++S++V F
Sbjct: 775 YEQLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQF 834
Query: 121 HEHRFPYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPSP-APISTELANPGPLSPQ 179
HE E FPL+ P S+ +P P+S+ + + SP
Sbjct: 835 HE---------------------EVFPLAKNPGSESSLKLFTPMVPVSSGIISDTTHSPS 873
Query: 180 SEASDLQSQPSPIPTGSGLASTSQPAEHASSESAHQEMATSSGVHAASSASTVAVPVNAH 239
S S + P I SS+ + A + H + S P+++
Sbjct: 874 SLPSQISDLPPQI----------------SSQRVRKPPAHLNDYHCNTMQSDHKYPISS- 916
Query: 240 PMQTRSKSGIIKPRLNPTLLLTHME-PTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVP 298
T S S I + +T + PT +A +W +A+ E A+ TW +
Sbjct: 917 ---TISYSKISPSHMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITT 973
Query: 299 LPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLI 358
LP +KAVGCKW++ +K DG++ +YKARLVAKGY+Q +G DY++TFSPV K T++L+
Sbjct: 974 LPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLL 1033
Query: 359 LSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHK-----DKTLVCKLHKALYGL 413
L ++ S+ W L+Q+DV+NAFLNG LEEE++M P G+ + +V +L +++YGL
Sbjct: 1034 LKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGL 1093
Query: 414 KQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQ 473
KQA R WF + LL GFK + D +LF + +L+YVDDI++ S Q
Sbjct: 1094 KQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQ 1153
Query: 474 LTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPM 533
LT +L F L+ LG L YFLG++V G + + Q KY +LL M P+S PM
Sbjct: 1154 LTEELDQRFKLRDLGDLKYFLGLEVARTTAG-ISICQRKYALELLQSTGMLACKPVSVPM 1212
Query: 534 QFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVK 593
K+ K G + D +YR +VG L Y TITRP+I++AVNK+CQF S P H A
Sbjct: 1213 IPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAY 1272
Query: 594 RILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWT 653
R+L+Y+KGT+ G+ S + L L F D+DW S D RRST+ +F+G +LISW
Sbjct: 1273 RVLQYIKGTVGQGLFY---SASSDLTLKGFADSDWASCQDSRRSTTSFTMFVGDSLISWR 1329
Query: 654 AKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVLCDNMSTVLLTHNPI 713
+KKQ V+RSS EAEYR+LA T E++W+ +LL L+ + VP + D+ + + + NP+
Sbjct: 1330 SKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASPPVPILYSDSTAAIYIATNPV 1389
Query: 714 LHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNVVD 772
H RTKH+++D VRE++ L + HV +E Q ADI TK L P +F ++ K+++++
Sbjct: 1390 FHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILTKPLFPYQFEHLKSKMSILN 1448
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 504 bits (1299), Expect = e-143
Identities = 299/797 (37%), Positives = 439/797 (54%), Gaps = 47/797 (5%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKHRH++ AL A++P+ FW LTA YLINR ++ L ++PY +L+ P
Sbjct: 549 VERKHRHLLNVARALRFQANLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPR 608
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
F L++FGS C+ R +K + S+ CVF+GY KG++ D + +VS+DV+F
Sbjct: 609 FDHLRVFGSLCYAHNRNRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVF 668
Query: 121 HEHRFPYTTLFPSEPFSPPTSSAEYFPL------STVPIISRSMPQPSPAPISTELANPG 174
E FP+ + + A + P+ V + + P P P +S+ ++ P
Sbjct: 669 SELEFPFR-ISHEQNVIEEEEEALWAPIVDGLIEEEVHLGQNAGPTP-PICVSSPIS-PS 725
Query: 175 PLSPQSE---ASDLQSQPSPIPTGSGLASTSQPAEHASSESAHQEMATSSGVHAASSAST 231
S +SE +S L ++ P P A+++ A SS + Q + S ++A
Sbjct: 726 ATSSRSEHSTSSPLDTEVVPTP-----ATSTTSASSPSSPTNLQFLPLSRA--KPTTAQA 778
Query: 232 VAVPVNAHPMQTRSKSGIIKPRLNPTLLLTHMEPTTVKQAM-------------RDD-KW 277
VA P P + +++ + P L + TTV Q RDD +
Sbjct: 779 VAPPAVPPPRRQSTRN-----KAPPVTLKDFVVNTTVCQESPSKLNSILYQLQKRDDTRR 833
Query: 278 LQAMKEEYNALMS---NGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGY 334
A Y A+ + N TW++ LP ++A+G +W+Y+VK N DGS+ +YKARLVA G
Sbjct: 834 FSASHTTYVAIDAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGN 893
Query: 335 SQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPG 394
Q +G DY ETF+PV K TVRL L +A+ R W + Q+DV+NAFL+G L EEVYM PPG
Sbjct: 894 KQKEGEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPG 953
Query: 395 FEHKDKTLVCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYM 454
FE VC+L KALYGLKQAPR WF +L L ++GF+ S D SLFT I +
Sbjct: 954 FEASHPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKI 1013
Query: 455 LIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYI 514
LIYVDD+I+TG+S QQ L S F +K LG L YFLG++V G + + Q KY
Sbjct: 1014 LIYVDDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTG-IYICQRKYA 1072
Query: 515 NDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAV 574
D++++ + P + P++ KL L DP YR +VG L Y +TR +++++V
Sbjct: 1073 LDIISETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSV 1132
Query: 575 NKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDD 634
+ + +F+ +P E+HW A R++RYLK GV L+ Q + +CD+DW DP
Sbjct: 1133 HILARFMQEPREDHWAAALRVVRYLKADPGQGVFLRRSGDFQ---ITGWCDSDWAGDPMS 1189
Query: 635 RRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFT 694
RRS +G V G + ISW KKQ V++SS EAEYR+++ +ELLW++ LL L ++
Sbjct: 1190 RRSVTGYFVQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHV 1249
Query: 695 VPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFT 753
P ++ CD+ S + + NP+ H RTKH+E+D FVR++ + +HV + Q ADIFT
Sbjct: 1250 QPMIMCCDSKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFT 1309
Query: 754 KALSPTRFLLMRDKLNV 770
K L F R KL +
Sbjct: 1310 KPLGRDCFSAFRIKLGI 1326
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 488 bits (1257), Expect = e-138
Identities = 287/793 (36%), Positives = 421/793 (52%), Gaps = 91/793 (11%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKH+HI+ ALL +++P+ +W TA +LINR+ + L SPY + PD
Sbjct: 734 VERKHQHILNVARALLFQSNIPMQYWSDCVTTAVFLINRLPSPLLNNKSPYELILNKQPD 793
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
+ LK FG CF + K + ++ CVFLGY S +KGYK LD +S + VS++V+F
Sbjct: 794 YSLLKNFGCLCFVSTNAHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSVTVSRNVVF 853
Query: 121 HEHRFPYTT---------LFPSE--PFSPPTSSAEYFPL----STVPIISRSMPQPSPAP 165
EH FP+ T +FP+ P P E PL S +P + S + A
Sbjct: 854 KEHVFPFKTSELLNKAVDMFPNSILPLPAPLHFVETMPLIDEDSLIPTTTDSRTADNHAS 913
Query: 166 ISTELANPGPLSPQSEASDLQSQPSPIPTGSGLASTSQPAE----HAS---SESAHQEMA 218
S+ A P + P S + +P +T P+ H S S S
Sbjct: 914 SSSS-ALPSIIPPSSNTETQDIDSNAVPITRSKRTTRAPSYLSEYHCSLVPSISTLPPTD 972
Query: 219 TSSGVHAASSASTVAVP--VNAHPMQTRSKSGIIKPRLNPTLLL--THMEPTTVKQAMRD 274
+S +H T + P +P+ T P + T EP T QAM+
Sbjct: 973 SSIPIHPLPEIFTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYNTETEPKTFSQAMKS 1032
Query: 275 DKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGY 334
+KW++ EE A+ N TWS+ LP ++ VGCKW++ +K NPDG++ +YKARLVA+G+
Sbjct: 1033 EKWIRVAVEELQAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDGTVERYKARLVAQGF 1092
Query: 335 SQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPG 394
+Q +G D+ +TFSPV K + +++L LA GW L Q+DV++AFL+G L+EE++M+ P G
Sbjct: 1093 TQQEGIDFLDTFSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLHGDLDEEIFMSLPQG 1152
Query: 395 FEHKDKTL-----VCKLHKALYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPL 449
+ T+ VC+L K++YGLKQA R W+ R
Sbjct: 1153 YTPPAGTILPPNPVCRLLKSIYGLKQASRQWYKRF------------------------- 1187
Query: 450 GCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLN 509
+ L+Y+DDI++ ++ ++ L A L S F +K LG +FLG
Sbjct: 1188 --VAALVYIDDIMIASNNDAEVENLKALLRSEFKIKDLGPARFFLG-------------- 1231
Query: 510 QTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPE 569
+ P S PM L + GT L +PT YR ++G L Y TITRP+
Sbjct: 1232 -------------LLGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKLIGRLLYLTITRPD 1278
Query: 570 ISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWG 629
I+YAV+++ QF+S P + H +A ++LRY+K G++ S + L F DADW
Sbjct: 1279 ITYAVHQLSQFISAPSDIHLQAAHKVLRYIKANPGQGLMY---SADYEICLNGFSDADWA 1335
Query: 630 SDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTEL 689
+ D RRS SG C++LG +LISW +KKQ + +RSSTE+EYRS+A T E++W++ LL +L
Sbjct: 1336 ACKDTRRSISGFCIYLGTSLISWKSKKQAVASRSSTESEYRSMAQATCEIIWLQQLLKDL 1395
Query: 690 KIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQR 748
I T P L CDN S + + NP+ H RTKH+E+D VR++++A +L HVP+E+Q
Sbjct: 1396 HIPLTCPAKLFCDNKSALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNLKALHVPTENQH 1455
Query: 749 ADIFTKALSPTRF 761
ADI TKAL P F
Sbjct: 1456 ADILTKALHPGPF 1468
>At1g70010 hypothetical protein
Length = 1315
Score = 486 bits (1250), Expect = e-137
Identities = 282/783 (36%), Positives = 430/783 (54%), Gaps = 48/783 (6%)
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKH+HI+ +L + +P+ +W LTA YLINR+ L+ P+ L P
Sbjct: 564 VERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDKCPFEVLTKTVPT 623
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD-QSGRIYVSKDVLF 120
+ +K+FG C+ P + +K S +K C F+GY S KGYK LD ++ I VS+ V+F
Sbjct: 624 YDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVF 683
Query: 121 HEHRFPYTTLFPSEPFSPPTSSAEYFP-LSTVPIISRSMPQPSPAPISTELANPGPLSPQ 179
HE FP+ S+ +FP L+ P + R + ++P
Sbjct: 684 HEELFPFLGSDLSQ------EEQNFFPDLNPTPPMQRQ--------------SSDHVNPS 723
Query: 180 SEASDLQSQPSPIPTGSGLASTSQPAEHASSESAHQEMATSSGVHAASSASTVAVPVNAH 239
+S ++ PS PT + + Q + + + A+ + V V++
Sbjct: 724 DSSSSVEILPSANPTNNVPEPSVQTSHRKAKKPAYLQDYYCHSV------------VSST 771
Query: 240 PMQTR---SKSGIIKPRLNPTLLLTHM-EPTTVKQAMRDDKWLQAMKEEYNALMSNGTWS 295
P + R S I P L L EP+ +A + W AM E++ L TW
Sbjct: 772 PHEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWE 831
Query: 296 LVPLPSNRKAVGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITV 355
+ LP++++ +GC+WI+++K N DGS+ +YKARLVA+GY+Q +G DY+ETFSPV K +V
Sbjct: 832 VCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSV 891
Query: 356 RLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKD-----KTLVCKLHKAL 410
+L+L +A L Q+D++NAFLNG L+EE+YM P G+ + VC+L K+L
Sbjct: 892 KLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSL 951
Query: 411 YGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVL 470
YGLKQA R W+ + LL GF S CD + F S + +L+Y+DDII+ ++
Sbjct: 952 YGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAA 1011
Query: 471 IQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPIS 530
+ L +++ S F L+ LG+L YFLG+++ G + ++Q KY DLL + P S
Sbjct: 1012 VDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKG-IHISQRKYALDLLDETGQLGCKPSS 1070
Query: 531 TPMQFGAKLTKHGGTSLHDPTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWK 590
PM G + YR ++G L Y ITRP+I++AVNK+ QF P + H +
Sbjct: 1071 IPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQ 1130
Query: 591 AVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLI 650
AV +IL+Y+KGTI G+ S T L L + +AD+ S D RRSTSG C+FLG +LI
Sbjct: 1131 AVYKILQYIKGTIGQGLFY---SATSELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLI 1187
Query: 651 SWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLLT 709
W ++KQ +V++SS EAEYRSL+ T EL+W+ + L EL++ + PT+L CDN + + +
Sbjct: 1188 CWKSRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIA 1247
Query: 710 HNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLN 769
+N + H RTKH+E D VRE++ + H+ +E Q AD FTK L P+ F + K+
Sbjct: 1248 NNHVFHERTKHIESDCHSVRERLLKGLFELYHINTELQIADPFTKPLYPSHFHRLISKMG 1307
Query: 770 VVD 772
+++
Sbjct: 1308 LLN 1310
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 466 bits (1199), Expect = e-131
Identities = 269/692 (38%), Positives = 392/692 (55%), Gaps = 31/692 (4%)
Query: 96 YSSSHKGYKCLD-QSGRIYVSKDVLFHEHRFPYTT---------LFPSE--PFSPPTSSA 143
Y S +KGYK LD +S I ++++V+FHE +FP+ T +FP+ P P
Sbjct: 335 YPSGYKGYKVLDLESHSISITRNVVFHETKFPFKTSKFLKESVDMFPNSILPLPAPLHFV 394
Query: 144 EYFPLSTVPIISRSMPQPSPAPISTELANPGPLSPQSEASD-LQSQPSPIPTGSGLASTS 202
E PL + S + S P P + ++ +D L + +P +
Sbjct: 395 ESMPLDDDLRADDNNASTSNSASSASSIPPLPSTVNTQNTDALDIDTNSVPIARPKRNAK 454
Query: 203 QPAE----HASSESAHQEMA--TSSGVHAASSASTVAVPVNAHPMQTRSKSGIIKPRLNP 256
PA H +S ++ TS+ + SS+ +PM T + P +
Sbjct: 455 APAYLSEYHCNSVPFLSSLSPTTSTSIETPSSSIPPKKITTPYPMSTAISYDKLTPLFHS 514
Query: 257 TLLLTHME--PTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKAVGCKWIYRV 314
+ ++E P QAM+ +KW +A EE +AL N TW + L + VGCKW++ +
Sbjct: 515 YICAYNVETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKWVFTI 574
Query: 315 KENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDV 374
K NPDGSI +YKARLVA+G++Q +G DY ETFSPV K +V+L+L LA + GW L Q+DV
Sbjct: 575 KYNPDGSIERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDV 634
Query: 375 NNAFLNGVLEEEVYMTQPPGFE-----HKDKTLVCKLHKALYGLKQAPRAWFHRLKEVLL 429
+NAFL+G L+EE+YM+ P G+ VC+L K+LYGLKQA R W+ RL V L
Sbjct: 635 SNAFLHGELDEEIYMSLPQGYTPPTGISLPSKPVCRLLKSLYGLKQASRQWYKRLSSVFL 694
Query: 430 QFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQ 489
F S D ++F S I +L+YVDD+++ + ++ L L S F +K LG
Sbjct: 695 GANFIQSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGP 754
Query: 490 LDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLHD 549
+FLG+++ + G + + Q KY +LL V ++ P S PM LTK GT L +
Sbjct: 755 ARFFLGLEIARSSEG-ISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLLPN 813
Query: 550 PTEYRSVVGALQYATITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLL 609
T YR +VG L Y ITRP+I++AV+ + QFLS P + H +A ++LRYLKG G++
Sbjct: 814 ATSYRELVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQGLMY 873
Query: 610 QPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNLISWTAKKQTLVARSSTEAEY 669
S + L L F DADWG+ D RRS +G C++LG +LI+W +KKQ++V+RSSTE+EY
Sbjct: 874 ---SASSELCLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEY 930
Query: 670 RSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFV 728
RSLA T E++W++ LL +L + T P L CDN S + L NP+ H RTKH+E+D V
Sbjct: 931 RSLAQATCEIIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDCHTV 990
Query: 729 REKVQAKSLVVQHVPSEHQRADIFTKALSPTR 760
R++++A L HVP+ +Q ADI TK L P +
Sbjct: 991 RDQIKAGKLKTLHVPTGNQLADILTKPLHPVQ 1022
>At1g60020 hypothetical protein
Length = 1194
Score = 451 bits (1161), Expect = e-127
Identities = 259/616 (42%), Positives = 349/616 (56%), Gaps = 62/616 (10%)
Query: 3 ERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPDF 62
ERKHRHIVETGL LL ASMP +W +AF A YLINRMS+ + G SPY +L+G P++
Sbjct: 583 ERKHRHIVETGLTLLGQASMPKSYWSYAFTIAIYLINRMSSDVIGGISPYKRLFGQAPNY 642
Query: 63 KSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLDQ-SGRIYVSKDVLFH 121
L++FG CFP+LRPY ++KL CVFLGYS + Y CL++ +GR+Y S+ V F
Sbjct: 643 LKLRVFGCLCFPWLRPYTTHKLDDRPAPCVFLGYSQTQSAYLCLNRTTGRVYTSRHVQFV 702
Query: 122 EHRFPYT--TLFP--------SEPFSPPTSSAEYFPLSTVPIISRSMPQPSPAPISTELA 171
E+ +P+T TL P + + S + L +VP +R QP P S
Sbjct: 703 ENTYPFTKPTLDPFTNLEESNNHSITTTVPSPPFVQLPSVPPPTRDPHQPPP---SQPAP 759
Query: 172 NPGPLSPQSEASDLQ--------------------------SQPSPIPTGSGLASTSQPA 205
+P PLSP S +S + S PS P +TS+
Sbjct: 760 SPSPLSPPSMSSPVMTSSPQFSSNRDSTTLHGDYSHVDYGLSSPSNPPGPITSPTTSKSP 819
Query: 206 EHASSESAHQEMATSSGVHAASSASTVAVPV----------------NAHPMQTRSKSGI 249
+S +H + ++ SS+S+ P+ N H M+TR+K+ I
Sbjct: 820 SEPTSSPSHSNQPNKTPPNSPSSSSSSPTPIPSPSPQSSNSPPPPPQNQHSMRTRAKNNI 879
Query: 250 IKPRLNPTLLLT----HMEPTTVKQAMRDDKWLQAMKEEYNALMSNGTWSLVPLPSNRKA 305
KP TL T PTTV +A+RD W AM EE+NA + N T+ LVP ++
Sbjct: 880 TKPIKKLTLAATPKGKSKIPTTVAEALRDPNWRNAMSEEFNAGLRNSTYDLVPPKPHQNF 939
Query: 306 VGCKWIYRVKENPDGSINKYKARLVAKGYSQVQGFDYSETFSPVVKPITVRLILSLAISR 365
VG +WI+ +K NPDGSIN+YKAR +AKG+ Q G DYS TFSPV+K TV+ +L +A+SR
Sbjct: 940 VGTRWIFTIKYNPDGSINRYKARFLAKGFHQQHGLDYSNTFSPVIKSTTVQTVLDIAVSR 999
Query: 366 GWPLQQIDVNNAFLNGVLEEEVYMTQPPGFEHKDK-TLVCKLHKALYGLKQAPRAWFHRL 424
W ++Q+D+NNAFL G L E+VY+ QPPGF + D+ VC L KAL+GLKQAPRAW+ L
Sbjct: 1000 SWDIRQLDINNAFLQGRLTEDVYVAQPPGFINPDRPNYVCHLKKALHGLKQAPRAWYQEL 1059
Query: 425 KEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIFAL 484
+ LL GF S + SLF IY+L+YVDD ++TG + LI Q L + F+L
Sbjct: 1060 RGFLLTCGFTNSVANTSLFIRQHNKDYIYILVYVDDFLITGSNSNLIAQFITCLANRFSL 1119
Query: 485 KQLGQLDYFLGVQVTHLANGSLLLNQTKYINDLLTKVNMADAAPISTPMQFGAKLTKHGG 544
K LGQL YFL ++ T G L L Q +Y+ DLLTK M DA +STPM KLT G
Sbjct: 1120 KDLGQLSYFLEIEATRTKAG-LHLMQRRYVLDLLTKTKMLDAKTVSTPMSPTPKLTLTSG 1178
Query: 545 TSLHDPTEYRSVVGAL 560
T + +P YR ++G+L
Sbjct: 1179 TPIDNPGGYRQILGSL 1194
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,566,450
Number of Sequences: 26719
Number of extensions: 773206
Number of successful extensions: 3734
Number of sequences better than 10.0: 234
Number of HSP's better than 10.0 without gapping: 136
Number of HSP's successfully gapped in prelim test: 104
Number of HSP's that attempted gapping in prelim test: 2570
Number of HSP's gapped (non-prelim): 604
length of query: 778
length of database: 11,318,596
effective HSP length: 107
effective length of query: 671
effective length of database: 8,459,663
effective search space: 5676433873
effective search space used: 5676433873
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0348.7


