

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.6
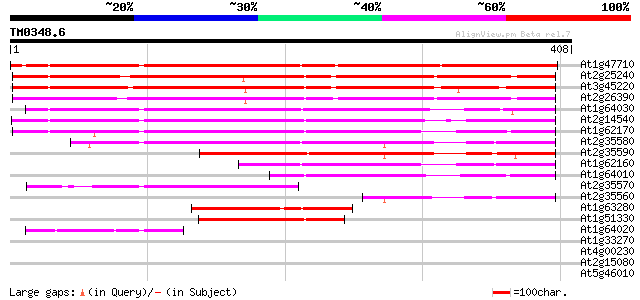
(408 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g47710 serpin, putative 342 3e-94
At2g25240 putative serpin 288 4e-78
At3g45220 serpin-like protein 281 5e-76
At2g26390 putative serpin 279 2e-75
At1g64030 hypothetical protein 272 2e-73
At2g14540 putative serpin 261 6e-70
At1g62170 hypothetical protein 259 2e-69
At2g35580 putative serpin 234 8e-62
At2g35590 putative serpin 199 3e-51
At1g62160 hypothetical protein 178 4e-45
At1g64010 hypothetical protein 164 6e-41
At2g35570 putative serpin 126 2e-29
At2g35560 putative serpin 86 4e-17
At1g63280 hypothetical protein 85 8e-17
At1g51330 hypothetical protein 80 3e-15
At1g64020 hypothetical protein 58 1e-08
At1g33270 unknown protein 29 4.1
At4g00230 subtilisin-type serine endopeptidase XSP1 29 5.3
At2g15080 putative disease resistance protein 29 5.3
At5g46010 unknown protein 28 7.0
>At1g47710 serpin, putative
Length = 391
Score = 342 bits (876), Expect = 3e-94
Identities = 186/399 (46%), Positives = 261/399 (64%), Gaps = 10/399 (2%)
Query: 1 MEIQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDEL 60
M++++S S Q+ V+++ KH+ + Q N+IFSP S+ LS++AAGS G T D++
Sbjct: 1 MDVRESISL--QNQVSMNLAKHVITTVS-QNSNVIFSPASINVVLSIIAAGSAGATKDQI 57
Query: 61 LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLL 120
LSFL+F S D LN++ S+ +S V +D P LS NG +ID ++S F++LL
Sbjct: 58 LSFLKFSSTDQLNSFSSEIVSAVLADGSANGGPK---LSVANGAWIDKSLSFKPSFKQLL 114
Query: 121 STHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQ 180
Y A+ DF + V+ +VNS E+E+NGLIT++LP G+ ++TKLIFANAL F+
Sbjct: 115 EDSYKAASNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFK 174
Query: 181 GMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D LT F+LL G V P+MT+ KK QYV A+DGFK+L LPY QG+D+ R
Sbjct: 175 GTWNEKFDESLTQEGEFHLLDGNKVTAPFMTS-KKKQYVSAYDGFKVLGLPYLQGQDK-R 232
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+FSM +LPDA +GLS L+ K+ S PGFL +P R+V+VR F++PKF SF F+ASNVL
Sbjct: 233 QFSMYFYLPDANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVL 292
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQ 359
K +G+ SPFS + T+MV NL V +IFHKA IEVNE+GT+A AA+ + +
Sbjct: 293 KGLGLTSPFSGEEG-LTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLR 351
Query: 360 CARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNPL 398
IDF+ADHPFL ++ E+ TG +LF+GQV++PL
Sbjct: 352 GLLMEEDEIDFVADHPFLLVVTENITGVVLFIGQVVDPL 390
>At2g25240 putative serpin
Length = 385
Score = 288 bits (737), Expect = 4e-78
Identities = 167/398 (41%), Positives = 245/398 (60%), Gaps = 19/398 (4%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLS 62
++ KS +DV + TKH+ + NL+FSP+S+ LS++AAGS T +++LS
Sbjct: 1 MELGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILS 59
Query: 63 FLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLST 122
FL S DHLN +Q I D + S LS NG++ID SL F+ LL
Sbjct: 60 FLMLPSTDHLNLVLAQII-------DGGTEKSDLRLSIANGVWIDKFFSLKLSFKDLLEN 112
Query: 123 HYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNL--TKLIFANALRFQ 180
Y A+ + +DF + V+ +VN+ E +NGLI Q+L ++ + + L+ ANA+ F+
Sbjct: 113 SYKATCSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFK 172
Query: 181 GMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D +T + F+LL GTSVKVP+MT ++ QY+R++DGFK+LRLPY + QR
Sbjct: 173 GAWSSKFDANMTKKNDFHLLDGTSVKVPFMTNYED-QYLRSYDGFKVLRLPYIED---QR 228
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+FSM I+LP+ ++GL+ L++K+ SEP F +P + V FR+PKF SF F AS VL
Sbjct: 229 QFSMYIYLPNDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVL 288
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQ 359
K++G+ SPF+ T+MV+ S D+L+V SI HKA IEV+E+GT+A A + ++
Sbjct: 289 KDMGLTSPFN-NGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVVSCT 347
Query: 360 CARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
R + DF+AD PFLF +RED +G ILF+GQVL+P
Sbjct: 348 SFRRNP---DFVADRPFLFTVREDKSGVILFMGQVLDP 382
>At3g45220 serpin-like protein
Length = 393
Score = 281 bits (719), Expect = 5e-76
Identities = 163/406 (40%), Positives = 249/406 (61%), Gaps = 27/406 (6%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLS 62
++ KS Q+DV + KH+ NL+FSP+S+ L ++AAGS T +++LS
Sbjct: 1 MELGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILS 59
Query: 63 FLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLST 122
F+ S D+LN ++ +S +D S HLS G++ID ++S F+ LL
Sbjct: 60 FIMLPSSDYLNAVLAKTVSVALND---GMERSDLHLSTAYGVWIDKSLSFKPSFKDLLEN 116
Query: 123 HYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTK--LIFANALRFQ 180
YNA+ +DF + V+++VN+ E +NGLI ++L ++ + + LI ANA+ F+
Sbjct: 117 SYNATCNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFK 176
Query: 181 GMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQR 239
G W D LT F+LL GT VKVP+MT +KK QY+ +DGFK+LRLPY + QR
Sbjct: 177 GAWSKKFDAKLTKSYDFHLLDGTMVKVPFMTNYKK-QYLEYYDGFKVLRLPYVED---QR 232
Query: 240 RFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVL 299
+F+M I+LP+ +DGL L++++SS+P FL +P +++ F++PKF SF F+AS+VL
Sbjct: 233 QFAMYIYLPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVL 292
Query: 300 KEVGVVSPFSPMDAHFTKMVNVNSPS--------DNLFVQSIFHKAFIEVNEKGTKATAA 351
KE+G+ PF+ H + V SPS +NLFV ++FHKA IEV+E+GT+A A
Sbjct: 293 KEMGLTLPFT----HGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAV 348
Query: 352 TWSALARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
+ +++ +D DF+ADHPFLF +RE+ +G ILF+GQVL+P
Sbjct: 349 SVASM----TKDMLLMGDFVADHPFLFTVREEKSGVILFMGQVLDP 390
>At2g26390 putative serpin
Length = 389
Score = 279 bits (714), Expect = 2e-75
Identities = 163/401 (40%), Positives = 240/401 (59%), Gaps = 21/401 (5%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLS 62
++ KS Q++V K + + N++FSP+S+ LS++AAGS T +E+LS
Sbjct: 1 MELGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILS 60
Query: 63 FLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLST 122
FL S DHLN ++ D + S LS +G++ID + L F+ LL
Sbjct: 61 FLMSPSTDHLNAVLAK-------IADGGTERSDLCLSTAHGVWIDKSSYLKPSFKELLEN 113
Query: 123 HYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTK-----LIFANAL 177
Y AS + +DF + V+ +VN + +NGLI Q+L + + LI ANA+
Sbjct: 114 SYKASCSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAV 173
Query: 178 RFQGMWKHTLDG-LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRD 236
F+ W D LT + F+LL G +VKVP+M ++K QY+R +DGF++LRLPY + +
Sbjct: 174 YFKAAWSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKD-QYLRGYDGFQVLRLPYVEDK- 231
Query: 237 RQRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEAS 296
R FSM I+LP+ +DGL+AL++K+S+EPGFL +P + V R+PK N SF F+AS
Sbjct: 232 --RHFSMYIYLPNDKDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKAS 289
Query: 297 NVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSAL 356
VLK++G+ SPF+ + T+MV+ S D L V SI HKA IEV+E+GT+A A + + +
Sbjct: 290 EVLKDMGLTSPFT-SKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIM 348
Query: 357 ARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
QC + DF+ADHPFLF +RED +G ILF+GQVL+P
Sbjct: 349 MPQCLMRNP---DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>At1g64030 hypothetical protein
Length = 385
Score = 272 bits (696), Expect = 2e-73
Identities = 157/392 (40%), Positives = 233/392 (59%), Gaps = 36/392 (9%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLD-ELLSFLRFDSID 70
Q+ VA+ + H+ S ++ N+IFSP S+ +A+++ AAG G + ++LSFLR SID
Sbjct: 10 QTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQILSFLRSSSID 68
Query: 71 HLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLAS 130
L T F + S V++D P ++ NG++ID ++ F+ L + A
Sbjct: 69 ELKTVFRELASVVYADRSATGGPK---ITAANGLWIDKSLPTDPKFKDLFENFFKAVYVP 125
Query: 131 LDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLDG- 189
+DF + V +VNS +E +N LI LLP G+VT+LT I+ANAL F+G WK +
Sbjct: 126 VDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWKRPFEKY 185
Query: 190 LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGR-DRQRRFSMCIFLP 248
T + F L++GTSV VP+M+++ + QYVRA+DGFK+LRLPY++G D R+FSM +LP
Sbjct: 186 YTRDNDFYLVNGTSVSVPFMSSY-ENQYVRAYDGFKVLRLPYQRGSDDTNRKFSMYFYLP 244
Query: 249 DAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPF 308
D +DGL L++K++S PGFL +P + + FR+PKF I F F ++VL +G+ S
Sbjct: 245 DKKDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRLGLRS-- 302
Query: 309 SPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDH---H 365
S++HKA +E++E+G +A AAT C+ D
Sbjct: 303 ----------------------MSMYHKACVEIDEEGAEAAAATADGDC-GCSLDFVEPP 339
Query: 366 PSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
IDF+ADHPFLFLIRE+ TGT+LFVGQ+ +P
Sbjct: 340 KKIDFVADHPFLFLIREEKTGTVLFVGQIFDP 371
>At2g14540 putative serpin
Length = 407
Score = 261 bits (666), Expect = 6e-70
Identities = 153/400 (38%), Positives = 233/400 (58%), Gaps = 34/400 (8%)
Query: 2 EIQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDE-L 60
+I ++ + Q++V+L + S + N +FSP S+ A L+V AA ++ +TL +
Sbjct: 28 KIDMQEAMKNQNEVSLLLVGKVISAVA-KNSNCVFSPASINAVLTVTAANTDNKTLRSFI 86
Query: 61 LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLL 120
LSFL+ S + N F + S VF D P ++ NG++++ ++S + + L
Sbjct: 87 LSFLKSSSTEETNAIFHELASVVFKDGSETGGPK---IAAVNGVWMEQSLSCNPDWEDLF 143
Query: 121 STHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQ 180
+ AS A +DF + + V DVN+ + +N LI ++LP G+VT+LT I+ NAL F+
Sbjct: 144 LNFFKASFAKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIYGNALYFK 203
Query: 181 GMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRD-RQ 238
G W+ D +T F+LL+G SV VP+M +++K Q++ A+DGFK+LRLPY+QGRD
Sbjct: 204 GAWEKAFDKSMTRDKPFHLLNGKSVSVPFMRSYEK-QFIEAYDGFKVLRLPYRQGRDDTN 262
Query: 239 RRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNV 298
R FSM ++LPD + L L+++++S PGFL +P +V V FR+PKF I F FEAS+V
Sbjct: 263 REFSMYLYLPDKKGELDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSV 322
Query: 299 LKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALAR 358
+ +NV S+ KA IE++E+GT+A AAT +
Sbjct: 323 FNDF---------------ELNV----------SLHQKALIEIDEEGTEAAAATTVVVVT 357
Query: 359 -QCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
C + IDF+ADHPFLFLIRED TGT+LF GQ+ +P
Sbjct: 358 GSCLWEPKKKIDFVADHPFLFLIREDKTGTLLFAGQIFDP 397
>At1g62170 hypothetical protein
Length = 433
Score = 259 bits (662), Expect = 2e-69
Identities = 158/402 (39%), Positives = 231/402 (57%), Gaps = 39/402 (9%)
Query: 3 IQKSKSTRCQSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDEL-- 60
I ++ + Q+DVA+ T + S + N +FSP S+ AAL+++AA S G +EL
Sbjct: 62 IDVGEAMKKQNDVAIFLTGIVISSVA-KNSNFVFSPASINAALTMVAASSGGEQGEELRS 120
Query: 61 --LSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRR 118
LSFL+ S D LN F + S V D P ++ NGM++D ++S++ +
Sbjct: 121 FILSFLKSSSTDELNAIFREIASVVLVDGSKKGGPK---IAVVNGMWMDQSLSVNPLSKD 177
Query: 119 LLSTHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALR 178
L ++A+ A +DF + + V +VN+ +NGLI LLP G+VT+LT ++ +AL
Sbjct: 178 LFKNFFSAAFAQVDFRSKAEEVRTEVNAWASSHTNGLIKDLLPRGSVTSLTDRVYGSALY 237
Query: 179 FQGMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDR 237
F+G W+ +T F LL+GTSV VP+M++F+K QY+ A+DGFK+LRLPY+QGRD
Sbjct: 238 FKGTWEEKYSKSMTKCKPFYLLNGTSVSVPFMSSFEK-QYIAAYDGFKVLRLPYRQGRDN 296
Query: 238 -QRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEAS 296
R F+M I+LPD + L L+++++S PGFL P R+V+V FR+PKF I F FEAS
Sbjct: 297 TNRNFAMYIYLPDKKGELDDLLERMTSTPGFLDSHNPERRVKVGKFRIPKFKIEFGFEAS 356
Query: 297 NVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKA-TAATWSA 355
+ SD S + K IE++EKGT+A T + +
Sbjct: 357 SAF-------------------------SDFELDVSFYQKTLIEIDEKGTEAVTFTAFRS 391
Query: 356 LARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
CA IDF+ADHPFLFLIRE+ TGT+LF GQ+ +P
Sbjct: 392 AYLGCAL--VKPIDFVADHPFLFLIREEQTGTVLFAGQIFDP 431
>At2g35580 putative serpin
Length = 374
Score = 234 bits (596), Expect = 8e-62
Identities = 139/359 (38%), Positives = 207/359 (56%), Gaps = 33/359 (9%)
Query: 45 LSVMAAGSEGRT--LDELLSFLRFDSIDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTN 102
LS++AA S G T D+++S L+ S D L+ S+ ++ V +D + P+ +S N
Sbjct: 40 LSIIAASSPGDTDTADKIVSLLQASSTDKLHAVSSEIVTTVLADSTASGGPT---ISAAN 96
Query: 103 GMFIDTTVSLSYPFRRLLSTHYNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPP 162
G++I+ T+++ F+ LL Y A+ +DF + D V +VNS +E+++NGLIT LLP
Sbjct: 97 GLWIEKTLNVEPSFKDLLLNSYKAAFNRVDFRTKADEVNREVNSWVEKQTNGLITNLLPS 156
Query: 163 GTVTN-LTKLIFANALRFQGMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRA 220
+ LT IFANAL F G W D LT S F+LL GT V+VP+MT +Y
Sbjct: 157 NPKSAPLTDHIFANALFFNGRWDSQFDPSLTKDSDFHLLDGTKVRVPFMTG-ASCRYTHV 215
Query: 221 FDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGK--LPHRKVR 278
++GFK++ L Y++GR+ R FSM I+LPD +DGL +++++L+S GFLK LP
Sbjct: 216 YEGFKVINLQYRRGREDSRSFSMQIYLPDEKDGLPSMLERLASTRGFLKDNEVLPSHSAV 275
Query: 279 VRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAF 338
++ ++P+F F FEAS LK G+V P S I HK+
Sbjct: 276 IKELKIPRFKFDFAFEASEALKGFGLVVPLS----------------------MIMHKSC 313
Query: 339 IEVNEKGTKATAATWSALARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
IEV+E G+KA AA + C R DF+ADHPFLF+++E +G +LF+GQV++P
Sbjct: 314 IEVDEVGSKAAAAA-AFRGIGCRRPPPEKHDFVADHPFLFIVKEYRSGLVLFLGQVMDP 371
>At2g35590 putative serpin
Length = 331
Score = 199 bits (505), Expect = 3e-51
Identities = 112/264 (42%), Positives = 161/264 (60%), Gaps = 31/264 (11%)
Query: 139 SVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD-GLTHVSSFN 197
S+ +VNS +E+++NGLIT LLPP + + LT LIFANAL F G W + LT S F+
Sbjct: 91 SLNKEVNSWVEEQTNGLITDLLPPNSASPLTDLIFANALFFNGRWDSQFNPSLTKESDFH 150
Query: 198 LLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSAL 257
LL GT V+VP+MT + + ++ FK+L LPY++GR+ R FSM I+LPD +DGL ++
Sbjct: 151 LLDGTKVRVPFMTGAHEDS-LDVYEAFKVLNLPYREGREDSRGFSMQIYLPDEKDGLPSM 209
Query: 258 IQKLSSEPGFLKGK--LPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAHF 315
++ L+S GFLK LP +K V+ ++P+F +F FEAS LK +G+ P S
Sbjct: 210 LESLASTRGFLKDNKVLPSQKAGVKELKIPRFKFAFDFEASKALKGLGLKVPLS------ 263
Query: 316 TKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHP--SIDFIAD 373
+I HK+ IEV+E G+KA AA A R C + P DF+AD
Sbjct: 264 ----------------TIIHKSCIEVDEVGSKAAAA---AALRSCGGCYFPPKKYDFVAD 304
Query: 374 HPFLFLIREDFTGTILFVGQVLNP 397
HPFLF+++E +G +LF+G V++P
Sbjct: 305 HPFLFIVKEYISGLVLFLGHVMDP 328
>At1g62160 hypothetical protein
Length = 265
Score = 178 bits (452), Expect = 4e-45
Identities = 101/233 (43%), Positives = 139/233 (59%), Gaps = 29/233 (12%)
Query: 167 NLTKLIFANALRFQGMWKHTLD-GLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFK 225
NLTK I+ NAL F+G W+ +T F L++G SV VP+M++ K QY+ A+DGFK
Sbjct: 58 NLTKRIYGNALYFKGTWEKKFSKSMTKRKPFYLVNGISVSVPFMSS-SKDQYIEAYDGFK 116
Query: 226 ILRLPYKQGRDR-QRRFSMCIFLPDAQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRV 284
+LRLPY+QGRD R FSM +LPD + L L+++++S PGFL P +V V FR+
Sbjct: 117 VLRLPYRQGRDNTNRNFSMYFYLPDKKGELDDLLKRMTSTPGFLDSHTPRERVEVDEFRI 176
Query: 285 PKFNISFTFEASNVLKEVGVVSPFSPMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEK 344
PKF I F FEAS+V SD S + KA IE++E+
Sbjct: 177 PKFKIEFGFEASSVF-------------------------SDFEIDVSFYQKALIEIDEE 211
Query: 345 GTKATAATWSALARQCARDHHPSIDFIADHPFLFLIREDFTGTILFVGQVLNP 397
GT+A AAT + + + ++DF+ADHPFLFLIRE+ TGT+LF GQ+ +P
Sbjct: 212 GTEAAAAT-AFVDNEDGCGFVETLDFVADHPFLFLIREEQTGTVLFAGQIFDP 263
>At1g64010 hypothetical protein
Length = 185
Score = 164 bits (416), Expect = 6e-41
Identities = 88/208 (42%), Positives = 125/208 (59%), Gaps = 27/208 (12%)
Query: 190 LTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPD 249
+T F+L++GTSV V M+++K QY+ A+DGFK+L+LP++QG D R FSM +LPD
Sbjct: 1 MTKDRDFHLINGTSVSVSLMSSYKD-QYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYLPD 59
Query: 250 AQDGLSALIQKLSSEPGFLKGKLPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFS 309
+DGL L++K++S GFL +P +KV+V F +PKF I F F AS +G
Sbjct: 60 EKDGLDNLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLG------ 113
Query: 310 PMDAHFTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSID 369
L +++ KA +E++E+G +A AAT CA ID
Sbjct: 114 ------------------LDEMALYQKACVEIDEEGAEAIAATAVVGGFGCA--FVKRID 153
Query: 370 FIADHPFLFLIREDFTGTILFVGQVLNP 397
F+ADHPFLF+IRED TGT+LFVGQ+ +P
Sbjct: 154 FVADHPFLFMIREDKTGTVLFVGQIFDP 181
>At2g35570 putative serpin
Length = 213
Score = 126 bits (316), Expect = 2e-29
Identities = 75/199 (37%), Positives = 112/199 (55%), Gaps = 20/199 (10%)
Query: 13 SDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTLDELLSFLRFDSIDHL 72
+D+ L T+H+ + + NL+FSP A LSV +LSF+ S D L
Sbjct: 25 NDIILRLTQHVIASTAGKTSNLVFSP----ALLSV------------ILSFVAASSPDEL 68
Query: 73 NTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNASLASLD 132
N S ++ + +D P+ P+ +S NG++I+ T+ + F+ LL Y A+ +D
Sbjct: 69 NAVSSGIVTTILADSTPSGGPT---ISAANGVWIEKTLYVEPSFKDLLLNSYKAAFNQVD 125
Query: 133 FNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD-GLT 191
F + D V +VNS +E+++NGLIT LLPP + ++ T LIFANAL F G W + LT
Sbjct: 126 FRTKADEVNKEVNSWVEEQTNGLITDLLPPNSASSWTDLIFANALFFNGRWDSQFNPSLT 185
Query: 192 HVSSFNLLSGTSVKVPYMT 210
S F+LL GT V+VP+MT
Sbjct: 186 KESDFHLLDGTKVRVPFMT 204
>At2g35560 putative serpin
Length = 121
Score = 85.9 bits (211), Expect = 4e-17
Identities = 52/143 (36%), Positives = 76/143 (52%), Gaps = 27/143 (18%)
Query: 257 LIQKLSSEPGFLKGK--LPHRKVRVRPFRVPKFNISFTFEASNVLKEVGVVSPFSPMDAH 314
++++L+S GFL GK +P V ++P+F F FEAS LK +G+ P
Sbjct: 1 MLERLASTRGFLNGKEDIPSHWADVGELKIPRFKFDFGFEASEALKGLGLEVP------- 53
Query: 315 FTKMVNVNSPSDNLFVQSIFHKAFIEVNEKGTKATAATWSALARQCARDHHPSIDFIADH 374
+I HK+ IEV+E G+KA AA + + C DF+ADH
Sbjct: 54 ----------------STIIHKSCIEVDEVGSKAAAA--AVIFVGCCGPAEKRYDFVADH 95
Query: 375 PFLFLIREDFTGTILFVGQVLNP 397
PFLFL++E +G ILF+GQV++P
Sbjct: 96 PFLFLVKEYRSGLILFLGQVMDP 118
>At1g63280 hypothetical protein
Length = 120
Score = 84.7 bits (208), Expect = 8e-17
Identities = 48/119 (40%), Positives = 73/119 (61%), Gaps = 5/119 (4%)
Query: 133 FNLRGDSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTLD-GLT 191
F L+ V ++N +NGLI LLP G+V + T ++ NAL F+G W++ D T
Sbjct: 5 FVLKAVQVRQELNKWASDHTNGLIIDLLPRGSVKSETVQVYGNALYFKGAWENKFDKSST 64
Query: 192 HVSSFNLLSGTSVKVPYMTTFKKTQYVRAFDGFKILRLPYKQG-RDRQRRFSMCIFLPD 249
+ F+ G V VP+M ++ ++QY+ A DGFK+L LPY+QG D +R+FS+ +LPD
Sbjct: 65 KDNEFH--QGKEVHVPFMRSY-ESQYIMACDGFKVLGLPYQQGLDDTKRKFSIYFYLPD 120
>At1g51330 hypothetical protein
Length = 193
Score = 79.7 bits (195), Expect = 3e-15
Identities = 45/109 (41%), Positives = 73/109 (66%), Gaps = 4/109 (3%)
Query: 138 DSVLHDVNSLIEQESNGLITQLLPPGTVTNLTKLIFANALRFQGMWKHTL-DGLTHVSSF 196
+ V +VNS + +NGLI LLPPG+VTN T I+ NAL F+G W++ +T F
Sbjct: 33 EEVRMEVNSWALRHTNGLIKNLLPPGSVTNQTIKIYGNALYFKGAWENKFGKSMTIHKPF 92
Query: 197 NLLSGTSVKVPYMTTFKKTQYVRAFDGFKILR-LPYK-QGRDRQRRFSM 243
+L++G V VP+M ++++ +Y++A++GFK+LR L Y+ +D R+FS+
Sbjct: 93 HLVNGKQVLVPFMKSYER-KYMKAYNGFKVLRILQYRVDYKDTSRQFSI 140
>At1g64020 hypothetical protein
Length = 121
Score = 57.8 bits (138), Expect = 1e-08
Identities = 39/116 (33%), Positives = 62/116 (52%), Gaps = 6/116 (5%)
Query: 12 QSDVALSFTKHLFSKEDYQEKNLIFSPLSLYAALSVMAAGSEGRTL-DELLSFLRFDSID 70
Q++VA+ + HLFS N +FSP S+ A ++MA+G + D++LSFL SID
Sbjct: 10 QNEVAMILSWHLFSTVAKHSNN-VFSPASITAVFTMMASGPGSSLISDQILSFLGSSSID 68
Query: 71 HLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTHYNA 126
LN+ F + I+ VF+D P+ + NG +ID + S+ + L + A
Sbjct: 69 ELNSVF-RVITTVFADGSNIGGPT---IKVANGAWIDQSFSIDSSSKNLFENFFKA 120
>At1g33270 unknown protein
Length = 369
Score = 29.3 bits (64), Expect = 4.1
Identities = 31/132 (23%), Positives = 57/132 (42%), Gaps = 13/132 (9%)
Query: 65 RFDS-IDHLNTYFSQGISPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLSYPFRRLLSTH 123
+FDS D ++ F+ P + PA+ F N + +D ++L P T
Sbjct: 235 QFDSKSDLIDAVFTSSFIPGYLAPRPATM-------FRNRLCVDGGLTLFMPPTAAAKTV 287
Query: 124 YNASLASLDFNLRGDSVLHDVNSLIEQESNGLITQLLPPG---TVTNLTKLIFANALRFQ 180
+ ++ +F L+G + D N L S L+ L P + L +L +A+A +
Sbjct: 288 RVCAFSASNFKLKGIEICPDCNPLNRATSRQLLNWALEPAEDEVLERLFELGYADAATWA 347
Query: 181 GMWKHTLDGLTH 192
M + ++GL +
Sbjct: 348 EM--NPVEGLVY 357
>At4g00230 subtilisin-type serine endopeptidase XSP1
Length = 749
Score = 28.9 bits (63), Expect = 5.3
Identities = 15/35 (42%), Positives = 21/35 (59%), Gaps = 2/35 (5%)
Query: 187 LDGLTHVSSFNLLSGTSVKVPYMTTFKKTQYVRAF 221
LDG T S F +LSGTS+ P++ YV++F
Sbjct: 520 LDGDTQFSKFTILSGTSMACPHVAGV--AAYVKSF 552
>At2g15080 putative disease resistance protein
Length = 983
Score = 28.9 bits (63), Expect = 5.3
Identities = 21/62 (33%), Positives = 33/62 (52%), Gaps = 10/62 (16%)
Query: 162 PGTVTNLTKLIFANALR--FQGMWKHTLDGLTHVSSFNL-LSGTSVKVP-------YMTT 211
P ++ NL+ LIF + F G +L L+H++SFNL + S +VP Y+TT
Sbjct: 153 PSSIGNLSHLIFVDFSHNNFSGQIPSSLGYLSHLTSFNLSYNNFSGRVPSSIGNLSYLTT 212
Query: 212 FK 213
+
Sbjct: 213 LR 214
>At5g46010 unknown protein
Length = 122
Score = 28.5 bits (62), Expect = 7.0
Identities = 14/33 (42%), Positives = 17/33 (51%)
Query: 81 SPVFSDEDPASSPSQHHLSFTNGMFIDTTVSLS 113
SPV SD P P+QH +FI TV+ S
Sbjct: 45 SPVVSDPKPEWKPNQHQAQILEELFIGGTVNPS 77
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,832,541
Number of Sequences: 26719
Number of extensions: 366563
Number of successful extensions: 870
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 780
Number of HSP's gapped (non-prelim): 20
length of query: 408
length of database: 11,318,596
effective HSP length: 102
effective length of query: 306
effective length of database: 8,593,258
effective search space: 2629536948
effective search space used: 2629536948
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0348.6


