

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.13
(381 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
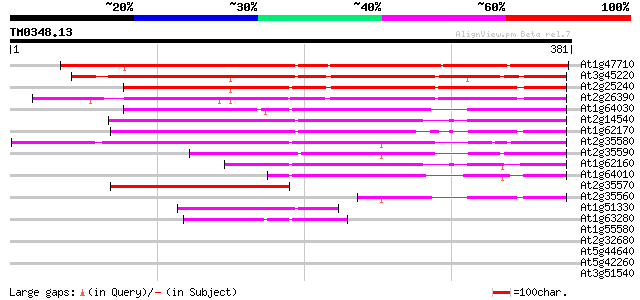
Score E
Sequences producing significant alignments: (bits) Value
At1g47710 serpin, putative 276 2e-74
At3g45220 serpin-like protein 237 9e-63
At2g25240 putative serpin 237 9e-63
At2g26390 putative serpin 234 6e-62
At1g64030 hypothetical protein 216 2e-56
At2g14540 putative serpin 215 3e-56
At1g62170 hypothetical protein 206 2e-53
At2g35580 putative serpin 202 2e-52
At2g35590 putative serpin 182 2e-46
At1g62160 hypothetical protein 171 5e-43
At1g64010 hypothetical protein 148 6e-36
At2g35570 putative serpin 95 6e-20
At2g35560 putative serpin 85 8e-17
At1g51330 hypothetical protein 78 9e-15
At1g63280 hypothetical protein 77 1e-14
At1g55580 unknown protein 31 0.99
At2g32680 putative disease resistance protein 31 1.3
At5g44640 beta-glucosidase 28 6.4
At5g42260 beta-glucosidase 28 6.4
At3g51540 unknown protein 28 6.4
>At1g47710 serpin, putative
Length = 391
Score = 276 bits (705), Expect = 2e-74
Identities = 159/349 (45%), Positives = 219/349 (62%), Gaps = 8/349 (2%)
Query: 35 AAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAADADAH---HLSFTNGMFVDKSVS 91
AA S + L+ S+ L F + AD A+ LS NG ++DKS+S
Sbjct: 46 AAGSAGATKDQILSFLKFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLS 105
Query: 92 LSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQLLPPGTITNLTRL 151
F+ LL Y A+ DF+++ V+ +VNS E ++NG IT++LP G+ ++T+L
Sbjct: 106 FKPSFKQLLEDSYKAASNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKL 165
Query: 152 IFANALRFQGLWKHKFDGP-RYEFPFDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILRLP 210
IFANAL F+G W KFD E F LL+G V PFMT KKK Y+ A+D FK+L LP
Sbjct: 166 IFANALYFKGTWNEKFDESLTQEGEFHLLDGNKVTAPFMTSKKK-QYVSAYDGFKVLGLP 224
Query: 211 YKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKVRVDALQIPKFNIS 270
Y QG+D KR+FSM +LPDA++GLS L+ K+ S PGFL +PRR+V+V +IPKF S
Sbjct: 225 YLQGQD-KRQFSMYFYLPDANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFS 283
Query: 271 FTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIFHKAFIEVNEKGTKATA 330
F F+ASNVLK +G+ SPFS + T+M+E NLCV +IFHKA IEVNE+GT+A A
Sbjct: 284 FGFDASNVLKGLGLTSPFSGEEG-LTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAA 342
Query: 331 ASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVLNPL 379
AS G + + I+F+A+HPFL ++ E+ TG +LF+GQV++PL
Sbjct: 343 ASAGVIKL-RGLLMEEDEIDFVADHPFLLVVTENITGVVLFIGQVVDPL 390
>At3g45220 serpin-like protein
Length = 393
Score = 237 bits (604), Expect = 9e-63
Identities = 142/345 (41%), Positives = 212/345 (61%), Gaps = 27/345 (7%)
Query: 43 PSSDLTPSTISTPSSLRGFPLCSPTKDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAY 102
PSSD + ++ S+ D + HLS G+++DKS+S F+ LL
Sbjct: 64 PSSDYLNAVLAKTVSVA-------LNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLEN 116
Query: 103 QYNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQLLPPGTITNL--TRLIFANALRFQ 160
YNA+ +DF + V+++VN+ E+ +NG I ++L +I + + LI ANA+ F+
Sbjct: 117 SYNATCNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFK 176
Query: 161 GLWKHKFDGPRYE-FPFDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKR 219
G W KFD + + F LL+GT VKVPFMT KK Y+ +D FK+LRLPY + + R
Sbjct: 177 GAWSKKFDAKLTKSYDFHLLDGTMVKVPFMTNYKK-QYLEYYDGFKVLRLPYVEDQ---R 232
Query: 220 RFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKVRVDALQIPKFNISFTFEASNVL 279
+F+M I+LP+ DGL L+ ++SS+P FL +PR+++ +A +IPKF SF F+AS+VL
Sbjct: 233 QFAMYIYLPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVL 292
Query: 280 KEVGVVSPFSPMDACFTKMLEINSPSNNLC------VESIFHKAFIEVNEKGTKATAASV 333
KE+G+ PF+ T+M+E S NLC V ++FHKA IEV+E+GT+A A SV
Sbjct: 293 KEMGLTLPFT--HGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSV 350
Query: 334 GFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVLNP 378
A +T+ G +F+A+HPFLF +RE+ +G ILF+GQVL+P
Sbjct: 351 --ASMTKDMLLMG---DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>At2g25240 putative serpin
Length = 385
Score = 237 bits (604), Expect = 9e-63
Identities = 134/304 (44%), Positives = 193/304 (63%), Gaps = 12/304 (3%)
Query: 78 LSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHIT 137
LS NG+++DK SL F+ LL Y A+ + +DF ++ V+ +VN+ E+ +NG I
Sbjct: 88 LSIANGVWIDKFFSLKLSFKDLLENSYKATCSQVDFASKPSEVIDEVNTWAEVHTNGLIK 147
Query: 138 QLLPPGTITNL--TRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVPFMTIKKK 194
Q+L +I + + L+ ANA+ F+G W KFD + F LL+GTSVKVPFMT +
Sbjct: 148 QILSRDSIDTIRSSTLVLANAVYFKGAWSSKFDANMTKKNDFHLLDGTSVKVPFMT-NYE 206
Query: 195 THYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPR 254
Y+R++D FK+LRLPY + + R+FSM I+LP+ +GL+ L+ K+ SEP F +P
Sbjct: 207 DQYLRSYDGFKVLRLPYIEDQ---RQFSMYIYLPNDKEGLAPLLEKIGSEPSFFDNHIPL 263
Query: 255 RKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIF 314
+ V A +IPKF SF F AS VLK++G+ SPF+ T+M++ S ++L V SI
Sbjct: 264 HCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPFN-NGGGLTEMVDSPSNGDDLYVSSIL 322
Query: 315 HKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQ 374
HKA IEV+E+GT+A A SVG T R P +F+A+ PFLF +RED +G ILF+GQ
Sbjct: 323 HKACIEVDEEGTEAAAVSVGVVSCTSFRRNP----DFVADRPFLFTVREDKSGVILFMGQ 378
Query: 375 VLNP 378
VL+P
Sbjct: 379 VLDP 382
>At2g26390 putative serpin
Length = 389
Score = 234 bits (597), Expect = 6e-62
Identities = 150/371 (40%), Positives = 213/371 (56%), Gaps = 30/371 (8%)
Query: 16 SIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTIS--TPSSLRGFPLCSPTKDAADA 73
SI+ LS + S P ++ S SPS+D + ++ LC
Sbjct: 38 SINVLLSLIAAGSNPVTKEEILSFLMSPSTDHLNAVLAKIADGGTERSDLC--------- 88
Query: 74 DAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSN 133
LS +G+++DKS L F+ LL Y AS + +DF + V+ +VN ++ +N
Sbjct: 89 ----LSTAHGVWIDKSSYLKPSFKELLENSYKASCSQVDFATKPVEVIDEVNIWADVHTN 144
Query: 134 GHITQLLP---PGTITNL--TRLIFANALRFQGLWKHKFDGP-RYEFPFDLLNGTSVKVP 187
G I Q+L TI + + LI ANA+ F+ W KFD + F LL+G +VKVP
Sbjct: 145 GLIKQILSRDCTDTIKEIRNSTLILANAVYFKAAWSRKFDAKLTKDNDFHLLDGNTVKVP 204
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKLSSEPGF 247
FM + K Y+R +D F++LRLPY + KR FSM I+LP+ DGL+AL+ K+S+EPGF
Sbjct: 205 FM-MSYKDQYLRGYDGFQVLRLPYVED---KRHFSMYIYLPNDKDGLAALLEKISTEPGF 260
Query: 248 LKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNN 307
L +P + VDAL+IPK N SF F+AS VLK++G+ SPF+ T+M++ S +
Sbjct: 261 LDSHIPLHRTPVDALRIPKLNFSFEFKASEVLKDMGLTSPFT-SKGNLTEMVDSPSNGDK 319
Query: 308 LCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDFTG 367
L V SI HKA IEV+E+GT+A A SV R P +F+A+HPFLF +RED +G
Sbjct: 320 LHVSSIIHKACIEVDEEGTEAAAVSVAIMMPQCLMRNP----DFVADHPFLFTVREDNSG 375
Query: 368 TILFVGQVLNP 378
ILF+GQVL+P
Sbjct: 376 VILFIGQVLDP 386
>At1g64030 hypothetical protein
Length = 385
Score = 216 bits (549), Expect = 2e-56
Identities = 121/306 (39%), Positives = 182/306 (58%), Gaps = 32/306 (10%)
Query: 78 LSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHIT 137
++ NG+++DKS+ F+ L + A +DF++ + V +VNS +E +N I
Sbjct: 93 ITAANGLWIDKSLPTDPKFKDLFENFFKAVYVPVDFRSEAEEVRKEVNSWVEHHTNNLIK 152
Query: 138 QLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRY---EFPFDLLNGTSVKVPFMTIKKK 194
LLP G++T+LT I+ANAL F+G WK F+ +Y + F L+NGTSV VPFM+ +
Sbjct: 153 DLLPDGSVTSLTNKIYANALSFKGAWKRPFE--KYYTRDNDFYLVNGTSVSVPFMS-SYE 209
Query: 195 THYIRAFDAFKILRLPYKQGR-DRKRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLP 253
Y+RA+D FK+LRLPY++G D R+FSM +LPD DGL L+ K++S PGFL +P
Sbjct: 210 NQYVRAYDGFKVLRLPYQRGSDDTNRKFSMYFYLPDKKDGLDDLLEKMASTPGFLDSHIP 269
Query: 254 RRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESI 313
+ ++ +IPKF I F F ++VL +G+ S S+
Sbjct: 270 TYRDELEKFRIPKFKIEFGFSVTSVLDRLGLRS------------------------MSM 305
Query: 314 FHKAFIEVNEKGTKATAASV-GFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFV 372
+HKA +E++E+G +A AA+ G + P I+F+A+HPFLFLIRE+ TGT+LFV
Sbjct: 306 YHKACVEIDEEGAEAAAATADGDCGCSLDFVEPPKKIDFVADHPFLFLIREEKTGTVLFV 365
Query: 373 GQVLNP 378
GQ+ +P
Sbjct: 366 GQIFDP 371
>At2g14540 putative serpin
Length = 407
Score = 215 bits (548), Expect = 3e-56
Identities = 123/313 (39%), Positives = 181/313 (57%), Gaps = 28/313 (8%)
Query: 68 KDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSL 127
KD ++ ++ NG+++++S+S + + L + AS A +DF+++ + V DVN+
Sbjct: 111 KDGSETGGPKIAAVNGVWMEQSLSCNPDWEDLFLNFFKASFAKVDFRHKAEEVRLDVNTW 170
Query: 128 IELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFD-GPRYEFPFDLLNGTSVKV 186
+N I ++LP G++T+LT I+ NAL F+G W+ FD + PF LLNG SV V
Sbjct: 171 ASRHTNDLIKEILPRGSVTSLTNWIYGNALYFKGAWEKAFDKSMTRDKPFHLLNGKSVSV 230
Query: 187 PFMTIKKKTHYIRAFDAFKILRLPYKQGR-DRKRRFSMCIFLPDASDGLSALVHKLSSEP 245
PFM +K +I A+D FK+LRLPY+QGR D R FSM ++LPD L L+ +++S P
Sbjct: 231 PFMRSYEK-QFIEAYDGFKVLRLPYRQGRDDTNREFSMYLYLPDKKGELDNLLERITSNP 289
Query: 246 GFLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPS 305
GFL +P +V V +IPKF I F FEAS+V + E+N
Sbjct: 290 GFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSVFND-----------------FELN--- 329
Query: 306 NNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDF 365
S+ KA IE++E+GT+A AA+ P I+F+A+HPFLFLIRED
Sbjct: 330 -----VSLHQKALIEIDEEGTEAAAATTVVVVTGSCLWEPKKKIDFVADHPFLFLIREDK 384
Query: 366 TGTILFVGQVLNP 378
TGT+LF GQ+ +P
Sbjct: 385 TGTLLFAGQIFDP 397
>At1g62170 hypothetical protein
Length = 433
Score = 206 bits (523), Expect = 2e-53
Identities = 124/313 (39%), Positives = 187/313 (59%), Gaps = 32/313 (10%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
D + ++ NGM++D+S+S++ + L ++A+ A +DF+++ + V +VN+
Sbjct: 148 DGSKKGGPKIAVVNGMWMDQSLSVNPLSKDLFKNFFSAAFAQVDFRSKAEEVRTEVNAWA 207
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVP 187
+NG I LLP G++T+LT ++ +AL F+G W+ K+ + PF LLNGTSV VP
Sbjct: 208 SSHTNGLIKDLLPRGSVTSLTDRVYGSALYFKGTWEEKYSKSMTKCKPFYLLNGTSVSVP 267
Query: 188 FMTIKKKTHYIRAFDAFKILRLPYKQGRDR-KRRFSMCIFLPDASDGLSALVHKLSSEPG 246
FM+ +K YI A+D FK+LRLPY+QGRD R F+M I+LPD L L+ +++S PG
Sbjct: 268 FMSSFEK-QYIAAYDGFKVLRLPYRQGRDNTNRNFAMYIYLPDKKGELDDLLERMTSTPG 326
Query: 247 FLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSN 306
FL P R+V+V +IPKF I F FEAS S FS + L++
Sbjct: 327 FLDSHNPERRVKVGKFRIPKFKIEFGFEAS---------SAFSDFE------LDV----- 366
Query: 307 NLCVESIFHKAFIEVNEKGTKA-TAASVGFAPVTQSARRPGPVINFIANHPFLFLIREDF 365
S + K IE++EKGT+A T + A + + +P I+F+A+HPFLFLIRE+
Sbjct: 367 -----SFYQKTLIEIDEKGTEAVTFTAFRSAYLGCALVKP---IDFVADHPFLFLIREEQ 418
Query: 366 TGTILFVGQVLNP 378
TGT+LF GQ+ +P
Sbjct: 419 TGTVLFAGQIFDP 431
>At2g35580 putative serpin
Length = 374
Score = 202 bits (515), Expect = 2e-52
Identities = 133/382 (34%), Positives = 205/382 (52%), Gaps = 35/382 (9%)
Query: 2 NIYSQKKIIKRRTSSIHPCLSTLLLASWPPVQKAAHSTSFSPSSDLTPSTISTPSSLRGF 61
++ +Q I+ R T+ + + +++ AS P A + T + S +
Sbjct: 20 SVGNQNDIVLRLTAPLINVILSIIAASSPGDTDTADKIVSLLQASSTDKLHAVSSEI--- 76
Query: 62 PLCSPTKDAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVL 121
+ + D+ + +S NG++++K++++ F+ LL Y A+ +DF+ + D V
Sbjct: 77 -VTTVLADSTASGGPTISAANGLWIEKTLNVEPSFKDLLLNSYKAAFNRVDFRTKADEVN 135
Query: 122 HDVNSLIELQSNGHITQLLPPGTITN-LTRLIFANALRFQGLWKHKFDGP-RYEFPFDLL 179
+VNS +E Q+NG IT LLP + LT IFANAL F G W +FD + F LL
Sbjct: 136 REVNSWVEKQTNGLITNLLPSNPKSAPLTDHIFANALFFNGRWDSQFDPSLTKDSDFHLL 195
Query: 180 NGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVH 239
+GT V+VPFMT Y ++ FK++ L Y++GR+ R FSM I+LPD DGL +++
Sbjct: 196 DGTKVRVPFMT-GASCRYTHVYEGFKVINLQYRRGREDSRSFSMQIYLPDEKDGLPSMLE 254
Query: 240 KLSSEPGFLKGK--LPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTK 297
+L+S GFLK LP + L+IP+F F FEAS LK G+V P S
Sbjct: 255 RLASTRGFLKDNEVLPSHSAVIKELKIPRFKFDFAFEASEALKGFGLVVPLS-------- 306
Query: 298 MLEINSPSNNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGP-VINFIANHP 356
I HK+ IEV+E G+KA AA+ F + RRP P +F+A+HP
Sbjct: 307 --------------MIMHKSCIEVDEVGSKA-AAAAAFRGI--GCRRPPPEKHDFVADHP 349
Query: 357 FLFLIREDFTGTILFVGQVLNP 378
FLF+++E +G +LF+GQV++P
Sbjct: 350 FLFIVKEYRSGLVLFLGQVMDP 371
>At2g35590 putative serpin
Length = 331
Score = 182 bits (463), Expect = 2e-46
Identities = 109/259 (42%), Positives = 153/259 (58%), Gaps = 28/259 (10%)
Query: 123 DVNSLIELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGP-RYEFPFDLLNG 181
+VNS +E Q+NG IT LLPP + + LT LIFANAL F G W +F+ E F LL+G
Sbjct: 95 EVNSWVEEQTNGLITDLLPPNSASPLTDLIFANALFFNGRWDSQFNPSLTKESDFHLLDG 154
Query: 182 TSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLSALVHKL 241
T V+VPFMT + + ++AFK+L LPY++GR+ R FSM I+LPD DGL +++ L
Sbjct: 155 TKVRVPFMTGAHEDS-LDVYEAFKVLNLPYREGREDSRGFSMQIYLPDEKDGLPSMLESL 213
Query: 242 SSEPGFLKGK--LPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACFTKML 299
+S GFLK LP +K V L+IP+F +F FEAS LK +G+ P S
Sbjct: 214 ASTRGFLKDNKVLPSQKAGVKELKIPRFKFAFDFEASKALKGLGLKVPLS---------- 263
Query: 300 EINSPSNNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIANHPFLF 359
+I HK+ IEV+E G+KA AA+ P +F+A+HPFLF
Sbjct: 264 ------------TIIHKSCIEVDEVGSKAAAAAA--LRSCGGCYFPPKKYDFVADHPFLF 309
Query: 360 LIREDFTGTILFVGQVLNP 378
+++E +G +LF+G V++P
Sbjct: 310 IVKEYISGLVLFLGHVMDP 328
>At1g62160 hypothetical protein
Length = 265
Score = 171 bits (434), Expect = 5e-43
Identities = 103/242 (42%), Positives = 137/242 (56%), Gaps = 46/242 (19%)
Query: 147 NLTRLIFANALRFQGLWKHKFDGPRYEF-PFDLLNGTSVKVPFMTIKKKTHYIRAFDAFK 205
NLT+ I+ NAL F+G W+ KF + PF L+NG SV VPFM+ K YI A+D FK
Sbjct: 58 NLTKRIYGNALYFKGTWEKKFSKSMTKRKPFYLVNGISVSVPFMS-SSKDQYIEAYDGFK 116
Query: 206 ILRLPYKQGRDR-KRRFSMCIFLPDASDGLSALVHKLSSEPGFLKGKLPRRKVRVDALQI 264
+LRLPY+QGRD R FSM +LPD L L+ +++S PGFL PR +V VD +I
Sbjct: 117 VLRLPYRQGRDNTNRNFSMYFYLPDKKGELDDLLKRMTSTPGFLDSHTPRERVEVDEFRI 176
Query: 265 PKFNISFTFEASNVLKEVGVVSPFSPMDACFTKMLEINSPSNNLCVESIFHKAFIEVNEK 324
PKF I F FEAS+V + EI+ S + KA IE++E+
Sbjct: 177 PKFKIEFGFEASSVFSD-----------------FEID--------VSFYQKALIEIDEE 211
Query: 325 GTKATAASV--------GFAPVTQSARRPGPVINFIANHPFLFLIREDFTGTILFVGQVL 376
GT+A AA+ GF ++F+A+HPFLFLIRE+ TGT+LF GQ+
Sbjct: 212 GTEAAAATAFVDNEDGCGFV----------ETLDFVADHPFLFLIREEQTGTVLFAGQIF 261
Query: 377 NP 378
+P
Sbjct: 262 DP 263
>At1g64010 hypothetical protein
Length = 185
Score = 148 bits (373), Expect = 6e-36
Identities = 86/208 (41%), Positives = 117/208 (55%), Gaps = 38/208 (18%)
Query: 176 FDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQGRDRKRRFSMCIFLPDASDGLS 235
F L+NGTSV V M+ K YI A+D FK+L+LP++QG D R FSM +LPD DGL
Sbjct: 7 FHLINGTSVSVSLMS-SYKDQYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYLPDEKDGLD 65
Query: 236 ALVHKLSSEPGFLKGKLPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDACF 295
LV K++S GFL +P +KV+V IPKF I F F AS +G
Sbjct: 66 NLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLG------------ 113
Query: 296 TKMLEINSPSNNLCVESIFHKAFIEVNEKGTKATAASV-----GFAPVTQSARRPGPVIN 350
L +++ KA +E++E+G +A AA+ G A V + I+
Sbjct: 114 ------------LDEMALYQKACVEIDEEGAEAIAATAVVGGFGCAFVKR--------ID 153
Query: 351 FIANHPFLFLIREDFTGTILFVGQVLNP 378
F+A+HPFLF+IRED TGT+LFVGQ+ +P
Sbjct: 154 FVADHPFLFMIREDKTGTVLFVGQIFDP 181
>At2g35570 putative serpin
Length = 213
Score = 95.1 bits (235), Expect = 6e-20
Identities = 51/123 (41%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Query: 69 DAADADAHHLSFTNGMFVDKSVSLSYPFRLLLAYQYNASLASLDFKNRGDRVLHDVNSLI 128
D+ + +S NG++++K++ + F+ LL Y A+ +DF+ + D V +VNS +
Sbjct: 82 DSTPSGGPTISAANGVWIEKTLYVEPSFKDLLLNSYKAAFNQVDFRTKADEVNKEVNSWV 141
Query: 129 ELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFD-GPRYEFPFDLLNGTSVKVP 187
E Q+NG IT LLPP + ++ T LIFANAL F G W +F+ E F LL+GT V+VP
Sbjct: 142 EEQTNGLITDLLPPNSASSWTDLIFANALFFNGRWDSQFNPSLTKESDFHLLDGTKVRVP 201
Query: 188 FMT 190
FMT
Sbjct: 202 FMT 204
>At2g35560 putative serpin
Length = 121
Score = 84.7 bits (208), Expect = 8e-17
Identities = 54/144 (37%), Positives = 78/144 (53%), Gaps = 28/144 (19%)
Query: 237 LVHKLSSEPGFLKGK--LPRRKVRVDALQIPKFNISFTFEASNVLKEVGVVSPFSPMDAC 294
++ +L+S GFL GK +P V L+IP+F F FEAS LK +G+ P
Sbjct: 1 MLERLASTRGFLNGKEDIPSHWADVGELKIPRFKFDFGFEASEALKGLGLEVP------- 53
Query: 295 FTKMLEINSPSNNLCVESIFHKAFIEVNEKGTKATAASVGFAPVTQSARRPGPVINFIAN 354
+I HK+ IEV+E G+KA AA+V F A + +F+A+
Sbjct: 54 ----------------STIIHKSCIEVDEVGSKAAAAAVIFVGCCGPAEKR---YDFVAD 94
Query: 355 HPFLFLIREDFTGTILFVGQVLNP 378
HPFLFL++E +G ILF+GQV++P
Sbjct: 95 HPFLFLVKEYRSGLILFLGQVMDP 118
>At1g51330 hypothetical protein
Length = 193
Score = 77.8 bits (190), Expect = 9e-15
Identities = 47/112 (41%), Positives = 68/112 (59%), Gaps = 4/112 (3%)
Query: 115 NRGDRVLHDVNSLIELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKF-DGPRYE 173
N + V +VNS +NG I LLPPG++TN T I+ NAL F+G W++KF
Sbjct: 30 NGAEEVRMEVNSWALRHTNGLIKNLLPPGSVTNQTIKIYGNALYFKGAWENKFGKSMTIH 89
Query: 174 FPFDLLNGTSVKVPFMTIKKKTHYIRAFDAFKILR-LPYK-QGRDRKRRFSM 223
PF L+NG V VPFM ++ Y++A++ FK+LR L Y+ +D R+FS+
Sbjct: 90 KPFHLVNGKQVLVPFMKSYER-KYMKAYNGFKVLRILQYRVDYKDTSRQFSI 140
>At1g63280 hypothetical protein
Length = 120
Score = 77.4 bits (189), Expect = 1e-14
Identities = 45/112 (40%), Positives = 66/112 (58%), Gaps = 3/112 (2%)
Query: 119 RVLHDVNSLIELQSNGHITQLLPPGTITNLTRLIFANALRFQGLWKHKFDGPRYEFPFDL 178
+V ++N +NG I LLP G++ + T ++ NAL F+G W++KFD + +
Sbjct: 11 QVRQELNKWASDHTNGLIIDLLPRGSVKSETVQVYGNALYFKGAWENKFDKSSTK-DNEF 69
Query: 179 LNGTSVKVPFMTIKKKTHYIRAFDAFKILRLPYKQG-RDRKRRFSMCIFLPD 229
G V VPFM ++ YI A D FK+L LPY+QG D KR+FS+ +LPD
Sbjct: 70 HQGKEVHVPFMR-SYESQYIMACDGFKVLGLPYQQGLDDTKRKFSIYFYLPD 120
>At1g55580 unknown protein
Length = 445
Score = 31.2 bits (69), Expect = 0.99
Identities = 30/101 (29%), Positives = 43/101 (41%), Gaps = 19/101 (18%)
Query: 39 TSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAADADAHHLS---FTNGMFVDKSVSLSYP 95
+S S S D T +T P PLC + AA + +HHL FT FV +S
Sbjct: 7 SSSSSSEDATATTTENPP-----PLCIASSSAATSASHHLRRLLFTAANFVSQS------ 55
Query: 96 FRLLLAYQYNASLASLDFKNRGD---RVLHDVNSLIELQSN 133
A Q S+ SL+ GD R++H + ++ N
Sbjct: 56 --NFTAAQNLLSILSLNSSPHGDSTERLVHLFTKALSVRIN 94
>At2g32680 putative disease resistance protein
Length = 890
Score = 30.8 bits (68), Expect = 1.3
Identities = 35/114 (30%), Positives = 51/114 (44%), Gaps = 9/114 (7%)
Query: 39 TSFSPSSDLTPSTISTPSSLRGFPLCSPTKDAADADAHHLSFTNGMFVDKSVSLSYPFRL 98
+SFS + L +S FPL + D + F+ + + S+ + R
Sbjct: 141 SSFSNLTMLAQLDLSYNKLTGSFPLVRGLRKLIVLDLSYNHFSGTLNPNSSLFELHQLRY 200
Query: 99 L-LAYQYNASLASLDFKNRGDRVLHDVNSLIELQSNGHITQLLPPGTITNLTRL 151
L LA+ +S F N LH + +LI L SNG Q+ P TI+NLTRL
Sbjct: 201 LNLAFNNFSSSLPSKFGN-----LHRLENLI-LSSNGFSGQV--PSTISNLTRL 246
>At5g44640 beta-glucosidase
Length = 507
Score = 28.5 bits (62), Expect = 6.4
Identities = 14/43 (32%), Positives = 23/43 (52%), Gaps = 2/43 (4%)
Query: 109 ASLDFKNRGDRVLH--DVNSLIELQSNGHITQLLPPGTITNLT 149
A + FKN GDRV H +N + + G++ ++ PG + T
Sbjct: 181 ADICFKNFGDRVKHWMTLNEPLTVVQQGYVAGVMAPGRCSKFT 223
>At5g42260 beta-glucosidase
Length = 507
Score = 28.5 bits (62), Expect = 6.4
Identities = 14/43 (32%), Positives = 23/43 (52%), Gaps = 2/43 (4%)
Query: 109 ASLDFKNRGDRVLH--DVNSLIELQSNGHITQLLPPGTITNLT 149
A + FKN GDRV H +N + + G++ ++ PG + T
Sbjct: 181 ADICFKNFGDRVKHWMTLNEPLTVVQQGYVAGVMAPGRCSKFT 223
>At3g51540 unknown protein
Length = 438
Score = 28.5 bits (62), Expect = 6.4
Identities = 23/64 (35%), Positives = 32/64 (49%), Gaps = 3/64 (4%)
Query: 13 RTSSIHPCLSTLLLASWPPVQ--KAAHSTSFSPSSDLTPSTISTPSSLRGFPLCSPT-KD 69
R+SS+ +T + + V K S S +PSS TPS STPS + +P+ K
Sbjct: 129 RSSSVPKKTTTTQIQASASVSSPKRTVSRSLTPSSRKTPSPTSTPSRISTTTSTTPSFKT 188
Query: 70 AADA 73
A DA
Sbjct: 189 AGDA 192
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,525,732
Number of Sequences: 26719
Number of extensions: 360478
Number of successful extensions: 931
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 851
Number of HSP's gapped (non-prelim): 24
length of query: 381
length of database: 11,318,596
effective HSP length: 101
effective length of query: 280
effective length of database: 8,619,977
effective search space: 2413593560
effective search space used: 2413593560
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0348.13


