

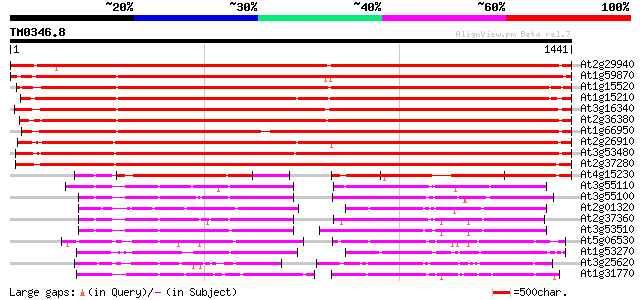
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.8
(1441 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g29940 putative ABC transporter 2111 0.0
At1g59870 putative ABC transporter (At1g59870) 1568 0.0
At1g15520 hypothetical protein 1543 0.0
At1g15210 putative ABC transporter 1536 0.0
At3g16340 putative ABC transporter 1525 0.0
At2g36380 putative ABC transporter 1457 0.0
At1g66950 ABC transporter, putative 1437 0.0
At2g26910 putative ABC transporter 1433 0.0
At3g53480 ABC transporter - like protein 1352 0.0
At2g37280 putative ABC transporter 1343 0.0
At4g15230 ABC transporter like protein 514 e-145
At3g55110 ABC transporter - like protein 233 8e-61
At3g55100 ABC transporter - like protein 229 1e-59
At2g01320 putative ABC transporter 218 2e-56
At2g37360 ABC transporter like protein 217 4e-56
At3g53510 ABC transporter -like protein 211 3e-54
At5g06530 ABC transporter like protein 207 5e-53
At1g53270 putative ABC transporter emb|AAD22683.1 204 3e-52
At3g25620 membrane transporter, putative 202 1e-51
At1g31770 unknown protein 195 2e-49
>At2g29940 putative ABC transporter
Length = 1443
Score = 2111 bits (5469), Expect = 0.0
Identities = 1031/1459 (70%), Positives = 1225/1459 (83%), Gaps = 37/1459 (2%)
Query: 2 AASDGSEYFEIG-SFRSESFARASNADTVEEDEEELQWAALSRLPSQKRINY-ALYRASS 59
AAS+GSEYFE ESFAR SNA+TVE+DEE+L+WAA+ RLPSQ++ + A+ R S
Sbjct: 3 AASNGSEYFEFDVETGRESFARPSNAETVEQDEEDLRWAAIGRLPSQRQGTHNAILRRSQ 62
Query: 60 SRRQPPPLGSPAGAGGRADNLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDR 119
++ Q + A G +DV+KL R+ RE++V++ALAT+DQDN++LLSAIKERLDR
Sbjct: 63 TQTQ-----TSGYADGNVVQTIDVKKLDRADREMLVRQALATSDQDNFKLLSAIKERLDR 117
Query: 120 -----------------IGIEVPNIEVRFSNLSVSADVQIGSRALPTLINYTRDALETIC 162
+G+EVP IEVRF NL++ ADVQ G+RALPTL+N +RD E
Sbjct: 118 FVTTLRILSVSNFREKKVGMEVPKIEVRFENLNIEADVQAGTRALPTLVNVSRDFFERCL 177
Query: 163 TNLGIYRPKRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSI 222
++L I +P++H L IL ++SG+IKPGRMTLLLG PG+GKS+LLLALAGKLD +LKKTG+I
Sbjct: 178 SSLRIIKPRKHKLNILKDISGIIKPGRMTLLLGPPGSGKSTLLLALAGKLDKSLKKTGNI 237
Query: 223 TYNGHEADEFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLEN 282
TYNG ++F VKRT AYISQTDNH AELTVRETLDFAARCQGA EGFA Y KD+ RLE
Sbjct: 238 TYNGENLNKFHVKRTSAYISQTDNHIAELTVRETLDFAARCQGASEGFAGYMKDLTRLEK 297
Query: 283 ERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKR 342
ER IRPS EIDAFMKA+SV G+KHSV+TDY+LKVLGLD+CS+T+VG+DM+RGVSGGQRKR
Sbjct: 298 ERGIRPSSEIDAFMKAASVKGEKHSVSTDYVLKVLGLDVCSDTMVGNDMMRGVSGGQRKR 357
Query: 343 VTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFEL 402
VTTGEM VGPRKTLFMDEISTGLDSSTTFQIVKCI+NFVHLMDATVLMALLQPAPETF+L
Sbjct: 358 VTTGEMTVGPRKTLFMDEISTGLDSSTTFQIVKCIRNFVHLMDATVLMALLQPAPETFDL 417
Query: 403 FDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSK 462
FDDL+LLSEG+++Y+GPRE+V+ FFES+GF+LPPRKG+ADFLQEV+S+KDQAQYWADPSK
Sbjct: 418 FDDLILLSEGYMVYQGPREDVIAFFESLGFRLPPRKGVADFLQEVTSKKDQAQYWADPSK 477
Query: 463 QYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAR 522
YQF+P +IA AFRNS++G +S P+DK PSAL RTK+A+S WE K CF R
Sbjct: 478 PYQFIPVSDIAAAFRNSKYGHAADSKLAAPFDKKSADPSALCRTKFAISGWENLKVCFVR 537
Query: 523 EALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFN 582
E LLI R +FLY F+TCQV FVG VT T+FL+TR+HPT E +GN Y+S LFFGLVHMMFN
Sbjct: 538 ELLLIKRHKFLYTFRTCQVGFVGLVTATVFLKTRLHPTSEQFGNEYLSCLFFGLVHMMFN 597
Query: 583 GFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAP 642
GFSEL LMI+RLPVFYKQRDN F+PAW+WS+ +W+LRVPYS++EAV+W+ +VY+TVG AP
Sbjct: 598 GFSELPLMISRLPVFYKQRDNSFHPAWSWSIASWLLRVPYSVLEAVVWSGVVYFTVGLAP 657
Query: 643 SAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMI 702
SAGRFFRYM +LF +HQMA+GLFRMMAS+ARDMV+ANTFGSAA+L++FLLGGF+IPK I
Sbjct: 658 SAGRFFRYMLLLFSVHQMALGLFRMMASLARDMVIANTFGSAAILIVFLLGGFVIPKADI 717
Query: 703 KPWWIWGYWLSPLTYGQRAITVNEFTASRWMKQSALGNNTIGYNILHAQSLPSEDYWYWV 762
KPWW+WG+W+SPL+YGQRAI VNEFTA+RWM SA+ + TIG N+L +S P+ DYWYW+
Sbjct: 718 KPWWVWGFWVSPLSYGQRAIAVNEFTATRWMTPSAISDTTIGLNLLKLRSFPTNDYWYWI 777
Query: 763 SVAVLVTYAIIFNIMVTLALAYLHPLQKPRTVIPQDDEPEKSSSRDANYVFSTRSTKDES 822
+AVL+ YAI+FN +VTLALAYL+PL+K R V+ D E + DAN V S +
Sbjct: 778 GIAVLIGYAILFNNVVTLALAYLNPLRKARAVVLDDPNEETALVADANQVISEK------ 831
Query: 823 NTKGMILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGS 882
KGMILPF+PLTMTFHNV+Y+VDMP+E+R QG+PETRLQLLSNVSGVFSPGVLTALVGS
Sbjct: 832 --KGMILPFKPLTMTFHNVNYYVDMPKEMRSQGVPETRLQLLSNVSGVFSPGVLTALVGS 889
Query: 883 SGAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESL 942
SGAGKTTLMDVLAGRKTGGY EGDI+ISG+PKEQ+TFARISGYVEQNDIHSPQVT+EESL
Sbjct: 890 SGAGKTTLMDVLAGRKTGGYTEGDIRISGHPKEQQTFARISGYVEQNDIHSPQVTVEESL 949
Query: 943 WFSASLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVEL 1002
WFSASLRLPKEI+ ++K+EFVEQVM+LVELD+LR ALVG+PG++GLSTEQRKRLTIAVEL
Sbjct: 950 WFSASLRLPKEITKEQKKEFVEQVMRLVELDTLRYALVGLPGTTGLSTEQRKRLTIAVEL 1009
Query: 1003 VANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMK 1062
VANPSIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCTIHQPSIDIFEAFD+LLLMK
Sbjct: 1010 VANPSIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFEAFDELLLMK 1069
Query: 1063 RGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTPSVEETIDADFAEI 1122
RGG+VIYGGKLG SQ+++DYFQGI G+ PI GYNPATW+LEVTTP++EE + +FA++
Sbjct: 1070 RGGQVIYGGKLGTHSQVLVDYFQGINGVPPISSGYNPATWMLEVTTPALEEKYNMEFADL 1129
Query: 1123 YNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQFYRCLWKQNLVYWRSPPYNA 1182
Y SDQ+R VEA+I + PP GSEP+ F + YSQ+ LSQF CLWKQNLVYWRSP YN
Sbjct: 1130 YKKSDQFREVEANIKQLSVPPEGSEPISFTSRYSQNQLSQFLLCLWKQNLVYWRSPEYNL 1189
Query: 1183 MRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIGVNNASTVQPIVSIER 1242
+R+ FTTI+A + GTVFWDIGSKR+S+Q+L VMGALY++CLF+GV+NAS+VQPIVSIER
Sbjct: 1190 VRLVFTTIAAFILGTVFWDIGSKRTSSQDLITVMGALYSACLFLGVSNASSVQPIVSIER 1249
Query: 1243 TVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMINFERTAGKFFLYLLFM 1302
TVFYREKAAGMY+PI YA AQGL+EIPYI Q +++G+ITYF I FERT KF LYL+FM
Sbjct: 1250 TVFYREKAAGMYAPIPYAAAQGLVEIPYILTQTILYGVITYFTIGFERTFSKFVLYLVFM 1309
Query: 1303 FLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPESHIPGWWIWFYYICP 1362
FLTFTYFTFYGMMAVGL+P QHLAAVISSAFYSLWNLLSGFL+ + IP WWIWFYYICP
Sbjct: 1310 FLTFTYFTFYGMMAVGLTPNQHLAAVISSAFYSLWNLLSGFLVQKPLIPVWWIWFYYICP 1369
Query: 1363 VQWTLRGVITSQLGDVETKIIGPGFEGTVKEYLSLNLGYDPKIMGISTVGLSVIVLFGFI 1422
V WTL+GVI SQLGDVE+ I P F GTVKE++ GY P ++G+S VL GF
Sbjct: 1370 VAWTLQGVILSQLGDVESMINEPLFHGTVKEFIEYYFGYKPNMIGVSAA-----VLVGFC 1424
Query: 1423 ILFFCSFVVSVKVLNFQKR 1441
LFF +F +SVK LNFQ+R
Sbjct: 1425 ALFFSAFALSVKYLNFQRR 1443
>At1g59870 putative ABC transporter (At1g59870)
Length = 1469
Score = 1568 bits (4060), Expect = 0.0
Identities = 776/1466 (52%), Positives = 1038/1466 (69%), Gaps = 50/1466 (3%)
Query: 2 AASDGSEYFEIGSFRSESFARASNADTVEEDEEELQWAALSRLPSQKRINYALYRASSSR 61
A+ + + F GS R++S V +DEE L+WAA+ +LP+ R+ L A
Sbjct: 28 ASRNIEDIFSSGSRRTQS---------VNDDEEALKWAAIEKLPTYSRLRTTLMNAV--- 75
Query: 62 RQPPPLGSPAGAGGRADNLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIG 121
+ +DV KL R+ + +QDN R+L+ ++ R+DR+G
Sbjct: 76 -----VEDDVYGNQLMSKEVDVTKLDGEDRQKFIDMVFKVAEQDNERILTKLRNRIDRVG 130
Query: 122 IEVPNIEVRFSNLSVSADVQIGSRALPTLINYTRDALETICTNLGIYRPKRHSLTILDNV 181
I++P +EVR+ +L++ AD G+R+LPTL+N R+ E+ +GI K+ LTIL ++
Sbjct: 131 IKLPTVEVRYEHLTIKADCYTGNRSLPTLLNVVRNMGESALGMIGIQFAKKAQLTILKDI 190
Query: 182 SGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYI 241
SGVIKPGRMTLLLG P +GK++LLLALAGKLD +L+ +G ITYNG++ DEF ++T AYI
Sbjct: 191 SGVIKPGRMTLLLGPPSSGKTTLLLALAGKLDKSLQVSGDITYNGYQLDEFVPRKTSAYI 250
Query: 242 SQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSV 301
SQ D H +TV+ETLDF+ARCQG + ++ R E + I P ++D FMKAS+
Sbjct: 251 SQNDLHVGIMTVKETLDFSARCQGVGTRYDLLN-ELARREKDAGIFPEADVDLFMKASAA 309
Query: 302 GGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEI 361
G K+S+ TDY LK+LGLDIC +TIVG DM+RG+SGGQ+KRVTTGEMIVGP KTLFMDEI
Sbjct: 310 QGVKNSLVTDYTLKILGLDICKDTIVGDDMMRGISGGQKKRVTTGEMIVGPTKTLFMDEI 369
Query: 362 STGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRE 421
STGLDSSTTFQIVKC++ VHL +ATVLM+LLQPAPETF+LFDD++L+SEG ++Y+GPR+
Sbjct: 370 STGLDSSTTFQIVKCLQQIVHLNEATVLMSLLQPAPETFDLFDDIILVSEGQIVYQGPRD 429
Query: 422 NVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRF 481
N+LEFFES GFK P RKG ADFLQEV+S+KDQ QYW +P++ Y ++P E A +++
Sbjct: 430 NILEFFESFGFKCPERKGTADFLQEVTSKKDQEQYWVNPNRPYHYIPVSEFASRYKSFHV 489
Query: 482 GSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREALLISRQRFLYIFKTCQV 541
G+ + + P+DKS+ H +AL KY+VS+ E+ K+C+ +E LL+ R F Y+FKT Q+
Sbjct: 490 GTKMSNELAVPFDKSRGHKAALVFDKYSVSKRELLKSCWDKEWLLMQRNAFFYVFKTVQI 549
Query: 542 AFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQR 601
+ +T T+FLRT M+ +E NLY+ AL FG++ MFNGF+E+++M++RLPVFYKQR
Sbjct: 550 VIIAAITSTLFLRTEMNTRNEGDANLYIGALLFGMIINMFNGFAEMAMMVSRLPVFYKQR 609
Query: 602 DNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMA 661
D LFYP+W +SL ++L +P SI+E+ W V+ YY++GFAP A RFF+ ++F++ QMA
Sbjct: 610 DLLFYPSWTFSLPTFLLGIPSSILESTAWMVVTYYSIGFAPDASRFFKQFLLVFLIQQMA 669
Query: 662 IGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRA 721
LFR++AS+ R M++ANT G+ LL++FLLGGF++PKG I WW W YW+SPLTY
Sbjct: 670 ASLFRLIASVCRTMMIANTGGALTLLLVFLLGGFLLPKGKIPDWWGWAYWVSPLTYAFNG 729
Query: 722 ITVNEFTASRWMKQSALGNNTI--GYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVT 779
+ VNE A RWM + A N+TI G +L+ + + WYW+SV L+ + +FNI+ T
Sbjct: 730 LVVNEMFAPRWMNKMASSNSTIKLGTMVLNTWDVYHQKNWYWISVGALLCFTALFNILFT 789
Query: 780 LALAYLHPLQKPRTVIPQDDEPEKSSSRDA-------------NYVFSTRSTKDES---- 822
LAL YL+PL K ++P+++ + +D V R ++D +
Sbjct: 790 LALTYLNPLGKKAGLLPEEENEDADQGKDPMRRSLSTADGNRRGEVAMGRMSRDSAAEAS 849
Query: 823 ----NTKGMILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTA 878
N KGM+LPF PL M+F +V YFVDMP E+R QG+ ETRLQLL V+G F PGVLTA
Sbjct: 850 GGAGNKKGMVLPFTPLAMSFDDVKYFVDMPGEMRDQGVTETRLQLLKGVTGAFRPGVLTA 909
Query: 879 LVGSSGAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTI 938
L+G SGAGKTTLMDVLAGRKTGGYIEGD++ISG+PK Q TFARISGY EQ DIHSPQVT+
Sbjct: 910 LMGVSGAGKTTLMDVLAGRKTGGYIEGDVRISGFPKVQETFARISGYCEQTDIHSPQVTV 969
Query: 939 EESLWFSASLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTI 998
ESL FSA LRLPKE+ D+K FV+QVM+LVELDSLR+++VG+PG +GLSTEQRKRLTI
Sbjct: 970 RESLIFSAFLRLPKEVGKDEKMMFVDQVMELVELDSLRDSIVGLPGVTGLSTEQRKRLTI 1029
Query: 999 AVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDL 1058
AVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFD+L
Sbjct: 1030 AVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDEL 1089
Query: 1059 LLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTPSVEETIDAD 1118
+LMKRGG+VIY G LG S +++YF+ G+ IP YNPATW+LE ++ + E + D
Sbjct: 1090 MLMKRGGQVIYAGPLGQNSHKVVEYFESFPGVSKIPEKYNPATWMLEASSLAAELKLSVD 1149
Query: 1119 FAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQFYRCLWKQNLVYWRSP 1178
FAE+YN S ++ +A + E PPAG+ L F T +SQ+ QF CLWKQ YWRSP
Sbjct: 1150 FAELYNQSALHQRNKALVKELSVPPAGASDLYFATQFSQNTWGQFKSCLWKQWWTYWRSP 1209
Query: 1179 PYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIGVNNASTVQPIV 1238
YN +R FT ++L+ GTVFW IG RS+ +L +V+GALYA+ +F+G+NN STVQP+V
Sbjct: 1210 DYNLVRFIFTLATSLLIGTVFWQIGGNRSNAGDLTMVIGALYAAIIFVGINNCSTVQPMV 1269
Query: 1239 SIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMINFERTAGKFFLY 1298
++ERTVFYRE+AAGMYS + YA++Q E+PY+ +Q + + LI Y M+ FE A KFF +
Sbjct: 1270 AVERTVFYRERAAGMYSAMPYAISQVTCELPYVLIQTVYYSLIVYAMVGFEWKAEKFFWF 1329
Query: 1299 LLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPESHIPGWWIWFY 1358
+ + +F Y+T+YGMM V L+P Q +A++ +SAFY ++NL SGF IP IP WWIW+Y
Sbjct: 1330 VFVSYFSFLYWTYYGMMTVSLTPNQQVASIFASAFYGIFNLFSGFFIPRPKIPKWWIWYY 1389
Query: 1359 YICPVQWTLRGVITSQLGDVETKI--IGPGFEGTVKEYLSLNLGYDPKIMG-ISTVGLSV 1415
+ICPV WT+ G+I SQ GDVET+I +G + TVK+Y+ + G+ MG ++ V ++
Sbjct: 1390 WICPVAWTVYGLIVSQYGDVETRIQVLGGAPDLTVKQYIEDHYGFQSDFMGPVAAVLIAF 1449
Query: 1416 IVLFGFIILFFCSFVVSVKVLNFQKR 1441
V F FI F ++ LNFQ R
Sbjct: 1450 TVFFAFI------FAFCIRTLNFQTR 1469
>At1g15520 hypothetical protein
Length = 1423
Score = 1543 bits (3995), Expect = 0.0
Identities = 772/1427 (54%), Positives = 1012/1427 (70%), Gaps = 33/1427 (2%)
Query: 18 ESFARASNADTVEEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPAGAGGRA 77
E F+R+S E+DEE L+WAAL +LP+ R+ + AS + G P
Sbjct: 27 EIFSRSSRE---EDDEEALRWAALEKLPTFDRLRKGILTASHA-------GGPI------ 70
Query: 78 DNLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRFSNLSVS 137
N +D++KL + ++++ + D ++ +LL +K+R+DR+GI++P IEVRF +L V
Sbjct: 71 -NEIDIQKLGFQDTKKLLERLIKVGDDEHEKLLWKLKKRIDRVGIDLPTIEVRFDHLKVE 129
Query: 138 ADVQIGSRALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMTLLLGAP 197
A+V +G RALPT +N+ + + L + ++ TIL++VSG++KPGRM LLLG P
Sbjct: 130 AEVHVGGRALPTFVNFISNFADKFLNTLHLVPNRKKKFTILNDVSGIVKPGRMALLLGPP 189
Query: 198 GAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAELTVRETL 257
+GK++LLLALAGKLD LK+TG +TYNGH +EF +RT AYI Q D H E+TVRET
Sbjct: 190 SSGKTTLLLALAGKLDQELKQTGRVTYNGHGMNEFVPQRTAAYIGQNDVHIGEMTVRETF 249
Query: 258 DFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVL 317
+AAR QG + T ++ R E E NI+P P+ID FMKA S G+K +V TDYILK+L
Sbjct: 250 AYAARFQGVGSRYDMLT-ELARREKEANIKPDPDIDIFMKAMSTAGEKTNVMTDYILKIL 308
Query: 318 GLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCI 377
GL++C++T+VG DMLRG+SGGQ+KRVTTGEM+VGP + LFMDEISTGLDSSTT+QIV +
Sbjct: 309 GLEVCADTMVGDDMLRGISGGQKKRVTTGEMLVGPSRALFMDEISTGLDSSTTYQIVNSL 368
Query: 378 KNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPR 437
+N+VH+ + T L++LLQPAPETF LFDD++L++EG +IYEGPR++V+EFFE++GFK PPR
Sbjct: 369 RNYVHIFNGTALISLLQPAPETFNLFDDIILIAEGEIIYEGPRDHVVEFFETMGFKCPPR 428
Query: 438 KGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSK 497
KG+ADFLQEV+S+KDQ QYWA + Y+F+ E AEAF++ G + P+DK+K
Sbjct: 429 KGVADFLQEVTSKKDQMQYWARRDEPYRFIRVREFAEAFQSFHVGRRIGDELALPFDKTK 488
Query: 498 CHPSALARTKYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRM 557
HP+AL KY V E+ K F+RE LL+ R F+Y FK Q+ + F+T T+F RT M
Sbjct: 489 SHPAALTTKKYGVGIKELVKTSFSREYLLMKRNSFVYYFKFGQLLVMAFLTMTLFFRTEM 548
Query: 558 HPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWV 617
E G+LY ALFF L+ +MFNG SELS+ IA+LPVFYKQRD LFYPAW +SL W+
Sbjct: 549 QKKTEVDGSLYTGALFFILMMLMFNGMSELSMTIAKLPVFYKQRDLLFYPAWVYSLPPWL 608
Query: 618 LRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVL 677
L++P S +EA + T I YY +GF P+ GR F+ +L +M+QMA LF+M+A++ R+M++
Sbjct: 609 LKIPISFMEAALTTFITYYVIGFDPNVGRLFKQYILLVLMNQMASALFKMVAALGRNMIV 668
Query: 678 ANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRWMKQSA 737
ANTFG+ A+LV F LGG ++ + IK WWIWGYW+SP+ YGQ AI NEF W +
Sbjct: 669 ANTFGAFAMLVFFALGGVVLSRDDIKKWWIWGYWISPIMYGQNAILANEFFGHSWSRAVE 728
Query: 738 LGNNTIGYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQKPRTVI-- 795
+ T+G L ++ YWYW+ L+ + ++FN TLAL +L+ L KP+ VI
Sbjct: 729 NSSETLGVTFLKSRGFLPHAYWYWIGTGALLGFVVLFNFGFTLALTFLNSLGKPQAVIAE 788
Query: 796 -PQDDEPEKSSSRDANYVFSTRSTKDESNTKGMILPFQPLTMTFHNVSYFVDMPQEIRKQ 854
P DE E S+R V + + K +GM+LPF+P ++TF NV Y VDMPQE+ +Q
Sbjct: 789 EPASDETELQSARSEGVVEAGANKK-----RGMVLPFEPHSITFDNVVYSVDMPQEMIEQ 843
Query: 855 GIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKISGYPK 914
G E RL LL V+G F PGVLTAL+G SGAGKTTLMDVLAGRKTGGYI+G+I ISGYPK
Sbjct: 844 GTQEDRLVLLKGVNGAFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGYIDGNITISGYPK 903
Query: 915 EQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLVELDS 974
Q+TFARISGY EQ DIHSP VT+ ESL +SA LRLPKE+ +K++ F+E+VM+LVEL
Sbjct: 904 NQQTFARISGYCEQTDIHSPHVTVYESLVYSAWLRLPKEVDKNKRKIFIEEVMELVELTP 963
Query: 975 LRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTV 1034
LR ALVG+PG SGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMR VRNTV
Sbjct: 964 LRQALVGLPGESGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRTVRNTV 1023
Query: 1035 DTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIP 1094
DTGRTVVCTIHQPSIDIFEAFD+L L+KRGG IY G LG +S +I+YF+ I+GI I
Sbjct: 1024 DTGRTVVCTIHQPSIDIFEAFDELFLLKRGGEEIYVGPLGHESTHLINYFESIQGINKIT 1083
Query: 1095 RGYNPATWVLEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTI 1154
GYNPATW+LEV+T S E + DFA++Y NS+ Y+ + I E P GS+ L F T
Sbjct: 1084 EGYNPATWMLEVSTTSQEAALGVDFAQVYKNSELYKRNKELIKELSQPAPGSKDLYFPTQ 1143
Query: 1155 YSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYV 1214
YSQS L+Q LWKQ+ YWR+PPY A+R FT AL+FGT+FWD+G K + Q+L
Sbjct: 1144 YSQSFLTQCMASLWKQHWSYWRNPPYTAVRFLFTIGIALMFGTMFWDLGGKTKTRQDLSN 1203
Query: 1215 VMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQ 1274
MG++Y + LF+G+ NA++VQP+V++ERTVFYRE+AAGMYS + YA AQ IEIPY+ VQ
Sbjct: 1204 AMGSMYTAVLFLGLQNAASVQPVVNVERTVFYREQAAGMYSAMPYAFAQVFIEIPYVLVQ 1263
Query: 1275 AMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFY 1334
A+V+GLI Y MI FE TA KFF YL FM+ +F FTFYGMMAV ++P H+A+V+SSAFY
Sbjct: 1264 AIVYGLIVYAMIGFEWTAVKFFWYLFFMYGSFLTFTFYGMMAVAMTPNHHIASVVSSAFY 1323
Query: 1335 SLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGFEGTVKEY 1394
+WNL SGFLIP +P WW W+Y++CPV WTL G+I SQ GD+ + +VK++
Sbjct: 1324 GIWNLFSGFLIPRPSMPVWWEWYYWLCPVAWTLYGLIASQFGDITEPMADSNM--SVKQF 1381
Query: 1395 LSLNLGYDPKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
+ GY +G+ ++VI F +LF F + +K NFQKR
Sbjct: 1382 IREFYGYREGFLGV-VAAMNVI----FPLLFAVIFAIGIKSFNFQKR 1423
>At1g15210 putative ABC transporter
Length = 1442
Score = 1536 bits (3977), Expect = 0.0
Identities = 762/1421 (53%), Positives = 1005/1421 (70%), Gaps = 28/1421 (1%)
Query: 28 TVEEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPAGAGGRADN-LMDVRKL 86
+V EDEE L+WA++ +LP+ R+ +L P LG G + N +DV KL
Sbjct: 43 SVNEDEEALKWASIEKLPTYNRLRTSLM---------PELGEDDVYGNQILNKAVDVTKL 93
Query: 87 SRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRFSNLSVSADVQIGSRA 146
R+ + +QDN R+L+ ++ R+DR+GI++P +EVR+ +L+V AD G R+
Sbjct: 94 DGEERQKFIDMVFKVAEQDNERILTKLRNRIDRVGIQLPTVEVRYDHLTVKADCYTGDRS 153
Query: 147 LPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLL 206
LP+L+N R+ E +GI K+ LTIL +VSG++KP RMTLLLG P +GK++LLL
Sbjct: 154 LPSLLNAVRNMGEAALGMIGIRLAKKAQLTILKDVSGIVKPSRMTLLLGPPSSGKTTLLL 213
Query: 207 ALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGA 266
ALAGKLD +L +G +TYNG+ +EF +T AYISQ D H +TV+ETLDF+ARCQG
Sbjct: 214 ALAGKLDKSLDVSGEVTYNGYRLNEFVPIKTSAYISQNDLHVGIMTVKETLDFSARCQGV 273
Query: 267 QEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETI 326
+ ++ R E + I P ++D FMKAS+ G K S+ TDY LK+LGLDIC +TI
Sbjct: 274 GTRYDLLN-ELARREKDAGIFPEADVDLFMKASAAQGVKSSLITDYTLKILGLDICKDTI 332
Query: 327 VGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDA 386
VG DM+RG+SGGQ+KRVTTGEMIVGP KTLFMDEISTGLDSSTTFQIVKC++ VHL +A
Sbjct: 333 VGDDMMRGISGGQKKRVTTGEMIVGPTKTLFMDEISTGLDSSTTFQIVKCLQQIVHLTEA 392
Query: 387 TVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQE 446
TVL++LLQPAPETF+LFDD++LLSEG ++Y+GPR+++LEFFES GFK P RKG ADFLQE
Sbjct: 393 TVLISLLQPAPETFDLFDDIILLSEGQIVYQGPRDHILEFFESFGFKCPERKGTADFLQE 452
Query: 447 VSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSKCHPSALART 506
V+S+KDQ QYW DP++ Y+++P E A +F+ GS + + + PYDKSK H +AL
Sbjct: 453 VTSKKDQEQYWVDPNRPYRYIPVSEFASSFKKFHVGSKLSNELSVPYDKSKSHKAALMFD 512
Query: 507 KYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGN 566
KY++ + E+ K+C+ +E +L+ R F Y+FKT Q+ + +T T++LRT MH +E N
Sbjct: 513 KYSIKKTELLKSCWDKEWMLMKRNSFFYVFKTVQIIIIAAITSTLYLRTEMHTRNEIDAN 572
Query: 567 LYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIE 626
+YV +L F ++ MFNG +E+++ I RLPVFYKQRD LF+P W ++L ++L +P SI E
Sbjct: 573 IYVGSLLFAMIVNMFNGLAEMAMTIQRLPVFYKQRDLLFHPPWTYTLPTFLLGIPISIFE 632
Query: 627 AVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAAL 686
+ W V+ YY++G+AP A RFF+ I+F++ QMA G+FR +AS R M +ANT G L
Sbjct: 633 STAWMVVTYYSIGYAPDAERFFKQFLIIFLIQQMAAGIFRFIASTCRTMTIANTGGVLVL 692
Query: 687 LVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRWMKQSALGNNT--IG 744
LV+FL GGF++P+ I WW W YW+SPL+Y AITVNE A RWM + + GN+T +G
Sbjct: 693 LVVFLTGGFLLPRSEIPVWWRWAYWISPLSYAFNAITVNELFAPRWMNKMS-GNSTTRLG 751
Query: 745 YNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQKPRTVIPQDDEPEKS 804
++L+ + + WYW+ V L+ + +IFN TLAL YL PL K + ++P++++ E
Sbjct: 752 TSVLNIWDVFDDKNWYWIGVGGLLGFTVIFNGFFTLALTYLDPLGKAQAILPKEEDEEAK 811
Query: 805 SSRDANYVFSTRSTKDESNTKGMILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQLL 864
+N S S KGM+LPF PL M+F +V YFVDMP E+R+QG+ ETRLQLL
Sbjct: 812 GKAGSNKETEMESV---SAKKGMVLPFTPLAMSFDDVKYFVDMPAEMREQGVQETRLQLL 868
Query: 865 SNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISG 924
V+ F PGVLTAL+G SGAGKTTLMDVLAGRKTGGYIEGD+++SG+PK+Q TFARISG
Sbjct: 869 KGVTSAFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGYIEGDVRVSGFPKKQETFARISG 928
Query: 925 YVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPG 984
Y EQ DIHSPQVT+ ESL FSA LRL KE+S + K FV+QVM+LVEL LR+A+VG+PG
Sbjct: 929 YCEQTDIHSPQVTVRESLIFSAFLRLAKEVSKEDKLMFVDQVMELVELVDLRDAIVGLPG 988
Query: 985 SSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTI 1044
+GLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTI
Sbjct: 989 VTGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTI 1048
Query: 1045 HQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVL 1104
HQPSIDIFEAFD+LLLMKRGG VIY G LG S +++YF+ G+ IP YNPATW+L
Sbjct: 1049 HQPSIDIFEAFDELLLMKRGGHVIYSGPLGRNSHKVVEYFESFPGVPKIPEKYNPATWML 1108
Query: 1105 EVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQFY 1164
E ++ + E + DFAE+Y S + +A + E PP G+ L F T +SQ+ QF
Sbjct: 1109 EASSLAAELKLGVDFAELYKASALCQRNKALVQELSVPPQGATDLYFATQFSQNTWGQFK 1168
Query: 1165 RCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCL 1224
CLWKQ YWRSP YN +R FT ++L+ G+VFW IG KRS+ Q+L +V+GA+YA+ +
Sbjct: 1169 SCLWKQWWTYWRSPDYNLVRFIFTLATSLMIGSVFWQIGGKRSNVQDLTMVIGAIYAAVV 1228
Query: 1225 FIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYF 1284
F+G+NN STVQP+V++ERTVFYREKAAGMYS I YA++Q E+PY+ +Q + LI Y
Sbjct: 1229 FVGINNCSTVQPMVAVERTVFYREKAAGMYSAIPYAISQVTCELPYVLIQTTYYSLIIYS 1288
Query: 1285 MINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFL 1344
M+ FE A KF ++ + +F Y+T+YGMM V L+P Q +A++ +SAFY ++NL SGF
Sbjct: 1289 MVGFEWKASKFLWFIFINYFSFLYWTYYGMMTVSLTPNQQVASIFASAFYGIFNLFSGFF 1348
Query: 1345 IPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKII----GPGFEGTVKEYLSLNLG 1400
IP IP WW+W+Y+ICPV WT+ G+ITSQ GDVET I PG TVK+Y+ G
Sbjct: 1349 IPRPKIPKWWVWYYWICPVAWTIYGLITSQYGDVETPIALLGGAPGL--TVKQYIKDQYG 1406
Query: 1401 YDPKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
++ MG G VL GF + F F +K LNFQ R
Sbjct: 1407 FESDYMG-PVAG----VLVGFTVFFAFIFAFCIKTLNFQSR 1442
>At3g16340 putative ABC transporter
Length = 1416
Score = 1525 bits (3949), Expect = 0.0
Identities = 754/1434 (52%), Positives = 1011/1434 (69%), Gaps = 37/1434 (2%)
Query: 12 IGSFRSESFARASNADTVEEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPA 71
+ S + F+R S + + DEE L+WAAL +LP+ R+ +
Sbjct: 16 LASNSNNHFSRRSGSTIDDHDEEALKWAALEKLPTFARLRTTIIHPH------------- 62
Query: 72 GAGGRADNLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRF 131
++L+DV KL R+ + ++DN + L + R+DR+ I++P +EVRF
Sbjct: 63 ------EDLVDVTKLGVDDRQKFIDSIFKVTEEDNEKFLKKFRNRIDRVRIKLPTVEVRF 116
Query: 132 SNLSVSADVQIGSRALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMT 191
+++ A+ IG RALPTL N + E LG K +TIL +VSG+IKP RMT
Sbjct: 117 EKVTIEANCHIGKRALPTLPNAALNIAERGLRLLGFNFTKTTKVTILRDVSGIIKPSRMT 176
Query: 192 LLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAEL 251
LLLG P +GK++LLLALAGKLD +LK TG +TYNGH +EF ++T AYISQ D H +
Sbjct: 177 LLLGPPSSGKTTLLLALAGKLDQSLKVTGRVTYNGHGLEEFVPQKTSAYISQNDVHVGVM 236
Query: 252 TVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTD 311
TV+ETLDF+ARCQG + + ++ R E + I P PE+D FMK+ + G K S+ TD
Sbjct: 237 TVQETLDFSARCQGVGTRYDLLS-ELVRREKDAGILPEPEVDLFMKSIAAGNVKSSLITD 295
Query: 312 YILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTF 371
Y L++LGLDIC +T+VG +M+RG+SGGQ+KRVTTGEMIVGP KTLFMDEISTGLDSSTT+
Sbjct: 296 YTLRILGLDICKDTVVGDEMIRGISGGQKKRVTTGEMIVGPTKTLFMDEISTGLDSSTTY 355
Query: 372 QIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIG 431
QIVKC++ V DATVLM+LLQPAPETFELFDD++LLSEG ++Y+GPR++VL FFE+ G
Sbjct: 356 QIVKCLQEIVRFTDATVLMSLLQPAPETFELFDDIILLSEGQIVYQGPRDHVLTFFETCG 415
Query: 432 FKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTH 491
FK P RKG ADFLQEV+SRKDQ QYWAD K Y ++ E ++ FR G+ +E +
Sbjct: 416 FKCPDRKGTADFLQEVTSRKDQEQYWADSKKPYSYISVSEFSKRFRTFHVGANLEKDLSV 475
Query: 492 PYDKSKCHPSALARTKYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTI 551
PYD+ K HP++L K++V + ++ K C+ RE LL+ R F YI KT Q+ + + T+
Sbjct: 476 PYDRFKSHPASLVFKKHSVPKSQLFKVCWDRELLLMKRNAFFYITKTVQIIIMALIASTV 535
Query: 552 FLRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAW 611
+LRT M +E+ G +Y+ AL F ++ MFNGF+EL+LMI RLPVFYKQRD LF+P W +
Sbjct: 536 YLRTEMGTKNESDGAVYIGALMFSMIVNMFNGFAELALMIQRLPVFYKQRDLLFHPPWTF 595
Query: 612 SLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASI 671
SL ++L +P SI E+V+W I YY +GFAP RF +++ ++F+ QMA G+FR +A+
Sbjct: 596 SLPTFLLGIPISIFESVVWVTITYYMIGFAPELSRFLKHLLVIFLTQQMAGGIFRFIAAT 655
Query: 672 ARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASR 731
R M+LANT G+ +L++FLLGGFI+P+G I WW W YW+SP+ Y A+TVNE A R
Sbjct: 656 CRSMILANTGGALVILLLFLLGGFIVPRGEIPKWWKWAYWVSPMAYTYDALTVNEMLAPR 715
Query: 732 WMKQSALGNNT-IGYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQK 790
W+ Q + N+T +G +L + ++ WYW+ V ++ + ++FNI+VTLAL +L+PL+K
Sbjct: 716 WINQPSSDNSTSLGLAVLEIFDIFTDPNWYWIGVGGILGFTVLFNILVTLALTFLNPLEK 775
Query: 791 PRTVIPQDDEPEKSSSRDANYVFSTRSTKDESNTKGMILPFQPLTMTFHNVSYFVDMPQE 850
+ V+ +++ E + + +K +GM+LPF PLTM+F NV+Y+VDMP+E
Sbjct: 776 QQAVVSKENTEENRAENGSK-------SKSIDVKRGMVLPFTPLTMSFDNVNYYVDMPKE 828
Query: 851 IRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKIS 910
+++QG+ + +LQLL V+GVF PGVLTAL+G SGAGKTTLMDVLAGRKTGGYIEGDI+IS
Sbjct: 829 MKEQGVSKDKLQLLKEVTGVFRPGVLTALMGVSGAGKTTLMDVLAGRKTGGYIEGDIRIS 888
Query: 911 GYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLV 970
G+PK Q TFARISGY EQNDIHSPQVT++ESL +SA LRLPKE++ +K FV++VM+LV
Sbjct: 889 GFPKRQETFARISGYCEQNDIHSPQVTVKESLIYSAFLRLPKEVTKYEKMRFVDEVMELV 948
Query: 971 ELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAV 1030
EL+SL++A+VG+PG +GLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMR V
Sbjct: 949 ELESLKDAVVGLPGITGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRTV 1008
Query: 1031 RNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGI 1090
RNTVDTGRTVVCTIHQPSIDIFEAFD+LLL+KRGG+VIY G LG S +I+YFQ I G+
Sbjct: 1009 RNTVDTGRTVVCTIHQPSIDIFEAFDELLLLKRGGQVIYAGPLGQNSHKIIEYFQAIHGV 1068
Query: 1091 RPIPRGYNPATWVLEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLK 1150
I YNPATW+LEV++ + E ++ DFAE Y S Y+ + + E PP G+ L
Sbjct: 1069 PKIKEKYNPATWMLEVSSMAAEAKLEIDFAEHYKTSSLYQQNKNLVKELSTPPQGASDLY 1128
Query: 1151 FDTIYSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQ 1210
F T +SQSLL QF CLWKQ + YWR+P YN R +FT +A++ G++FW +G+KR +
Sbjct: 1129 FSTRFSQSLLGQFKSCLWKQWITYWRTPDYNLARFFFTLAAAVMLGSIFWKVGTKRENAN 1188
Query: 1211 ELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPY 1270
+L V+GA+YA+ LF+GVNN+S+VQP++++ER+VFYRE+AA MYS + YA+AQ + EIPY
Sbjct: 1189 DLTKVIGAMYAAVLFVGVNNSSSVQPLIAVERSVFYRERAAEMYSALPYALAQVVCEIPY 1248
Query: 1271 IAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVIS 1330
+ +Q + LI Y M+ FE T KFF + F++F YFT+YGMM V L+P Q +AAV +
Sbjct: 1249 VLIQTTYYTLIIYAMMCFEWTLAKFFWFYFVSFMSFLYFTYYGMMTVALTPNQQVAAVFA 1308
Query: 1331 SAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGF--E 1388
AFY L+NL SGF+IP IP WWIW+Y+ICPV WT+ G+I SQ GDVE I PG +
Sbjct: 1309 GAFYGLFNLFSGFVIPRPRIPKWWIWYYWICPVAWTVYGLIVSQYGDVEDTIKVPGMAND 1368
Query: 1389 GTVKEYLSLNLGYDPKIM-GISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
T+K Y+ + GYD M I+T VL GF + F F ++ LNFQ+R
Sbjct: 1369 PTIKWYIENHYGYDADFMIPIAT------VLVGFTLFFAFMFAFGIRTLNFQQR 1416
>At2g36380 putative ABC transporter
Length = 1450
Score = 1457 bits (3771), Expect = 0.0
Identities = 743/1423 (52%), Positives = 969/1423 (67%), Gaps = 28/1423 (1%)
Query: 26 ADTVEEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPAGAGGRADNLMDVRK 85
+D EED+ EL+WAAL RLP+ R+ R+ P G G D +DV
Sbjct: 49 SDRREEDDVELRWAALERLPTYDRL----------RKGMLPQTMVNGKIGLED--VDVTN 96
Query: 86 LSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRFSNLSVSADVQIGSR 145
L+ ++ +++ L ++DN + L ++ER DR+GIEVP IEVR+ NLSV DV+ SR
Sbjct: 97 LAPKEKKHLMEMILKFVEEDNEKFLRRLRERTDRVGIEVPKIEVRYENLSVEGDVRSASR 156
Query: 146 ALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLL 205
ALPTL N T + +E+I + K+ + IL ++SG+IKP RMTLLLG P +GK++LL
Sbjct: 157 ALPTLFNVTLNTIESILGLFHLLPSKKRKIEILKDISGIIKPSRMTLLLGPPSSGKTTLL 216
Query: 206 LALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAELTVRETLDFAARCQG 265
ALAGKLD L+ +G ITY GHE EF ++TCAYISQ D H E+TVRE+LDF+ RC G
Sbjct: 217 QALAGKLDDTLQMSGRITYCGHEFREFVPQKTCAYISQHDLHFGEMTVRESLDFSGRCLG 276
Query: 266 AQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSET 325
+ T ++ R E E I+P PEIDAFMK+ ++ G++ S+ TDY+LK+LGLDIC++T
Sbjct: 277 VGTRYQLLT-ELSRREREAGIKPDPEIDAFMKSIAISGQETSLVTDYVLKLLGLDICADT 335
Query: 326 IVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMD 385
+VG M RG+SGGQRKR+TTGEM+VGP LFMDEISTGLDSSTTFQI K ++ VH+ D
Sbjct: 336 LVGDVMRRGISGGQRKRLTTGEMLVGPATALFMDEISTGLDSSTTFQICKFMRQLVHIAD 395
Query: 386 ATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQ 445
T++++LLQPAPETFELFDD++LLSEG ++Y+G R+NVLEFFE +GFK P RKGIADFLQ
Sbjct: 396 VTMVISLLQPAPETFELFDDIILLSEGQIVYQGSRDNVLEFFEYMGFKCPERKGIADFLQ 455
Query: 446 EVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSKCHPSALAR 505
EV+S+KDQ QYW Y +V + + F + G + S PYDK+K HP+AL
Sbjct: 456 EVTSKKDQEQYWNRREHPYSYVSVHDFSSGFNSFHAGQQLASEFRVPYDKAKTHPAALVT 515
Query: 506 TKYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYG 565
KY +S ++ KACF RE LL+ R F+Y+FKT Q+ + + T++ RT MH G
Sbjct: 516 QKYGISNKDLFKACFDREWLLMKRNSFVYVFKTVQITIMSLIAMTVYFRTEMHVGTVQDG 575
Query: 566 NLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSII 625
+ ALFF L+++MFNG +EL+ + RLPVF+KQRD LFYP WA++L ++L++P S+I
Sbjct: 576 QKFYGALFFSLINLMFNGMAELAFTVMRLPVFFKQRDFLFYPPWAFALPGFLLKIPLSLI 635
Query: 626 EAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAA 685
E+VIW + YYT+GFAPSA RFFR + F ++QMA+ LFR + ++ R V+AN+ G+ A
Sbjct: 636 ESVIWIALTYYTIGFAPSAARFFRQLLAYFCVNQMALSLFRFLGALGRTEVIANSGGTLA 695
Query: 686 LLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRW---MKQSALGNNT 742
LLV+F+LGGFII K I W W Y+ SP+ YGQ A+ +NEF RW + + T
Sbjct: 696 LLVVFVLGGFIISKDDIPSWLTWCYYTSPMMYGQTALVINEFLDERWGSPNNDTRINAKT 755
Query: 743 IGYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPL--QKPRTVIPQDDE 800
+G +L ++ +E YW+W+ + L+ + ++FN +AL YL+PL K TV+ + +
Sbjct: 756 VGEVLLKSRGFFTEPYWFWICIGALLGFTVLFNFCYIIALMYLNPLGNSKATTVVEEGKD 815
Query: 801 PEKSSSRDANYVFSTRSTKDESNTKGMILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETR 860
K S + ST KGM+LPFQPL++ F+NV+Y+VDMP E++ QG+ R
Sbjct: 816 KHKGSHSGTGVELT--STSSHGPKKGMVLPFQPLSLAFNNVNYYVDMPAEMKAQGVEGDR 873
Query: 861 LQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFA 920
LQLL +V G F PGVLTALVG SGAGKTTLMDVLAGRKTGGY+EG I ISGYPK Q TFA
Sbjct: 874 LQLLRDVGGAFRPGVLTALVGVSGAGKTTLMDVLAGRKTGGYVEGSINISGYPKNQATFA 933
Query: 921 RISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLVELDSLRNALV 980
R+SGY EQNDIHSP VT+ ESL +SA LRL +I T + FVE+VM+LVEL LRN++V
Sbjct: 934 RVSGYCEQNDIHSPHVTVYESLIYSAWLRLSADIDTKTREMFVEEVMELVELKPLRNSIV 993
Query: 981 GMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTV 1040
G+PG GLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTV
Sbjct: 994 GLPGVDGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTV 1053
Query: 1041 VCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPA 1100
VCTIHQPSIDIFE+FD+LLLMKRGG+VIY G LG SQ +++YF+ I G+ I GYNPA
Sbjct: 1054 VCTIHQPSIDIFESFDELLLMKRGGQVIYAGTLGHHSQKLVEYFEAIEGVPKIKDGYNPA 1113
Query: 1101 TWVLEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLL 1160
TW+L+VTTPS+E + DFA+I+ NS R + I E PP GS L F T Y+Q
Sbjct: 1114 TWMLDVTTPSMESQMSVDFAQIFVNSSVNRRNQELIKELSTPPPGSNDLYFRTKYAQPFS 1173
Query: 1161 SQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALY 1220
+Q C WK WR P YNA+R T + ++FG +FW G+K Q+L GA+Y
Sbjct: 1174 TQTKACFWKMYWSNWRYPQYNAIRFLMTVVIGVLFGLLFWQTGTKIEKEQDLNNFFGAMY 1233
Query: 1221 ASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGL 1280
A+ LF+G NA+TVQP V+IERTVFYREKAAGMYS I YA++Q +EI Y +Q V+ L
Sbjct: 1234 AAVLFLGATNAATVQPAVAIERTVFYREKAAGMYSAIPYAISQVAVEIMYNTIQTGVYTL 1293
Query: 1281 ITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLL 1340
I Y MI ++ T KFF + +M F YFT YGMM V L+P +A + S F S WNL
Sbjct: 1294 ILYSMIGYDWTVVKFFWFYYYMLTCFVYFTLYGMMLVALTPNYQIAGICLSFFLSFWNLF 1353
Query: 1341 SGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVET--KIIGPGFEGTVKEYLSLN 1398
SGFLIP IP WW W+Y+ PV WTL G+ITSQ+GD ++ I G G + ++K L
Sbjct: 1354 SGFLIPRPQIPIWWRWYYWASPVAWTLYGIITSQVGDRDSIVHITGVG-DMSLKTLLKNG 1412
Query: 1399 LGYDPKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
G+D + + V V +I++F +F +K LNFQ+R
Sbjct: 1413 FGFDYDFLPVVAV-----VHIAWILIFLFAFAYGIKFLNFQRR 1450
>At1g66950 ABC transporter, putative
Length = 1434
Score = 1437 bits (3721), Expect = 0.0
Identities = 722/1419 (50%), Positives = 965/1419 (67%), Gaps = 46/1419 (3%)
Query: 30 EEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPAGAGGRAD-NLMDVRKLSR 88
EED+ EL+WAA+ RLP+ R+ + +S A G+ + +D+ +L
Sbjct: 55 EEDDMELRWAAIERLPTFDRLRKGMLPQTS-------------ANGKIELEDIDLTRLEP 101
Query: 89 SHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRFSNLSVSADVQIGSRALP 148
++ +++ L+ ++DN + L ++ER DR+GIEVP IEVR+ N+SV DV+ SRALP
Sbjct: 102 KDKKHLMEMILSFVEEDNEKFLRDLRERTDRVGIEVPKIEVRYENISVEGDVRSASRALP 161
Query: 149 TLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLLAL 208
TL N T + LE+I + KR + IL ++SG++KP RMTLLLG P +GK++LL AL
Sbjct: 162 TLFNVTLNTLESILGFFHLLPSKRKKIQILKDISGIVKPSRMTLLLGPPSSGKTTLLQAL 221
Query: 209 AGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGAQE 268
AGKLD L+ +G ITY GHE EF ++TCAYISQ D H E+TVRE LDF+ RC G
Sbjct: 222 AGKLDDTLQMSGRITYCGHEFREFVPQKTCAYISQHDLHFGEMTVREILDFSGRCLGVGS 281
Query: 269 GFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVG 328
+ + ++ R E E I+P P+IDAFMK+ ++ G++ S+ TDY+LK+LGLDIC++ + G
Sbjct: 282 RYQLMS-ELSRREKEEGIKPDPKIDAFMKSIAISGQETSLVTDYVLKILGLDICADILAG 340
Query: 329 SDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATV 388
M RG+SGGQ+KR+TTGEM+VGP + LFMDEISTGLDSSTTFQI K ++ VH+ D T+
Sbjct: 341 DVMRRGISGGQKKRLTTGEMLVGPARALFMDEISTGLDSSTTFQICKFMRQLVHISDVTM 400
Query: 389 LMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVS 448
+++LLQPAPETFELFDD++LLSEG ++Y+GPR+NVLEFFE GF+ P RKG+ADFLQEV+
Sbjct: 401 IISLLQPAPETFELFDDIILLSEGQIVYQGPRDNVLEFFEYFGFQCPERKGVADFLQEVT 460
Query: 449 SRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSKCHPSALARTKY 508
S+KDQ QYW + Y +V + + F G + S PYDK+K H +AL KY
Sbjct: 461 SKKDQEQYWNKREQPYNYVSVSDFSSGFSTFHTGQKLTSEFRVPYDKAKTHSAALVTQKY 520
Query: 509 AVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLY 568
+S WE+ KACF RE LL+ R F+Y+FKT Q+ + +T T++LRT MH G +
Sbjct: 521 GISNWELFKACFDREWLLMKRNSFVYVFKTVQITIMSLITMTVYLRTEMHVGTVRDGQKF 580
Query: 569 VSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAV 628
A+FF L+++MFNG +EL+ + RLPVFYKQRD LFYP WA++L W+L++P S+IE+
Sbjct: 581 YGAMFFSLINVMFNGLAELAFTVMRLPVFYKQRDFLFYPPWAFALPAWLLKIPLSLIESG 640
Query: 629 IWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLV 688
IW + YYT+GFAPSA RF + +I R V++N+ G+ LL+
Sbjct: 641 IWIGLTYYTIGFAPSAARF--------------------LGAIGRTEVISNSIGTFTLLI 680
Query: 689 IFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRWMK---QSALGNNTIGY 745
+F LGGFII K I+PW W Y++SP+ YGQ AI +NEF RW + + T+G
Sbjct: 681 VFTLGGFIIAKDDIRPWMTWAYYMSPMMYGQTAIVMNEFLDERWSSPNYDTRINAKTVGE 740
Query: 746 NILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQKPR-TVIPQDDEPEKS 804
+L ++ +E YW+W+ + L+ ++++FN+ LAL YL+PL + TV+ + + +K
Sbjct: 741 VLLKSRGFFTEPYWFWICIVALLGFSLLFNLFYILALMYLNPLGNSKATVVEEGKDKQKG 800
Query: 805 SSRDAN-YVFSTRSTKDESNTKGMILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQL 863
+R V S+ ++ +GM+LPFQPL++ F+NV+Y+VDMP E++ QG+ RLQL
Sbjct: 801 ENRGTEGSVVELNSSSNKGPKRGMVLPFQPLSLAFNNVNYYVDMPSEMKAQGVEGDRLQL 860
Query: 864 LSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARIS 923
L +V G F PG+LTALVG SGAGKTTLMDVLAGRKTGGYIEG I ISGYPK Q TFAR+S
Sbjct: 861 LRDVGGAFRPGILTALVGVSGAGKTTLMDVLAGRKTGGYIEGSISISGYPKNQTTFARVS 920
Query: 924 GYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMP 983
GY EQNDIHSP VT+ ESL +SA LRL +I + FVE+VM+LVEL LRN++VG+P
Sbjct: 921 GYCEQNDIHSPHVTVYESLIYSAWLRLSTDIDIKTRELFVEEVMELVELKPLRNSIVGLP 980
Query: 984 GSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCT 1043
G GLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCT
Sbjct: 981 GVDGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCT 1040
Query: 1044 IHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWV 1103
IHQPSIDIFE+FD+LLLMKRGG+VIY G LG SQ +++YF+ + G+ I GYNPATW+
Sbjct: 1041 IHQPSIDIFESFDELLLMKRGGQVIYAGSLGHHSQKLVEYFEAVEGVPKINDGYNPATWM 1100
Query: 1104 LEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQF 1163
L+VTTPS+E + DFA+I++NS YR + I + PP GS+ + F T Y+QS +Q
Sbjct: 1101 LDVTTPSMESQMSLDFAQIFSNSSLYRRNQELIKDLSTPPPGSKDVYFKTKYAQSFSTQT 1160
Query: 1164 YRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASC 1223
C WKQ YWR P YNA+R T + ++FG +FW IG+K + Q+L GA+YA+
Sbjct: 1161 KACFWKQYWSYWRHPQYNAIRFLMTVVIGVLFGLIFWQIGTKTENEQDLNNFFGAMYAAV 1220
Query: 1224 LFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITY 1283
LF+G NA+TVQP ++IERTVFYREKAAGMYS I YA++Q +EI Y +Q V+ LI Y
Sbjct: 1221 LFLGALNAATVQPAIAIERTVFYREKAAGMYSAIPYAISQVAVEIMYNTIQTGVYTLILY 1280
Query: 1284 FMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGF 1343
MI T KF + +M +F YFT YGMM + L+P +A + S F SLWNL SGF
Sbjct: 1281 SMIGCNWTMAKFLWFYYYMLTSFIYFTLYGMMLMALTPNYQIAGICMSFFLSLWNLFSGF 1340
Query: 1344 LIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGF-EGTVKEYLSLNLGYD 1402
LIP IP WW W+Y+ PV WTL G+ITSQ+GD ++ + G + +K L G++
Sbjct: 1341 LIPRPQIPIWWRWYYWATPVAWTLYGLITSQVGDKDSMVHISGIGDIDLKTLLKEGFGFE 1400
Query: 1403 PKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
+ + V V +I+LF F +K LNFQ+R
Sbjct: 1401 HDFLPVVAV-----VHIAWILLFLFVFAYGIKFLNFQRR 1434
>At2g26910 putative ABC transporter
Length = 1420
Score = 1433 bits (3709), Expect = 0.0
Identities = 710/1438 (49%), Positives = 992/1438 (68%), Gaps = 40/1438 (2%)
Query: 19 SFARASNADTVEEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPAGAGGRAD 78
+F+R+++ EDEEEL+WAAL RLP+ RI ++R +G P
Sbjct: 8 AFSRSTSFKDEIEDEEELRWAALQRLPTYSRIRRGIFRDM--------VGEPKE------ 53
Query: 79 NLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRFSNLSVSA 138
+ + L S + L++ + + + + D + + +++R D + ++ P IEVRF NL V +
Sbjct: 54 --IQIGNLEASEQRLLLDRLVNSVENDPEQFFARVRKRFDAVDLKFPKIEVRFQNLMVES 111
Query: 139 DVQIGSRALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMTLLLGAPG 198
V +GSRALPT+ N+ + E + N+ + KR+ LTILD +SGVI+P R+TLLLG P
Sbjct: 112 FVHVGSRALPTIPNFIINMAEGLLRNIHVIGGKRNKLTILDGISGVIRPSRLTLLLGPPS 171
Query: 199 AGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAELTVRETLD 258
+GK++LLLALAG+L +NL+ +G ITYNG++ E RT AY+SQ D H AE+TVR+TL+
Sbjct: 172 SGKTTLLLALAGRLGTNLQTSGKITYNGYDLKEIIAPRTSAYVSQQDWHVAEMTVRQTLE 231
Query: 259 FAARCQGAQEGFAA-YTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVL 317
FA RCQG GF ++ R E I P ++D FMK+ ++GG + S+ +Y++K+L
Sbjct: 232 FAGRCQGV--GFKYDMLLELARREKLAGIVPDEDLDIFMKSLALGGMETSLVVEYVMKIL 289
Query: 318 GLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCI 377
GLD C++T+VG +M++G+SGGQ+KR+TTGE++VGP + LFMDEIS GLDSSTT QI+ +
Sbjct: 290 GLDTCADTLVGDEMIKGISGGQKKRLTTGELLVGPARVLFMDEISNGLDSSTTHQIIMYM 349
Query: 378 KNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPR 437
++ H ++ T +++LLQP+PET+ELFDD++L+SEG +IY+GPR+ VL+FF S+GF P R
Sbjct: 350 RHSTHALEGTTVISLLQPSPETYELFDDVILMSEGQIIYQGPRDEVLDFFSSLGFTCPDR 409
Query: 438 KGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSK 497
K +ADFLQEV+S+KDQ QYW+ P + Y++VP G+ AEAFR+ G + P+DK
Sbjct: 410 KNVADFLQEVTSKKDQQQYWSVPFRPYRYVPPGKFAEAFRSYPTGKKLAKKLEVPFDKRF 469
Query: 498 CHPSALARTKYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRM 557
H +AL+ ++Y V + E+ K FA + L+ + F+Y+FK Q+ V +T T+F RT M
Sbjct: 470 NHSAALSTSQYGVKKSELLKINFAWQKQLMKQNAFIYVFKFVQLLLVALITMTVFCRTTM 529
Query: 558 HPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWV 617
H GN+Y+ +L+F +V ++FNGF+E+ +++A+LPV YK RD FYP+WA++L +W+
Sbjct: 530 HHNTIDDGNIYLGSLYFSMVIILFNGFTEVPMLVAKLPVLYKHRDLHFYPSWAYTLPSWL 589
Query: 618 LRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVL 677
L +P SIIE+ W + YYT+G+ P RF + + F +HQM++GLFR+M S+ R M++
Sbjct: 590 LSIPTSIIESATWVAVTYYTIGYDPLFSRFLQQFLLYFSLHQMSLGLFRVMGSLGRHMIV 649
Query: 678 ANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRWMKQSA 737
ANTFGS A+LV+ LGGFII + I WWIWGYW+SPL Y Q A +VNEF W K +
Sbjct: 650 ANTFGSFAMLVVMTLGGFIISRDSIPSWWIWGYWISPLMYAQNAASVNEFLGHNWQKTA- 708
Query: 738 LGNNT---IGYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQKPRTV 794
GN+T +G +L +SL S +YWYW+ VA L+ Y ++FNI+ TL LA+L+P K + V
Sbjct: 709 -GNHTSDSLGLALLKERSLFSGNYWYWIGVAALLGYTVLFNILFTLFLAHLNPWGKFQAV 767
Query: 795 IPQDDEPEKSSSRDAN-YVFSTRSTKDESNT--------KGMILPFQPLTMTFHNVSYFV 845
+ +++ E+ R + +V R S + +GM+LPFQPL+++F N++Y+V
Sbjct: 768 VSREELDEREKKRKGDEFVVELREYLQHSGSIHGKYFKNRGMVLPFQPLSLSFSNINYYV 827
Query: 846 DMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEG 905
D+P +++QGI E RLQLL N++G F PGVLTALVG SGAGKTTLMDVLAGRKTGG IEG
Sbjct: 828 DVPLGLKEQGILEDRLQLLVNITGAFRPGVLTALVGVSGAGKTTLMDVLAGRKTGGTIEG 887
Query: 906 DIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQ 965
D+ ISG+PK Q TFARISGY EQND+HSP +T+ ESL FSA LRLP +I ++ +R FV +
Sbjct: 888 DVYISGFPKRQETFARISGYCEQNDVHSPCLTVVESLLFSACLRLPADIDSETQRAFVHE 947
Query: 966 VMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAI 1025
VM+LVEL SL ALVG+PG GLSTEQRKRLTIAVELVANPSI+FMDEPTSGLDARAAAI
Sbjct: 948 VMELVELTSLSGALVGLPGVDGLSTEQRKRLTIAVELVANPSIVFMDEPTSGLDARAAAI 1007
Query: 1026 VMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQ 1085
VMR VRN V+TGRT+VCTIHQPSIDIFE+FD+LL MKRGG +IY G LG +S +I YF+
Sbjct: 1008 VMRTVRNIVNTGRTIVCTIHQPSIDIFESFDELLFMKRGGELIYAGPLGQKSCELIKYFE 1067
Query: 1086 GIRGIRPIPRGYNPATWVLEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAG 1145
I G++ I G+NPA W+L+VT + E + DFAEIY NS+ + + I P
Sbjct: 1068 SIEGVQKIKPGHNPAAWMLDVTASTEEHRLGVDFAEIYRNSNLCQRNKELIEVLSKPSNI 1127
Query: 1146 SEPLKFDTIYSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSK 1205
++ ++F T YSQSL SQF CLWKQNL YWR+P Y A+R ++T + +L+ GT+ W GSK
Sbjct: 1128 AKEIEFPTRYSQSLYSQFVACLWKQNLSYWRNPQYTAVRFFYTVVISLMLGTICWKFGSK 1187
Query: 1206 RSSTQELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGL 1265
R + Q+L+ MG++YA+ LFIG+ NA+ QP+VSIER V YRE+AAGMYS + +A AQ
Sbjct: 1188 RDTQQQLFNAMGSMYAAVLFIGITNATAAQPVVSIERFVSYRERAAGMYSALPFAFAQVF 1247
Query: 1266 IEIPYIAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHL 1325
IE PY+ Q+ ++ I Y M FE +A KF YL FM+ + YFTFYGMM ++P ++
Sbjct: 1248 IEFPYVLAQSTIYSTIFYAMAAFEWSAVKFLWYLFFMYFSIMYFTFYGMMTTAITPNHNV 1307
Query: 1326 AAVISSAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKI-IG 1384
A++I++ FY LWNL SGF+IP IP WW W+Y+ PV WTL G++ SQ GD E + +
Sbjct: 1308 ASIIAAPFYMLWNLFSGFMIPYKRIPLWWRWYYWANPVAWTLYGLLVSQYGDDERSVKLS 1367
Query: 1385 PGF-EGTVKEYLSLNLGYDPKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
G + VK+ L +GY +G+ S I++ F + F F ++K NFQ+R
Sbjct: 1368 DGIHQVMVKQLLEDVMGYKHDFLGV-----SAIMVVAFCVFFSLVFAFAIKAFNFQRR 1420
>At3g53480 ABC transporter - like protein
Length = 1450
Score = 1352 bits (3500), Expect = 0.0
Identities = 694/1437 (48%), Positives = 950/1437 (65%), Gaps = 29/1437 (2%)
Query: 14 SFRSES--FARASNADTVEEDEE-ELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSP 70
SFRS S + ++ D + D E LQWA + RLP+ KR+ L S
Sbjct: 34 SFRSSSSIYEVENDGDVNDHDAEYALQWAEIERLPTVKRMRSTLLDDGDE--------SM 85
Query: 71 AGAGGRADNLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVR 130
G R ++DV KL R L+++K + + DN +LL I+ R+DR+G+E+P IEVR
Sbjct: 86 TEKGRR---VVDVTKLGAVERHLMIEKLIKHIENDNLKLLKKIRRRIDRVGMELPTIEVR 142
Query: 131 FSNLSVSADVQI-GSRALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGR 189
+ +L V A+ ++ +ALPTL N + L + G + + I+++V+G+IKPGR
Sbjct: 143 YESLKVVAECEVVEGKALPTLWNTAKRVLSELVKLTGA-KTHEAKINIINDVNGIIKPGR 201
Query: 190 MTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTA 249
+TLLLG P GK++LL AL+G L++NLK +G I+YNGH DEF ++T AYISQ D H A
Sbjct: 202 LTLLLGPPSCGKTTLLKALSGNLENNLKCSGEISYNGHRLDEFVPQKTSAYISQYDLHIA 261
Query: 250 ELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVN 309
E+TVRET+DF+ARCQG ++ + E E+ I P E+DA+MKA SV G + S+
Sbjct: 262 EMTVRETVDFSARCQGVGSR-TDIMMEVSKREKEKGIIPDTEVDAYMKAISVEGLQRSLQ 320
Query: 310 TDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSST 369
TDYILK+LGLDIC+E ++G M RG+SGGQ+KR+TT EMIVGP K LFMDEI+ GLDSST
Sbjct: 321 TDYILKILGLDICAEILIGDVMRRGISGGQKKRLTTAEMIVGPTKALFMDEITNGLDSST 380
Query: 370 TFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFES 429
FQIVK ++ F H+ ATVL++LLQPAPE+++LFDD++L+++G ++Y GPR VL FFE
Sbjct: 381 AFQIVKSLQQFAHISSATVLVSLLQPAPESYDLFDDIMLMAKGRIVYHGPRGEVLNFFED 440
Query: 430 IGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQ 489
GF+ P RKG+ADFLQEV S+KDQAQYW Y FV +++ F++ G +E
Sbjct: 441 CGFRCPERKGVADFLQEVISKKDQAQYWWHEDLPYSFVSVEMLSKKFKDLSIGKKIEDTL 500
Query: 490 THPYDKSKCHPSALARTKYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTC 549
+ PYD+SK H AL+ + Y++ WE+ AC +RE LL+ R F+YIFKT Q+ F+T
Sbjct: 501 SKPYDRSKSHKDALSFSVYSLPNWELFIACISREYLLMKRNYFVYIFKTAQLVMAAFITM 560
Query: 550 TIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAW 609
T+F+RTRM D +GN Y+SALFF L+ ++ +GF ELS+ RL VFYKQ+ FYPAW
Sbjct: 561 TVFIRTRMG-IDIIHGNSYMSALFFALIILLVDGFPELSMTAQRLAVFYKQKQLCFYPAW 619
Query: 610 AWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMA 669
A+++ VL+VP S E+++WT + YY +G+ P A RFF+ +LF +H +I +FR +A
Sbjct: 620 AYAIPATVLKVPLSFFESLVWTCLSYYVIGYTPEASRFFKQFILLFAVHFTSISMFRCLA 679
Query: 670 SIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTA 729
+I + +V + T GS +L F+ GF+IP + W WG+W +PL+YG+ ++VNEF A
Sbjct: 680 AIFQTVVASITAGSFGILFTFVFAGFVIPPPSMPAWLKWGFWANPLSYGEIGLSVNEFLA 739
Query: 730 SRWMKQSALGNNTIGYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQ 789
RW Q N T+G IL + + Y YWVS+ L+ + ++FNI+ TLAL +L
Sbjct: 740 PRW-NQMQPNNFTLGRTILQTRGMDYNGYMYWVSLCALLGFTVLFNIIFTLALTFLKSPT 798
Query: 790 KPRTVIPQDDEPEKSSSRDANYVFSTRSTKDESNTKG-----MILPFQPLTMTFHNVSYF 844
R +I QD E + + S R +S K M+LPF+PLT+TF +++YF
Sbjct: 799 SSRAMISQDKLSELQGTEKSTEDSSVRKKTTDSPVKTEEEDKMVLPFKPLTVTFQDLNYF 858
Query: 845 VDMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIE 904
VDMP E+R QG + +LQLLS+++G F PG+LTAL+G SGAGKTTL+DVLAGRKT GYIE
Sbjct: 859 VDMPVEMRDQGYDQKKLQLLSDITGAFRPGILTALMGVSGAGKTTLLDVLAGRKTSGYIE 918
Query: 905 GDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVE 964
GDI+ISG+PK Q TFAR+SGY EQ DIHSP +T+EES+ +SA LRL EI K +FV+
Sbjct: 919 GDIRISGFPKVQETFARVSGYCEQTDIHSPNITVEESVIYSAWLRLAPEIDATTKTKFVK 978
Query: 965 QVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAA 1024
QV++ +ELD ++++LVG+ G SGLSTEQRKRLTIAVELVANPSIIFMDEPT+GLDARAAA
Sbjct: 979 QVLETIELDEIKDSLVGVTGVSGLSTEQRKRLTIAVELVANPSIIFMDEPTTGLDARAAA 1038
Query: 1025 IVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYF 1084
IVMRAV+N DTGRT+VCTIHQPSIDIFEAFD+L+L+KRGGR+IY G LG S+ +I+YF
Sbjct: 1039 IVMRAVKNVADTGRTIVCTIHQPSIDIFEAFDELVLLKRGGRMIYTGPLGQHSRHIIEYF 1098
Query: 1085 QGIRGIRPIPRGYNPATWVLEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPA 1144
+ + I I +NPATW+L+V++ SVE + DFA+IY++S Y+ + + P +
Sbjct: 1099 ESVPEIPKIKDNHNPATWMLDVSSQSVEIELGVDFAKIYHDSALYKRNSELVKQLSQPDS 1158
Query: 1145 GSEPLKFDTIYSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGS 1204
GS ++F ++QS QF LWK NL YWRSP YN MRM T +S+L+FG +FW G
Sbjct: 1159 GSSDIQFKRTFAQSWWGQFKSILWKMNLSYWRSPSYNLMRMMHTLVSSLIFGALFWKQGQ 1218
Query: 1205 KRSSTQELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQG 1264
+ Q ++ V GA+Y LF+G+NN ++ ER V YRE+ AGMYS AYA+ Q
Sbjct: 1219 NLDTQQSMFTVFGAIYGLVLFLGINNCASALQYFETERNVMYRERFAGMYSATAYALGQV 1278
Query: 1265 LIEIPYIAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQH 1324
+ EIPYI +QA F ++TY MI F +A K F L MF + F + M V ++P
Sbjct: 1279 VTEIPYIFIQAAEFVIVTYPMIGFYPSAYKVFWSLYSMFCSLLTFNYLAMFLVSITPNFM 1338
Query: 1325 LAAVISSAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIG 1384
+AA++ S FY +NL SGFLIP++ +PGWWIW YY+ P WTL G I+SQ GD+ +I
Sbjct: 1339 VAAILQSLFYVGFNLFSGFLIPQTQVPGWWIWLYYLTPTSWTLNGFISSQYGDIHEEINV 1398
Query: 1385 PGFEGTVKEYLSLNLGYDPKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
G TV +L G+ ++ ++ V V F I F V LNFQ+R
Sbjct: 1399 FGQSTTVARFLKDYFGFHHDLLAVTAV-----VQIAFPIALASMFAFFVGKLNFQRR 1450
>At2g37280 putative ABC transporter
Length = 1413
Score = 1343 bits (3475), Expect = 0.0
Identities = 688/1432 (48%), Positives = 945/1432 (65%), Gaps = 26/1432 (1%)
Query: 14 SFRSESFARASNADTVEEDEEELQWAALSRLPSQKRINYALYRASSSRRQPPPLGSPAGA 73
SFRS S +R + D +E E LQWA + RLP+ KR+ +L +
Sbjct: 4 SFRSSS-SRNEHEDGGDEAEHALQWAEIQRLPTFKRLRSSLVDKYGEGTE---------- 52
Query: 74 GGRADNLMDVRKLSRSHRELVVKKALATNDQDNYRLLSAIKERLDRIGIEVPNIEVRFSN 133
+ ++DV KL R L+++K + + DN +LL I+ R++R+G+E P+IEVR+ +
Sbjct: 53 --KGKKVVDVTKLGAMERHLMIEKLIKHIENDNLKLLKKIRRRMERVGVEFPSIEVRYEH 110
Query: 134 LSVSADVQI-GSRALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSGVIKPGRMTL 192
L V A ++ +ALPTL N + + G+ R ++ IL +VSG+I PGR+TL
Sbjct: 111 LGVEAACEVVEGKALPTLWNSLKHVFLDLLKLSGV-RTNEANIKILTDVSGIISPGRLTL 169
Query: 193 LLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQTDNHTAELT 252
LLG PG GK++LL AL+G L++NLK G I+YNGH +E ++T AYISQ D H AE+T
Sbjct: 170 LLGPPGCGKTTLLKALSGNLENNLKCYGEISYNGHGLNEVVPQKTSAYISQHDLHIAEMT 229
Query: 253 VRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDY 312
RET+DF+ARCQG ++ + E + I P PEIDA+MKA SV G K S+ TDY
Sbjct: 230 TRETIDFSARCQGVGSR-TDIMMEVSKREKDGGIIPDPEIDAYMKAISVKGLKRSLQTDY 288
Query: 313 ILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQ 372
ILK+LGLDIC+ET+VG+ M RG+SGGQ+KR+TT EMIVGP K LFMDEI+ GLDSST FQ
Sbjct: 289 ILKILGLDICAETLVGNAMKRGISGGQKKRLTTAEMIVGPTKALFMDEITNGLDSSTAFQ 348
Query: 373 IVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGF 432
I+K ++ H+ +ATV ++LLQPAPE+++LFDD+VL++EG ++Y GPR++VL+FFE GF
Sbjct: 349 IIKSLQQVAHITNATVFVSLLQPAPESYDLFDDIVLMAEGKIVYHGPRDDVLKFFEECGF 408
Query: 433 KLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHP 492
+ P RKG+ADFLQEV S+KDQ QYW + + FV +++ F++ G +E + P
Sbjct: 409 QCPERKGVADFLQEVISKKDQGQYWLHQNLPHSFVSVDTLSKRFKDLEIGRKIEEALSKP 468
Query: 493 YDKSKCHPSALARTKYAVSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIF 552
YD SK H AL+ Y++ +WE+ +AC +RE LL+ R F+Y+FKT Q+ +T T+F
Sbjct: 469 YDISKTHKDALSFNVYSLPKWELFRACISREFLLMKRNYFVYLFKTFQLVLAAIITMTVF 528
Query: 553 LRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWS 612
+RTRM D +GN Y+S LFF V ++ +G ELS+ + RL VFYKQ+ FYPAWA++
Sbjct: 529 IRTRMD-IDIIHGNSYMSCLFFATVVLLVDGIPELSMTVQRLSVFYKQKQLCFYPAWAYA 587
Query: 613 LTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIA 672
+ VL++P S E+++WT + YY +G+ P RFFR ILF +H +I +FR +A+I
Sbjct: 588 IPATVLKIPLSFFESLVWTCLTYYVIGYTPEPYRFFRQFMILFAVHFTSISMFRCIAAIF 647
Query: 673 RDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRW 732
+ V A T GS +L+ F+ GF IP + W WG+W++P++Y + ++VNEF A RW
Sbjct: 648 QTGVAAMTAGSFVMLITFVFAGFAIPYTDMPGWLKWGFWVNPISYAEIGLSVNEFLAPRW 707
Query: 733 MKQSALGNNTIGYNILHAQSLPSEDYWYWVSVAVLVTYAIIFNIMVTLALAYLHPLQKPR 792
K N T+G IL ++ L +DY YWVS++ L+ IIFN + TLAL++L R
Sbjct: 708 QKMQPT-NVTLGRTILESRGLNYDDYMYWVSLSALLGLTIIFNTIFTLALSFLKSPTSSR 766
Query: 793 TVIPQDDEPEKSSSRDANYVFST---RSTKDESNTKGMILPFQPLTMTFHNVSYFVDMPQ 849
+I QD E ++D++ + S K + MILPF+PLT+TF +++Y+VD+P
Sbjct: 767 PMISQDKLSELQGTKDSSVKKNKPLDSSIKTNEDPGKMILPFKPLTITFQDLNYYVDVPV 826
Query: 850 EIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKI 909
E++ QG E +LQLLS ++G F PGVLTAL+G SGAGKTTL+DVLAGRKT GYIEG+I+I
Sbjct: 827 EMKGQGYNEKKLQLLSEITGAFRPGVLTALMGISGAGKTTLLDVLAGRKTSGYIEGEIRI 886
Query: 910 SGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKL 969
SG+ K Q TFAR+SGY EQ DIHSP +T+EESL +SA LRL EI+ K FV+QV++
Sbjct: 887 SGFLKVQETFARVSGYCEQTDIHSPSITVEESLIYSAWLRLVPEINPQTKIRFVKQVLET 946
Query: 970 VELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRA 1029
+EL+ +++ALVG+ G SGLSTEQRKRLT+AVELVANPSIIFMDEPT+GLDARAAAIVMRA
Sbjct: 947 IELEEIKDALVGVAGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTTGLDARAAAIVMRA 1006
Query: 1030 VRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRG 1089
V+N +TGRT+VCTIHQPSI IFEAFD+L+L+KRGGR+IY G LG S +I+YFQ I G
Sbjct: 1007 VKNVAETGRTIVCTIHQPSIHIFEAFDELVLLKRGGRMIYSGPLGQHSSCVIEYFQNIPG 1066
Query: 1090 IRPIPRGYNPATWVLEVTTPSVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPL 1149
+ I YNPATW+LEVT+ SVE +D DFA+IYN SD Y+ + E P GS L
Sbjct: 1067 VAKIRDKYNPATWMLEVTSESVETELDMDFAKIYNESDLYKNNSELVKELSKPDHGSSDL 1126
Query: 1150 KFDTIYSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSST 1209
F ++Q+ QF CLWK +L YWRSP YN MR+ T IS+ +FG +FW+ G K +
Sbjct: 1127 HFKRTFAQNWWEQFKSCLWKMSLSYWRSPSYNLMRIGHTFISSFIFGLLFWNQGKKIDTQ 1186
Query: 1210 QELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIP 1269
Q L+ V+GA+Y LF+G+NN ++ ER V YRE+ AGMYS AYA+AQ + EIP
Sbjct: 1187 QNLFTVLGAIYGLVLFVGINNCTSALQYFETERNVMYRERFAGMYSAFAYALAQVVTEIP 1246
Query: 1270 YIAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVI 1329
YI +Q+ F ++ Y MI F + K F L MF F + M + ++P +AA++
Sbjct: 1247 YIFIQSAEFVIVIYPMIGFYASFSKVFWSLYAMFCNLLCFNYLAMFLISITPNFMVAAIL 1306
Query: 1330 SSAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGFEG 1389
S F++ +N+ +GFLIP+ IP WW+WFYYI P WTL +SQ GD+ KI G
Sbjct: 1307 QSLFFTTFNIFAGFLIPKPQIPKWWVWFYYITPTSWTLNLFFSSQYGDIHQKINAFGETK 1366
Query: 1390 TVKEYLSLNLGYDPKIMGISTVGLSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
TV +L G+ + ++ I+L F I + V LNFQKR
Sbjct: 1367 TVASFLEDYFGFHH-----DRLMITAIILIAFPIALATMYAFFVAKLNFQKR 1413
>At4g15230 ABC transporter like protein
Length = 979
Score = 514 bits (1325), Expect = e-145
Identities = 275/629 (43%), Positives = 386/629 (60%), Gaps = 73/629 (11%)
Query: 827 MILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAG 886
+ILPF+PLT+TF NV Y+++ PQ +TR QLLS+++G PGVLT+L+G SGAG
Sbjct: 410 IILPFKPLTVTFQNVQYYIETPQG-------KTR-QLLSDITGALKPGVLTSLMGVSGAG 461
Query: 887 KTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSA 946
KTTL+DVL+GRKT G I+G+IK+ GYPK Q TFAR+SGY EQ DIHSP +T+EESL +SA
Sbjct: 462 KTTLLDVLSGRKTRGIIKGEIKVGGYPKVQETFARVSGYCEQFDIHSPNITVEESLKYSA 521
Query: 947 SLRLPKEISTDKKR--------------EFVEQVMKLVELDSLRNALVGMPGSSGLSTEQ 992
LRLP I + K E V++V++ VELD +++++VG+PG SGLS EQ
Sbjct: 522 WLRLPYNIDSKTKNVRNYTLKTNRLKEIELVKEVLETVELDDIKDSVVGLPGISGLSIEQ 581
Query: 993 RKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIF 1052
RKRLTIAVELVANPSIIFMDEPT+GLDARAAAIVMRAV+N +TGRTVVCTIHQPSIDIF
Sbjct: 582 RKRLTIAVELVANPSIIFMDEPTTGLDARAAAIVMRAVKNVAETGRTVVCTIHQPSIDIF 641
Query: 1053 EAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTPSVE 1112
E FD+L+LMK GG+++Y G G S +I+YF+
Sbjct: 642 ETFDELILMKNGGQLVYYGPPGQNSSKVIEYFEN-------------------------- 675
Query: 1113 ETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQFYRCLWKQNL 1172
+ + + GSE L+F + +SQ+ Q CLWKQ+
Sbjct: 676 --------------------KMVVEQLSSASLGSEALRFPSQFSQTAWVQLKACLWKQHY 715
Query: 1173 VYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIGVNNAS 1232
YWR+P +N R+ F + + + G +FW ++ Q+L + G++Y +F G+NN +
Sbjct: 716 SYWRNPSHNITRIVFILLDSTLCGLLFWQKAEDINNQQDLISIFGSMYTLVVFPGMNNCA 775
Query: 1233 TVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMINFERTA 1292
V ++ ER VFYRE+ A MYS AY+ +Q LIE+PY +Q+++ +I Y I + +
Sbjct: 776 AVINFIAAERNVFYRERFARMYSSWAYSFSQVLIEVPYSLLQSLLCTIIVYPTIGYHMSV 835
Query: 1293 GKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPESHIPG 1352
K F L +F + F + GM+ V L+P H+A + S+F+S+ NL +GF+IP+ IP
Sbjct: 836 YKMFWSLYSIFCSLLIFNYSGMLMVALTPNIHMAVTLRSSFFSMLNLFAGFVIPKQKIPK 895
Query: 1353 WWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGFEGTVKEYLSLNLGYDPKIMGISTVG 1412
WWIW YY+ P W L G+++SQ GDV+ +I+ G + V +L GY + ++
Sbjct: 896 WWIWMYYLSPTSWVLEGLLSSQYGDVDKEILVFGEKKRVSAFLEDYFGYKHE-----SLA 950
Query: 1413 LSVIVLFGFIILFFCSFVVSVKVLNFQKR 1441
+ VL + I+ F + L+FQK+
Sbjct: 951 VVAFVLIAYPIIVATLFAFFMSKLSFQKK 979
Score = 330 bits (845), Expect = 5e-90
Identities = 159/349 (45%), Positives = 226/349 (64%), Gaps = 19/349 (5%)
Query: 275 KDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRG 334
K+I R+E + I P P +DA+MK +LGLDIC++T VG G
Sbjct: 2 KEISRMEKLQEIIPDPAVDAYMK------------------ILGLDICADTRVGDATRPG 43
Query: 335 VSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQ 394
+SGG+++R+TTGE++VGP TLFMDEIS GLDSSTTFQIV C++ H+ +AT+L++LLQ
Sbjct: 44 ISGGEKRRLTTGELVVGPATTLFMDEISNGLDSSTTFQIVSCLQQLAHIAEATILISLLQ 103
Query: 395 PAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQA 454
PAPETFELFDD++L+ EG +IY PR ++ FFE GFK P RKG+ADFLQE+ S+KDQ
Sbjct: 104 PAPETFELFDDVILMGEGKIIYHAPRADICRFFEEFGFKCPERKGVADFLQEIMSKKDQE 163
Query: 455 QYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWE 514
QYW K Y ++ F+ S G ++ + P++KS+ L KY++ +WE
Sbjct: 164 QYWCHRDKPYSYISVDSFINKFKESNLGLLLKEELSKPFNKSQTRKDGLCYKKYSLGKWE 223
Query: 515 ISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFF 574
+ KAC RE LL+ R F+Y+FK+ + F VT T+FL+ TD +GN + +LF
Sbjct: 224 MLKACSRREFLLMKRNSFIYLFKSALLVFNALVTMTVFLQVGA-TTDSLHGNYLMGSLFT 282
Query: 575 GLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYS 623
L ++ +G EL+L I+RL VF KQ+D FYPAWA+++ + +L++P S
Sbjct: 283 ALFRLLADGLPELTLTISRLGVFCKQKDLYFYPAWAYAIPSIILKIPLS 331
Score = 171 bits (432), Expect = 4e-42
Identities = 140/557 (25%), Positives = 255/557 (45%), Gaps = 82/557 (14%)
Query: 167 IYRPKRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNG 226
I P+ + +L +++G +KPG +T L+G GAGK++LL L+G+ + K G I G
Sbjct: 428 IETPQGKTRQLLSDITGALKPGVLTSLMGVSGAGKTTLLDVLSGRKTRGIIK-GEIKVGG 486
Query: 227 HEADEFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNI 286
+ + R Y Q D H+ +TV E+L ++A + Y D + +N RN
Sbjct: 487 YPKVQETFARVSGYCEQFDIHSPNITVEESLKYSAWLR------LPYNID-SKTKNVRNY 539
Query: 287 RPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTG 346
+K + + K +L+ + LD +++VG + G+S QRKR+T
Sbjct: 540 T--------LKTNRL---KEIELVKEVLETVELDDIKDSVVGLPGISGLSIEQRKRLTIA 588
Query: 347 EMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDL 406
+V +FMDE +TGLD+ +++ +KN TV+ + QP+ + FE FD+L
Sbjct: 589 VELVANPSIIFMDEPTTGLDARAAAIVMRAVKNVAET-GRTVVCTIHQPSIDIFETFDEL 647
Query: 407 VLLSEG-HVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQ 465
+L+ G ++Y GP + SK +
Sbjct: 648 ILMKNGGQLVYYGPPGQ------------------------------------NSSKVIE 671
Query: 466 FVPSGEIAEAFRNSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREAL 525
+ + + E ++ GS E+L+ PS ++T W KAC ++
Sbjct: 672 YFENKMVVEQLSSASLGS--EALR---------FPSQFSQTA-----WVQLKACLWKQHY 715
Query: 526 LISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEA-----YGNLYVSALFFGLVHMM 580
R I + + + +F + ++ +G++Y +F G M
Sbjct: 716 SYWRNPSHNITRIVFILLDSTLCGLLFWQKAEDINNQQDLISIFGSMYTLVVFPG----M 771
Query: 581 FNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGF 640
N + ++ + A VFY++R Y +WA+S + ++ VPYS++++++ T+IVY T+G+
Sbjct: 772 NNCAAVINFIAAERNVFYRERFARMYSSWAYSFSQVLIEVPYSLLQSLLCTIIVYPTIGY 831
Query: 641 APSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKG 700
S + F ++ +F + +M ++ ++ +A T S+ ++ L GF+IPK
Sbjct: 832 HMSVYKMFWSLYSIFCSLLIFNYSGMLMVALTPNIHMAVTLRSSFFSMLNLFAGFVIPKQ 891
Query: 701 MIKPWWIWGYWLSPLTY 717
I WWIW Y+LSP ++
Sbjct: 892 KIPKWWIWMYYLSPTSW 908
Score = 66.2 bits (160), Expect = 1e-10
Identities = 78/337 (23%), Positives = 151/337 (44%), Gaps = 28/337 (8%)
Query: 952 KEISTDKKRE------FVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVAN 1005
KEIS +K + V+ MK++ LD + VG G+S +++RLT +V
Sbjct: 2 KEISRMEKLQEIIPDPAVDAYMKILGLDICADTRVGDATRPGISGGEKRRLTTGELVVGP 61
Query: 1006 PSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGR-TVVCTIHQPSIDIFEAFDDLLLMKRG 1064
+ +FMDE ++GLD+ ++ ++ T++ ++ QP+ + FE FDD++LM
Sbjct: 62 ATTLFMDEISNGLDSSTTFQIVSCLQQLAHIAEATILISLLQPAPETFELFDDVILMGE- 120
Query: 1065 GRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTPSVEETI----DADFA 1120
G++IY + ++ G + P A ++ E+ + +E D ++
Sbjct: 121 GKIIYHAPRADICRFFEEF--GFK----CPERKGVADFLQEIMSKKDQEQYWCHRDKPYS 174
Query: 1121 EIYNNS--DQYRGVEASIL---EFEHPPAGSEPLKFDTIYSQSLLSQF---YRCLWKQNL 1172
I +S ++++ +L E P S+ K Y + L ++ C ++ L
Sbjct: 175 YISVDSFINKFKESNLGLLLKEELSKPFNKSQTRKDGLCYKKYSLGKWEMLKACSRREFL 234
Query: 1173 VYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIGVNNAS 1232
+ R+ + +ALV TVF +G+ S Y +MG+L+ + + +
Sbjct: 235 LMKRNSFIYLFKSALLVFNALVTMTVFLQVGATTDSLHGNY-LMGSLFTALFRLLADGLP 293
Query: 1233 TVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIP 1269
+ +S VF ++K Y AYA+ +++IP
Sbjct: 294 ELTLTIS-RLGVFCKQKDLYFYPAWAYAIPSIILKIP 329
>At3g55110 ABC transporter - like protein
Length = 708
Score = 233 bits (593), Expect = 8e-61
Identities = 157/570 (27%), Positives = 281/570 (48%), Gaps = 40/570 (7%)
Query: 833 PLTMTFHNVSYFVDMPQEI----RKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKT 888
P ++F+N+SY V + + RK +T LL +++G G + A++G SGAGK+
Sbjct: 60 PFLLSFNNLSYNVVLRRRFDFSRRKTASVKT---LLDDITGEARDGEILAVLGGSGAGKS 116
Query: 889 TLMDVLAGRKTGGYIEGDIKISGYPKEQ-RTFARISGYVEQNDIHSPQVTIEESLWFSAS 947
TL+D LAGR ++G + ++G Q R IS YV Q+D+ P +T++E+L F++
Sbjct: 117 TLIDALAGRVAEDSLKGTVTLNGEKVLQSRLLKVISAYVMQDDLLFPMLTVKETLMFASE 176
Query: 948 LRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPS 1007
RLP+ + KK E VE ++ + L + + ++G G G+S +R+R++I ++++ +P
Sbjct: 177 FRLPRSLPKSKKMERVETLIDQLGLRNAADTVIGDEGHRGVSGGERRRVSIGIDIIHDPI 236
Query: 1008 IIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRV 1067
++F+DEPTSGLD+ A +V++ ++ +G V+ +IHQPS I D L+++ G V
Sbjct: 237 LLFLDEPTSGLDSTNAFMVVQVLKRIAQSGSVVIMSIHQPSARIIGLLDRLIILSHGKSV 296
Query: 1068 IYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTP---SVEETID-ADFAEIY 1123
G + + S +F RPIP N + L+V S E T D +F E +
Sbjct: 297 FNGSPVSLPS-----FFSSFG--RPIPEKENITEFALDVIRELEGSSEGTRDLVEFNEKW 349
Query: 1124 NNSDQYRGVEASILEFEHPPA-------------GSEPLKFDTI--YSQSLLSQFYRCLW 1168
+ R S + + A G+ P+ +T+ Y+ L++ + L
Sbjct: 350 QQNQTARATTQSRVSLKEAIAASVSRGKLVSGSSGANPISMETVSSYANPPLAETF-ILA 408
Query: 1169 KQNLVYW-RSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIG 1227
K+ + W R+P MR+ ++ L+ TV+W + + QE ++ +
Sbjct: 409 KRYIKNWIRTPELIGMRIGTVMVTGLLLATVYWRLDNTPRGAQERMGFFAFGMSTMFYCC 468
Query: 1228 VNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMIN 1287
+N P+ ER +F RE Y +Y ++ L+ +P + ++ F T++ +
Sbjct: 469 ADNI----PVFIQERYIFLRETTHNAYRTSSYVISHALVSLPQLLALSIAFAATTFWTVG 524
Query: 1288 FERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPE 1347
FF Y L ++ F + GL P ++ +++ A+ S LL GF I
Sbjct: 525 LSGGLESFFYYCLIIYAAFWSGSSIVTFISGLIPNVMMSYMVTIAYLSYCLLLGGFYINR 584
Query: 1348 SHIPGWWIWFYYICPVQWTLRGVITSQLGD 1377
IP +WIWF+YI +++ V+ ++ D
Sbjct: 585 DRIPLYWIWFHYISLLKYPYEAVLINEFDD 614
Score = 167 bits (423), Expect = 4e-41
Identities = 141/598 (23%), Positives = 268/598 (44%), Gaps = 55/598 (9%)
Query: 144 SRALPTLINYTRDALETICTN-LGIYRPKRHSL-TILDNVSGVIKPGRMTLLLGAPGAGK 201
+R++P L+++ + + R K S+ T+LD+++G + G + +LG GAGK
Sbjct: 56 TRSVPFLLSFNNLSYNVVLRRRFDFSRRKTASVKTLLDDITGEARDGEILAVLGGSGAGK 115
Query: 202 SSLLLALAGKLDSNLKKTGSITYNGHEA-DEFFVKRTCAYISQTDNHTAELTVRETLDFA 260
S+L+ ALAG++ + K G++T NG + +K AY+ Q D LTV+ETL FA
Sbjct: 116 STLIDALAGRVAEDSLK-GTVTLNGEKVLQSRLLKVISAYVMQDDLLFPMLTVKETLMFA 174
Query: 261 ARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLD 320
+ F S+ K + ++ LGL
Sbjct: 175 SE--------------------------------FRLPRSLPKSKKMERVETLIDQLGLR 202
Query: 321 ICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNF 380
++T++G + RGVSGG+R+RV+ G I+ LF+DE ++GLDS+ F +V+ +K
Sbjct: 203 NAADTVIGDEGHRGVSGGERRRVSIGIDIIHDPILLFLDEPTSGLDSTNAFMVVQVLKRI 262
Query: 381 VHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGI 440
+ V+M++ QP+ L D L++LS G ++ G ++ FF S G +P ++ I
Sbjct: 263 AQ-SGSVVIMSIHQPSARIIGLLDRLIILSHGKSVFNGSPVSLPSFFSSFGRPIPEKENI 321
Query: 441 ADF----LQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAFRNSRFGSYVESLQTHPYDKS 496
+F ++E+ + + + ++++Q + A A SR S E++
Sbjct: 322 TEFALDVIRELEGSSEGTRDLVEFNEKWQ---QNQTARATTQSRV-SLKEAIAASVSRGK 377
Query: 497 KCHPSALARTKYAVSRWEISKACFAREALLISRQRFLYI-------FKTCQVAFVGFVTC 549
S+ A + + A +L R +I + V G +
Sbjct: 378 LVSGSSGANPISMETVSSYANPPLAETFILAKRYIKNWIRTPELIGMRIGTVMVTGLLLA 437
Query: 550 TIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAW 609
T++ R P + FG+ M + + + I +F ++ + Y
Sbjct: 438 TVYWRLDNTPRG---AQERMGFFAFGMSTMFYCCADNIPVFIQERYIFLRETTHNAYRTS 494
Query: 610 AWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMA 669
++ +++ ++ +P + ++ + ++TVG + FF Y I++ + ++
Sbjct: 495 SYVISHALVSLPQLLALSIAFAATTFWTVGLSGGLESFFYYCLIIYAAFWSGSSIVTFIS 554
Query: 670 SIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGYWLSPLTYGQRAITVNEF 727
+ +++++ A L LLGGF I + I +WIW +++S L Y A+ +NEF
Sbjct: 555 GLIPNVMMSYMVTIAYLSYCLLLGGFYINRDRIPLYWIWFHYISLLKYPYEAVLINEF 612
>At3g55100 ABC transporter - like protein
Length = 662
Score = 229 bits (583), Expect = 1e-59
Identities = 153/591 (25%), Positives = 287/591 (47%), Gaps = 39/591 (6%)
Query: 829 LPFQPLTMTFHNVSYFVDMPQE--IRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAG 886
+P P + F++++Y V + Q +R P LL+ ++G G + A++G+SGAG
Sbjct: 15 IPPIPFVLAFNDLTYNVTLQQRFGLRFGHSPAKIKTLLNGITGEAKEGEILAILGASGAG 74
Query: 887 KTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSA 946
K+TL+D LAG+ G ++G + ++G + R IS YV Q D+ P +T+EE+L F+A
Sbjct: 75 KSTLIDALAGQIAEGSLKGTVTLNGEALQSRLLRVISAYVMQEDLLFPMLTVEETLMFAA 134
Query: 947 SLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANP 1006
RLP+ +S KKR VE ++ + L +++N ++G G G+S +R+R++I +++ +P
Sbjct: 135 EFRLPRSLSKSKKRNRVETLIDQLGLTTVKNTVIGDEGHRGVSGGERRRVSIGTDIIHDP 194
Query: 1007 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGR 1066
++F+DEPTSGLD+ +A +V++ ++ +G V+ +IHQPS I E D ++++ G+
Sbjct: 195 IVLFLDEPTSGLDSTSAFMVVQVLKKIARSGSIVIMSIHQPSGRIMEFLDRVIVLS-SGQ 253
Query: 1067 VIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTPSVEETIDADFAEIYNNS 1126
+++ S + F G PIP N A + L++ +N +
Sbjct: 254 IVFS-----DSPATLPLFFSEFG-SPIPEKENIAEFTLDLIKDLEGSPEGTRGLVEFNRN 307
Query: 1127 DQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQFYRCL------W--------KQNL 1172
Q+R + S + + E + +I L+S YR + W K+ +
Sbjct: 308 WQHRKLRVSQEPHHNSSSLGEAIN-ASISRGKLVSTSYRSIPSYVNPWWVETVILAKRYM 366
Query: 1173 VYW-RSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQE----LYVVMGALYASCLFIG 1227
+ W R+P R++ ++ + TV+W + QE M ++ SC
Sbjct: 367 INWTRTPELIGTRVFIVMMTGFLLATVYWKVDDSPRGVQERLSFFSFAMATMFYSC---- 422
Query: 1228 VNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMIN 1287
+ P ER +F RE A Y +Y ++ L+ +P++ ++ F T++ +
Sbjct: 423 ----ADGLPAFIQERYIFLRETAHNAYRRSSYVISHSLVTLPHLFALSIGFAATTFWFVG 478
Query: 1288 FERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPE 1347
F YL+ +F +F + G+ P ++ +++ + S L SGF +
Sbjct: 479 LNGGLAGFIYYLMIIFASFWSGCSFVTFVSGVIPNVMMSYMVTFGYLSYCLLFSGFYVNR 538
Query: 1348 SHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKII--GPGFEGTVKEYLS 1396
I +WIW +YI +++ V+ ++ D + F+ T+ E +S
Sbjct: 539 DRIHLYWIWIHYISLLKYPYEAVLHNEFDDPSRCFVRGNQVFDNTIMEGVS 589
Score = 163 bits (412), Expect = 8e-40
Identities = 128/557 (22%), Positives = 243/557 (42%), Gaps = 45/557 (8%)
Query: 176 TILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVK 235
T+L+ ++G K G + +LGA GAGKS+L+ ALAG++ K G++T NG ++
Sbjct: 50 TLLNGITGEAKEGEILAILGASGAGKSTLIDALAGQIAEGSLK-GTVTLNGEALQSRLLR 108
Query: 236 RTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAF 295
AY+ Q D LTV ETL FAA F
Sbjct: 109 VISAYVMQEDLLFPMLTVEETLMFAAE--------------------------------F 136
Query: 296 MKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKT 355
S+ K + ++ LGL T++G + RGVSGG+R+RV+ G I+
Sbjct: 137 RLPRSLSKSKKRNRVETLIDQLGLTTVKNTVIGDEGHRGVSGGERRRVSIGTDIIHDPIV 196
Query: 356 LFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVI 415
LF+DE ++GLDS++ F +V+ +K + V+M++ QP+ E D +++LS G ++
Sbjct: 197 LFLDEPTSGLDSTSAFMVVQVLKKIAR-SGSIVIMSIHQPSGRIMEFLDRVIVLSSGQIV 255
Query: 416 YEGPRENVLEFFESIGFKLPPRKGIADF----LQEVSSRKDQAQYWADPSKQYQFVPSGE 471
+ + FF G +P ++ IA+F ++++ + + + ++ +Q
Sbjct: 256 FSDSPATLPLFFSEFGSPIPEKENIAEFTLDLIKDLEGSPEGTRGLVEFNRNWQHRKLRV 315
Query: 472 IAEAFRNSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREALL-ISRQ 530
E NS S E++ + K ++ V+ W + A+ ++ +R
Sbjct: 316 SQEPHHNS--SSLGEAINA-SISRGKLVSTSYRSIPSYVNPWWVETVILAKRYMINWTRT 372
Query: 531 RFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFSELSLM 590
L + V GF+ T++ + P +S F + M ++ L
Sbjct: 373 PELIGTRVFIVMMTGFLLATVYWKVDDSPRGV---QERLSFFSFAMATMFYSCADGLPAF 429
Query: 591 IARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRY 650
I +F ++ + Y ++ +++ ++ +P+ ++ + ++ VG F Y
Sbjct: 430 IQERYIFLRETAHNAYRRSSYVISHSLVTLPHLFALSIGFAATTFWFVGLNGGLAGFIYY 489
Query: 651 MFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGY 710
+ I+F ++ + +++++ L L GF + + I +WIW +
Sbjct: 490 LMIIFASFWSGCSFVTFVSGVIPNVMMSYMVTFGYLSYCLLFSGFYVNRDRIHLYWIWIH 549
Query: 711 WLSPLTYGQRAITVNEF 727
++S L Y A+ NEF
Sbjct: 550 YISLLKYPYEAVLHNEF 566
>At2g01320 putative ABC transporter
Length = 725
Score = 218 bits (556), Expect = 2e-56
Identities = 145/529 (27%), Positives = 268/529 (50%), Gaps = 23/529 (4%)
Query: 863 LLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGG---YIEGDIKISGYPKEQRTF 919
LL NVSG PG L A++G SG+GKTTL++VLAG+ + ++ G ++++G P + +
Sbjct: 90 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLSLSPRLHLSGLLEVNGKPSSSKAY 149
Query: 920 ARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLVELDSLRNAL 979
+V Q D+ Q+T+ E+L F+A L+LP+ S +++ E+V ++ + L S ++
Sbjct: 150 KL--AFVRQEDLFFSQLTVRETLSFAAELQLPEISSAEERDEYVNNLLLKLGLVSCADSC 207
Query: 980 VGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRT 1039
VG G+S ++KRL++A EL+A+PS+IF DEPT+GLDA A VM ++ G T
Sbjct: 208 VGDAKVRGISGGEKKRLSLACELIASPSVIFADEPTTGLDAFQAEKVMETLQKLAQDGHT 267
Query: 1040 VVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNP 1099
V+C+IHQP ++ FDD++L+ G ++Y G G + + YF + P NP
Sbjct: 268 VICSIHQPRGSVYAKFDDIVLLTE-GTLVYAGPAGKEP---LTYFGNFGFL--CPEHVNP 321
Query: 1100 ATWVLEVTTP--SVEETIDADFAEIYNNSDQYRGVEASILEF------EHPPAGSEPLKF 1151
A ++ ++ + S ET+ + ++ D + +S+L E G P +
Sbjct: 322 AEFLADLISVDYSSSETVYSSQKRVHALVDAFSQRSSSVLYATPLSMKEETKNGMRPRRK 381
Query: 1152 DTI-YSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQ 1210
+ + QF+ L + + R P N +R + SA++FG+VFW +G ++S Q
Sbjct: 382 AIVERTDGWWRQFFLLLKRAWMQASRDGPTNKVRARMSVASAVIFGSVFWRMGKSQTSIQ 441
Query: 1211 ELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPY 1270
+ MG L + + + + + ER + RE++ G YS Y +++ + EIP
Sbjct: 442 DR---MGLLQVAAINTAMAALTKTVGVFPKERAIVDRERSKGSYSLGPYLLSKTIAEIPI 498
Query: 1271 IAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVIS 1330
A ++FG + Y M T +F + + + + G+ + P+ A +
Sbjct: 499 GAAFPLMFGAVLYPMARLNPTLSRFGKFCGIVTVESFAASAMGLTVGAMVPSTEAAMAVG 558
Query: 1331 SAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVE 1379
+ +++ + G+ + + P + W ++W +G+ ++ ++
Sbjct: 559 PSLMTVFIVFGGYYVNADNTPIIFRWIPRASLIRWAFQGLCINEFSGLK 607
Score = 135 bits (339), Expect = 2e-31
Identities = 137/574 (23%), Positives = 239/574 (40%), Gaps = 53/574 (9%)
Query: 177 ILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSN--LKKTGSITYNGHEADEFFV 234
+L NVSG KPGR+ ++G G+GK++LL LAG+L + L +G + NG +
Sbjct: 90 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLSLSPRLHLSGLLEVNGKPSSSKAY 149
Query: 235 KRTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDA 294
K A++ Q D ++LTVRETL FAA Q PEI
Sbjct: 150 K--LAFVRQEDLFFSQLTVRETLSFAAELQ------------------------LPEI-- 181
Query: 295 FMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRK 354
SS + VN +L LGL C+++ VG +RG+SGG++KR++ ++
Sbjct: 182 ----SSAEERDEYVNN--LLLKLGLVSCADSCVGDAKVRGISGGEKKRLSLACELIASPS 235
Query: 355 TLFMDEISTGLDSSTTFQIVKCIKNFVHLMD--ATVLMALLQPAPETFELFDDLVLLSEG 412
+F DE +TGLD+ FQ K ++ L TV+ ++ QP + FDD+VLL+EG
Sbjct: 236 VIFADEPTTGLDA---FQAEKVMETLQKLAQDGHTVICSIHQPRGSVYAKFDDIVLLTEG 292
Query: 413 HVIYEGPR-ENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGE 471
++Y GP + L +F + GF P A+FL ++ S Y + +
Sbjct: 293 TLVYAGPAGKEPLTYFGNFGFLCPEHVNPAEFLADLIS----VDYSSSETVYSSQKRVHA 348
Query: 472 IAEAF-RNSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREALLISRQ 530
+ +AF + S Y L K+ P A + W R + SR
Sbjct: 349 LVDAFSQRSSSVLYATPLSMKEETKNGMRPRRKAIVERTDGWWRQFFLLLKRAWMQASRD 408
Query: 531 RFLYIFKTCQVAFVGFVTCTIFLRTRMHPT--DEAYGNLYVSALFFGLVHMMFNGFSELS 588
+ + ++F R T + G L V+A+ + + +
Sbjct: 409 GPTNKVRARMSVASAVIFGSVFWRMGKSQTSIQDRMGLLQVAAINTAMAALT----KTVG 464
Query: 589 LMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFF 648
+ + ++R Y + L+ + +P +++ ++Y P+ RF
Sbjct: 465 VFPKERAIVDRERSKGSYSLGPYLLSKTIAEIPIGAAFPLMFGAVLYPMARLNPTLSRFG 524
Query: 649 RYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIW 708
++ I+ V A + + ++ A G + + V + GG+ + + W
Sbjct: 525 KFCGIVTVESFAASAMGLTVGAMVPSTEAAMAVGPSLMTVFIVFGGYYVNADNTPIIFRW 584
Query: 709 GYWLSPLTYGQRAITVNEFTASRWMKQSALGNNT 742
S + + + + +NEF+ ++ Q+ T
Sbjct: 585 IPRASLIRWAFQGLCINEFSGLKFDHQNTFDVQT 618
>At2g37360 ABC transporter like protein
Length = 755
Score = 217 bits (552), Expect = 4e-56
Identities = 154/576 (26%), Positives = 271/576 (46%), Gaps = 46/576 (7%)
Query: 833 PLTMTFHNVSYFVDMPQEI------RKQGIPE---TRLQLLSNVSGVFSPGVLTALVGSS 883
P ++F +++Y V + ++ R+ G T++ LL+ +SG G + A++G+S
Sbjct: 95 PFVLSFTDLTYSVKIQKKFNPLACCRRSGNDSSVNTKI-LLNGISGEAREGEMMAVLGAS 153
Query: 884 GAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLW 943
G+GK+TL+D LA R + G I ++G E IS YV Q+D+ P +T+EE+L
Sbjct: 154 GSGKSTLIDALANRIAKDSLRGSITLNGEVLESSMQKVISAYVMQDDLLFPMLTVEETLM 213
Query: 944 FSASLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELV 1003
FSA RLP+ +S KK+ V+ ++ + L S ++G G G+S +R+R++I +++
Sbjct: 214 FSAEFRLPRSLSKKKKKARVQALIDQLGLRSAAKTVIGDEGHRGVSGGERRRVSIGNDII 273
Query: 1004 ANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKR 1063
+P I+F+DEPTSGLD+ +A +V++ ++ +G V+ +IHQPS I D L+ + +
Sbjct: 274 HDPIILFLDEPTSGLDSTSAYMVIKVLQRIAQSGSIVIMSIHQPSYRIMGLLDQLIFLSK 333
Query: 1064 GGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEV----------TTPSVE- 1112
G V G + +F + PIP N + L++ T P VE
Sbjct: 334 GNTVYSGSPTHLP-----QFFSEFK--HPIPENENKTEFALDLIRELEYSTEGTKPLVEF 386
Query: 1113 -ETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQS-----LLSQFYRC 1166
+ A A YNN+++ +S+ E L + S F
Sbjct: 387 HKQWRAKQAPSYNNNNKRNTNVSSLKEAITASISRGKLVSGATNNNSSNLTPSFQTFANP 446
Query: 1167 LWKQNLVYW--------RSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGA 1218
W + +V R P MR+ ++ ++ T+F ++ + QE
Sbjct: 447 FWIEMIVIGKRAILNSRRQPELLGMRLGAVMVTGIILATMFTNLDNSPKGAQERL----G 502
Query: 1219 LYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVF 1278
+A + + P+ ER +F RE A Y +Y ++Q +I IP + V + F
Sbjct: 503 FFAFAMSTTFYTCAEAIPVFLQERYIFMRETAYNAYRRSSYVLSQSIISIPALIVLSASF 562
Query: 1279 GLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWN 1338
T++ + + A FF + + +F + + G+ P L + A + +
Sbjct: 563 AATTFWAVGLDGGANGFFFFYFTILASFWAGSSFVTFLSGVIPNVMLGFTVVVAILAYFL 622
Query: 1339 LLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQ 1374
L SGF I IP +W+WF+YI V++ GV+ ++
Sbjct: 623 LFSGFFISRDRIPVYWLWFHYISLVKYPYEGVLQNE 658
Score = 160 bits (406), Expect = 4e-39
Identities = 131/566 (23%), Positives = 238/566 (41%), Gaps = 54/566 (9%)
Query: 177 ILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKR 236
+L+ +SG + G M +LGA G+GKS+L+ ALA ++ + + GSIT NG + K
Sbjct: 133 LLNGISGEAREGEMMAVLGASGSGKSTLIDALANRIAKDSLR-GSITLNGEVLESSMQKV 191
Query: 237 TCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFM 296
AY+ Q D LTV ETL F+A F
Sbjct: 192 ISAYVMQDDLLFPMLTVEETLMFSAE--------------------------------FR 219
Query: 297 KASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTL 356
S+ KK ++ LGL ++T++G + RGVSGG+R+RV+ G I+ L
Sbjct: 220 LPRSLSKKKKKARVQALIDQLGLRSAAKTVIGDEGHRGVSGGERRRVSIGNDIIHDPIIL 279
Query: 357 FMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIY 416
F+DE ++GLDS++ + ++K ++ + V+M++ QP+ L D L+ LS+G+ +Y
Sbjct: 280 FLDEPTSGLDSTSAYMVIKVLQRIAQ-SGSIVIMSIHQPSYRIMGLLDQLIFLSKGNTVY 338
Query: 417 EGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEIAEAF 476
G ++ +FF +P + +F ++ + + P ++ + A ++
Sbjct: 339 SGSPTHLPQFFSEFKHPIPENENKTEFALDLIRELEYSTEGTKPLVEFHKQWRAKQAPSY 398
Query: 477 -----RNSRFGSYVESLQTHPYDKSKCHPSALARTK---------YAVSRWEISKACFAR 522
RN+ S E++ T + K A +A W I +
Sbjct: 399 NNNNKRNTNVSSLKEAI-TASISRGKLVSGATNNNSSNLTPSFQTFANPFW-IEMIVIGK 456
Query: 523 EALLIS-RQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFGLVHMMF 581
A+L S RQ L + V G + T+F P + F + +
Sbjct: 457 RAILNSRRQPELLGMRLGAVMVTGIILATMFTNLDNSPKG---AQERLGFFAFAMSTTFY 513
Query: 582 NGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFA 641
+ + + +F ++ Y ++ L+ ++ +P I+ + + ++ VG
Sbjct: 514 TCAEAIPVFLQERYIFMRETAYNAYRRSSYVLSQSIISIPALIVLSASFAATTFWAVGLD 573
Query: 642 PSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGM 701
A FF + F + ++ + +++L T A L L GF I +
Sbjct: 574 GGANGFFFFYFTILASFWAGSSFVTFLSGVIPNVMLGFTVVVAILAYFLLFSGFFISRDR 633
Query: 702 IKPWWIWGYWLSPLTYGQRAITVNEF 727
I +W+W +++S + Y + NEF
Sbjct: 634 IPVYWLWFHYISLVKYPYEGVLQNEF 659
>At3g53510 ABC transporter -like protein
Length = 739
Score = 211 bits (536), Expect = 3e-54
Identities = 153/606 (25%), Positives = 271/606 (44%), Gaps = 38/606 (6%)
Query: 797 QDDEPEKSSSRD----ANYVFSTRSTKDESNTKGMILPFQPLTMTFHNVSYFVDMP---- 848
+D+ ++S + D +N+ S S++ S++ +L F+ LT + F P
Sbjct: 53 EDEGDDQSRAMDIAVASNFWSSVPSSRVPSSSP-FVLSFKDLTYSVKIKKKFKPFPCCGN 111
Query: 849 QEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIK 908
+ LL+ +SG G + A++G+SG+GK+TL+D LA R + + GDI
Sbjct: 112 SPFDGNDMEMNTKVLLNGISGEAREGEMMAVLGASGSGKSTLIDALANRISKESLRGDIT 171
Query: 909 ISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMK 968
++G E IS YV Q+D+ P +T+EE+L FSA RLP +S KK+ V+ ++
Sbjct: 172 LNGEVLESSLHKVISAYVMQDDLLFPMLTVEETLMFSAEFRLPSSLSKKKKKARVQALID 231
Query: 969 LVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMR 1028
+ L + ++G G G+S +R+R++I +++ +P I+F+DEPTSGLD+ +A +V++
Sbjct: 232 QLGLRNAAKTVIGDEGHRGVSGGERRRVSIGTDIIHDPIILFLDEPTSGLDSTSAYMVVK 291
Query: 1029 AVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIR 1088
++ +G V+ +IHQPS I D L+ + R G +Y G Q ++
Sbjct: 292 VLQRIAQSGSIVIMSIHQPSYRILGLLDKLIFLSR-GNTVYSGSPTHLPQFFSEFG---- 346
Query: 1089 GIRPIPRGYNPATWVLEVT---------TPSVEETIDADFAEIYNNSDQYRGVEASILEF 1139
PIP N + L++ T S+ E + +S R S+ +
Sbjct: 347 --HPIPENENKPEFALDLIRELEDSPEGTKSLVE-FHKQWRAKQTSSQSRRNTNVSLKDA 403
Query: 1140 EHPPAGSEPLKFDTIYSQSLLSQFYRCLWKQNLVYW--------RSPPYNAMRMYFTTIS 1191
L +S F W + LV R P +R+ ++
Sbjct: 404 ISASISRGKLVSGATNLRSSFQTFANPFWTEMLVIGKRSILNSRRQPELFGIRLGAVLVT 463
Query: 1192 ALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAA 1251
++ T+FW + + QE +A + + P+ ER +F RE A
Sbjct: 464 GMILATIFWKLDNSPRGIQERL----GFFAFAMSTTFYTCAEAIPVFLQERYIFMRETAY 519
Query: 1252 GMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTF 1311
Y +Y +A +I IP + + + F T+ + + F + + F +
Sbjct: 520 NAYRRSSYVLAHTIISIPALIILSAAFAASTFSAVGLAGGSEGFLFFFFTILTAFWAGSS 579
Query: 1312 YGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVI 1371
+ G+ + + A + + L SGF I IP +WIWF+Y+ V++ GV+
Sbjct: 580 FVTFLSGVVSHVMIGFTVVVAILAYFLLFSGFFISRDRIPLYWIWFHYLSLVKYPYEGVL 639
Query: 1372 TSQLGD 1377
++ D
Sbjct: 640 QNEFED 645
Score = 162 bits (409), Expect = 2e-39
Identities = 131/557 (23%), Positives = 230/557 (40%), Gaps = 45/557 (8%)
Query: 177 ILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKR 236
+L+ +SG + G M +LGA G+GKS+L+ ALA ++ + G IT NG + K
Sbjct: 126 LLNGISGEAREGEMMAVLGASGSGKSTLIDALANRISKESLR-GDITLNGEVLESSLHKV 184
Query: 237 TCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFM 296
AY+ Q D LTV ETL F+A F
Sbjct: 185 ISAYVMQDDLLFPMLTVEETLMFSAE--------------------------------FR 212
Query: 297 KASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTL 356
SS+ KK ++ LGL ++T++G + RGVSGG+R+RV+ G I+ L
Sbjct: 213 LPSSLSKKKKKARVQALIDQLGLRNAAKTVIGDEGHRGVSGGERRRVSIGTDIIHDPIIL 272
Query: 357 FMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIY 416
F+DE ++GLDS++ + +VK ++ + V+M++ QP+ L D L+ LS G+ +Y
Sbjct: 273 FLDEPTSGLDSTSAYMVVKVLQRIAQ-SGSIVIMSIHQPSYRILGLLDKLIFLSRGNTVY 331
Query: 417 EGPRENVLEFFESIGFKLPPRKG----IADFLQEVSSRKDQAQYWADPSKQYQFVPSGEI 472
G ++ +FF G +P + D ++E+ + + + KQ++ +
Sbjct: 332 SGSPTHLPQFFSEFGHPIPENENKPEFALDLIRELEDSPEGTKSLVEFHKQWRAKQTSSQ 391
Query: 473 AEAFRNSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREALLISRQRF 532
+ N + + + S + +A W R L RQ
Sbjct: 392 SRRNTNVSLKDAISASISRGKLVSGATNLRSSFQTFANPFWTEMLVIGKRSILNSRRQPE 451
Query: 533 LYIFKTCQVAFVGFVTCTIFLRTRMHP--TDEAYGNLYVSALFFGLVHMMFNGFSELSLM 590
L+ + V G + TIF + P E G F + + + +
Sbjct: 452 LFGIRLGAVLVTGMILATIFWKLDNSPRGIQERLG-----FFAFAMSTTFYTCAEAIPVF 506
Query: 591 IARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAGRFFRY 650
+ +F ++ Y ++ L + ++ +P II + + + VG A + F +
Sbjct: 507 LQERYIFMRETAYNAYRRSSYVLAHTIISIPALIILSAAFAASTFSAVGLAGGSEGFLFF 566
Query: 651 MFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPWWIWGY 710
F + ++ + +++ T A L L GF I + I +WIW +
Sbjct: 567 FFTILTAFWAGSSFVTFLSGVVSHVMIGFTVVVAILAYFLLFSGFFISRDRIPLYWIWFH 626
Query: 711 WLSPLTYGQRAITVNEF 727
+LS + Y + NEF
Sbjct: 627 YLSLVKYPYEGVLQNEF 643
>At5g06530 ABC transporter like protein
Length = 627
Score = 207 bits (526), Expect = 5e-53
Identities = 166/628 (26%), Positives = 300/628 (47%), Gaps = 64/628 (10%)
Query: 830 PFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTT 889
P P+ + F +V+Y V + K+ ++L+ +SG +PG + AL+G SG+GKTT
Sbjct: 27 PTLPIFLKFRDVTYKV-----VIKKLTSSVEKEILTGISGSVNPGEVLALMGPSGSGKTT 81
Query: 890 LMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLR 949
L+ +LAGR + G + + P + ++I G+V Q+D+ P +T++E+L ++A LR
Sbjct: 82 LLSLLAGRISQSSTGGSVTYNDKPYSKYLKSKI-GFVTQDDVLFPHLTVKETLTYAARLR 140
Query: 950 LPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSII 1009
LPK ++ ++K++ V++ + L+ ++ ++G G+S +RKR++I E++ NPS++
Sbjct: 141 LPKTLTREQKKQRALDVIQELGLERCQDTMIGGAFVRGVSGGERKRVSIGNEIIINPSLL 200
Query: 1010 FMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIY 1069
+DEPTSGLD+ A + + + + G+TV+ TIHQPS +F FD L+L+ RG + +
Sbjct: 201 LLDEPTSGLDSTTALRTILMLHDIAEAGKTVITTIHQPSSRLFHRFDKLILLGRGSLLYF 260
Query: 1070 GGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTTP-----SVEETIDADFAEIYN 1124
G +S +DYF I G P+ NPA ++L++ SV +D D ++ N
Sbjct: 261 G-----KSSEALDYFSSI-GCSPLI-AMNPAEFLLDLANGNINDISVPSELD-DRVQVGN 312
Query: 1125 NSDQYRGVEAS--------ILEFEHPPAGSE------PLKFDT---IYSQSLLSQFYRCL 1167
+ + + + S + +E A E P+ D S L Q+ C
Sbjct: 313 SGRETQTGKPSPAAVHEYLVEAYETRVAEQEKKKLLDPVPLDEEAKAKSTRLKRQWGTCW 372
Query: 1168 WKQNLVYW-------RSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALY 1220
W+Q + + R ++ +R+ +A++ G ++W S + L G L+
Sbjct: 373 WEQYCILFCRGLKERRHEYFSWLRVTQVLSTAVILGLLWWQ--SDIRTPMGLQDQAGLLF 430
Query: 1221 ASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGL 1280
+F G T ER + +E+AA MY AY +A+ ++P + +F L
Sbjct: 431 FIAVFWGFFPVFTAIFAFPQERAMLNKERAADMYRLSAYFLARTTSDLPLDFILPSLFLL 490
Query: 1281 ITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLL 1340
+ YFM + FFL +L +FL G+ + A ++S + L
Sbjct: 491 VVYFMTGLRISPYPFFLSMLTVFLCIIAAQGLGLAIGAILMDLKKATTLASVTVMTFMLA 550
Query: 1341 SGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGFEGTVKEYLSLNLG 1400
GF + + +P + W Y+ T + ++ Q D I G +
Sbjct: 551 GGFFVKK--VPVFISWIRYLSFNYHTYKLLLKVQYQDFAVSINGMRIDN----------- 597
Query: 1401 YDPKIMGISTVGLSVIVLFGFIILFFCS 1428
G++ V V+++FG+ +L + S
Sbjct: 598 ------GLTEVAALVVMIFGYRLLAYLS 619
Score = 163 bits (412), Expect = 8e-40
Identities = 166/660 (25%), Positives = 275/660 (41%), Gaps = 84/660 (12%)
Query: 133 NLSVSA-DVQIGSR--------ALPTLINYTRDALETICTNLGIYRPKRHSLTILDNVSG 183
N++VSA D++ G + LP + + + + L K IL +SG
Sbjct: 6 NVTVSAEDIEAGKKKPKFQAEPTLPIFLKFRDVTYKVVIKKLTSSVEKE----ILTGISG 61
Query: 184 VIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEADEFFVKRTCAYISQ 243
+ PG + L+G G+GK++LL LAG++ S GS+TYN ++ +K +++Q
Sbjct: 62 SVNPGEVLALMGPSGSGKTTLLSLLAGRI-SQSSTGGSVTYNDKPYSKY-LKSKIGFVTQ 119
Query: 244 TDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSPEIDAFMKASSVGG 303
D LTV+ETL +AAR + K + R + ++
Sbjct: 120 DDVLFPHLTVKETLTYAARLR--------LPKTLTREQKKQRALD--------------- 156
Query: 304 KKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIVGPRKTLFMDEIST 363
+++ LGL+ C +T++G +RGVSGG+RKRV+ G I+ L +DE ++
Sbjct: 157 ---------VIQELGLERCQDTMIGGAFVRGVSGGERKRVSIGNEIIINPSLLLLDEPTS 207
Query: 364 GLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLSEGHVIYEGPRENV 423
GLDS+T + + + + TV+ + QP+ F FD L+LL G ++Y G
Sbjct: 208 GLDSTTALRTILMLHDIAE-AGKTVITTIHQPSSRLFHRFDKLILLGRGSLLYFGKSSEA 266
Query: 424 LEFFESIG-----------FKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSGEI 472
L++F SIG F L G + + S D+ Q + PS
Sbjct: 267 LDYFSSIGCSPLIAMNPAEFLLDLANGNINDISVPSELDDRVQVGNSGRETQTGKPSPAA 326
Query: 473 AEAFRNSRFGSYV------ESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREALL 526
+ + + V + L P D+ S + ++ WE F R L
Sbjct: 327 VHEYLVEAYETRVAEQEKKKLLDPVPLDEEAKAKSTRLKRQWGTCWWEQYCILFCR-GLK 385
Query: 527 ISRQRFLYIFKTCQV----AFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFN 582
R + + QV +G + +RT M D+A G L+ A+F+G F
Sbjct: 386 ERRHEYFSWLRVTQVLSTAVILGLLWWQSDIRTPMGLQDQA-GLLFFIAVFWG----FFP 440
Query: 583 GFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAP 642
F+ + + K+R Y A+ L +P I ++ ++VY+ G
Sbjct: 441 VFTAIFAFPQERAMLNKERAADMYRLSAYFLARTTSDLPLDFILPSLFLLVVYFMTGLRI 500
Query: 643 SAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMI 702
S FF M +F+ A GL + +I D+ A T S ++ L GGF + K +
Sbjct: 501 SPYPFFLSMLTVFLCIIAAQGLGLAIGAILMDLKKATTLASVTVMTFMLAGGFFVKKVPV 560
Query: 703 KPWWI----WGY----WLSPLTYGQRAITVNEFTASRWMKQ-SALGNNTIGYNILHAQSL 753
WI + Y L + Y A+++N + + +AL GY +L SL
Sbjct: 561 FISWIRYLSFNYHTYKLLLKVQYQDFAVSINGMRIDNGLTEVAALVVMIFGYRLLAYLSL 620
>At1g53270 putative ABC transporter emb|AAD22683.1
Length = 590
Score = 204 bits (519), Expect = 3e-52
Identities = 156/572 (27%), Positives = 286/572 (49%), Gaps = 46/572 (8%)
Query: 863 LLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARI 922
+L +VS +TA+ G SGAGKTTL+++LAG+ + G + G + ++G P + + R+
Sbjct: 50 ILKDVSCDARSAEITAIAGPSGAGKTTLLEILAGKVSHGKVSGQVLVNGRPMDGPEYRRV 109
Query: 923 SGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKREFVEQVMKLVE---LDSLRNAL 979
SG+V Q D P +T++E+L +SA LRL K+++ +V +L++ L+ + ++
Sbjct: 110 SGFVPQEDALFPFLTVQETLTYSALLRL-----KTKRKDAAAKVKRLIQELGLEHVADSR 164
Query: 980 VGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRN-TVDTGR 1038
+G SG+S +R+R++I VELV +P++I +DEPTSGLD+ +A V+ +++ T+ G+
Sbjct: 165 IGQGSRSGISGGERRRVSIGVELVHDPNVILIDEPTSGLDSASALQVVTLLKDMTIKQGK 224
Query: 1039 TVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYN 1098
T+V TIHQP I E D ++L+ G V G + +I Q IPR N
Sbjct: 225 TIVLTIHQPGFRILEQIDRIVLLSNGMVVQNGSVYSLHQKIKFSGHQ-------IPRRVN 277
Query: 1099 PATWVLEVTTPSVEETIDADFAEI--YNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYS 1156
+ +++ S+E EI Y +S ++ S AG E + D+ +S
Sbjct: 278 VLEYAIDI-AGSLEPIRTQSCREISCYGHSKTWKSCYIS--------AGGELHQSDS-HS 327
Query: 1157 QSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVM 1216
S+L + + +R+ R +I+ L+ G+++ ++G+++ +E V+
Sbjct: 328 NSVLEEVQILGQRSCKNIFRTKQLFTTRALQASIAGLILGSIYLNVGNQK---KEAKVLR 384
Query: 1217 GALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAM 1276
+A L +++ + PI +R + RE + Y ++Y +A LI IP++ + +M
Sbjct: 385 TGFFAFILTFLLSSTTEGLPIFLQDRRILMRETSRRAYRVLSYVLADTLIFIPFLLIISM 444
Query: 1277 VFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSL 1336
+F Y+++ R F + L +++ + L P + + S
Sbjct: 445 LFATPVYWLVGLRRELDGFLYFSLVIWIVLLMSNSFVACFSALVPNFIMGTSVISGLMGS 504
Query: 1337 WNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQL-GDVETKIIGPGFEGTVKEYL 1395
+ L SG+ I + IP +W + +Y+ ++ ++ ++ GDV K + +KE
Sbjct: 505 FFLFSGYFIAKDRIPVYWEFMHYLSLFKYPFECLMINEYRGDVFLK------QQDLKESQ 558
Query: 1396 S-LNLGYDPKIMGISTVGLSVIVLFGFIILFF 1426
NLG IM VG V+ GF IL++
Sbjct: 559 KWSNLG----IMASFIVGYRVL---GFFILWY 583
Score = 138 bits (348), Expect = 2e-32
Identities = 125/573 (21%), Positives = 244/573 (41%), Gaps = 67/573 (11%)
Query: 171 KRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEAD 230
++ IL +VS + +T + G GAGK++LL LAGK+ S+ K +G + NG D
Sbjct: 44 EKEEKVILKDVSCDARSAEITAIAGPSGAGKTTLLEILAGKV-SHGKVSGQVLVNGRPMD 102
Query: 231 EFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSP 290
+R ++ Q D LTV+ETL ++A + RL+ +R
Sbjct: 103 GPEYRRVSGFVPQEDALFPFLTVQETLTYSA---------------LLRLKTKRK----- 142
Query: 291 EIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTGEMIV 350
DA K +++ LGL+ +++ +G G+SGG+R+RV+ G +V
Sbjct: 143 --DAAAKVKR------------LIQELGLEHVADSRIGQGSRSGISGGERRRVSIGVELV 188
Query: 351 GPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLLS 410
+ +DE ++GLDS++ Q+V +K+ T+++ + QP E D +VLLS
Sbjct: 189 HDPNVILIDEPTSGLDSASALQVVTLLKDMTIKQGKTIVLTIHQPGFRILEQIDRIVLLS 248
Query: 411 EGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPSG 470
G V+ G ++ + + G ++P R + ++ +
Sbjct: 249 NGMVVQNGSVYSLHQKIKFSGHQIPRRVNVLEY-------------------------AI 283
Query: 471 EIAEAFRNSRFGSYVE-SLQTHPYDKSKCHPSALAR----TKYAVSRWEISKACFAREAL 525
+IA + R S E S H C+ SA ++ S E + R
Sbjct: 284 DIAGSLEPIRTQSCREISCYGHSKTWKSCYISAGGELHQSDSHSNSVLEEVQILGQRSCK 343
Query: 526 LISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFGLVHMMFNGFS 585
I R + L+ + Q + G + +I+L + L F L ++ +
Sbjct: 344 NIFRTKQLFTTRALQASIAGLILGSIYLNVGNQKKEAKV--LRTGFFAFILTFLLSSTTE 401
Query: 586 ELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTVGFAPSAG 645
L + + + ++ Y ++ L + ++ +P+ +I ++++ VY+ VG
Sbjct: 402 GLPIFLQDRRILMRETSRRAYRVLSYVLADTLIFIPFLLIISMLFATPVYWLVGLRRELD 461
Query: 646 RFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIPKGMIKPW 705
F + +++++ M+ +++ + ++ + S + FL G+ I K I +
Sbjct: 462 GFLYFSLVIWIVLLMSNSFVACFSALVPNFIMGTSVISGLMGSFFLFSGYFIAKDRIPVY 521
Query: 706 WIWGYWLSPLTYGQRAITVNEFTASRWMKQSAL 738
W + ++LS Y + +NE+ ++KQ L
Sbjct: 522 WEFMHYLSLFKYPFECLMINEYRGDVFLKQQDL 554
>At3g25620 membrane transporter, putative
Length = 646
Score = 202 bits (514), Expect = 1e-51
Identities = 172/617 (27%), Positives = 294/617 (46%), Gaps = 38/617 (6%)
Query: 787 PLQKPRTVIPQDDEP--EKSSSRDANYVFSTRSTKDESNTKGMILPFQPLTMTFHNV--- 841
P+Q+ R P P + + D S +S+ + + +IL F+ LT + +
Sbjct: 23 PVQENRFSSPSHVNPCLDDDNDHDGPSHQSRQSSVLRQSLRPIILKFEELTYSIKSQTGK 82
Query: 842 -SYFVDMPQEIRKQGIPETRLQLLSNVSGVFSPGVLTALVGSSGAGKTTLMDVLAGRKTG 900
SY+ + P+ +L VSG+ PG L A++G SG+GKTTL+ LAGR G
Sbjct: 83 GSYWFGSQE-------PKPNRLVLKCVSGIVKPGELLAMLGPSGSGKTTLVTALAGRLQG 135
Query: 901 GYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWFSASLRLPKEISTDKKR 960
+ G + +G P + R +G+V Q+D+ P +T+ E+L ++A LRLPKE++ +K
Sbjct: 136 K-LSGTVSYNGEPFTS-SVKRKTGFVTQDDVLYPHLTVMETLTYTALLRLPKELTRKEKL 193
Query: 961 EFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDA 1020
E VE V+ + L N+++G G+S +RKR++I E++ NPS++ +DEPTSGLD+
Sbjct: 194 EQVEMVVSDLGLTRCCNSVIGGGLIRGISGGERKRVSIGQEMLVNPSLLLLDEPTSGLDS 253
Query: 1021 RAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRGGRVIYGGKLGVQSQIM 1080
AA ++ +R+ GRTVV TIHQPS ++ FD +L++ G IY G G +
Sbjct: 254 TTAARIVATLRSLARGGRTVVTTIHQPSSRLYRMFDKVLVLSE-GCPIYSGDSG----RV 308
Query: 1081 IDYFQGIRGIRPIPRGYNPATWVLEVTTPSVEETIDADFAEIYNNSD---QYRGVEASIL 1137
++YF I G +P NPA +VL++ +T D E D + V+ S++
Sbjct: 309 MEYFGSI-GYQPGSSFVNPADFVLDLANGITSDTKQYDQIETNGRLDRLEEQNSVKQSLI 367
Query: 1138 EFEHPPAGSEPLKFDTIYSQSLLSQFYRCLWKQNLVYWRSPPYNAMRMYFTTISALVFGT 1197
+ PLK + S++ ++ + R P + M F+ + L G
Sbjct: 368 S-SYKKNLYPPLKEEV--SRTFPQDQTNARLRKKAITNRWP--TSWWMQFSVL--LKRGL 420
Query: 1198 VFWDIGSKRSSTQELYVVMGALYASCLFIGVNNASTVQPIVSIERTVFYREKAAGMYSPI 1257
++W S L +G L+ +F G ER + +E+++G+Y
Sbjct: 421 LWW-----HSRVAHLQDQVGLLFFFSIFWGFFPLFNAIFTFPQERPMLIKERSSGIYRLS 475
Query: 1258 AYAVAQGLIEIPYIAVQAMVFGLITYFMINFERTAGKFFLYLLFMFLTFTYFTFYGMMAV 1317
+Y +A+ + ++P + +F ITY+M + + F + L+ + G+
Sbjct: 476 SYYIARTVGDLPMELILPTIFVTITYWMGGLKPSLTTFIMTLMIVLYNVLVAQGVGLALG 535
Query: 1318 GLSPTQHLAAVISSAFYSLWNLLSGFLIPESHIPGWWIWFYYICPVQWTLRGVITSQLGD 1377
+ AA +SS ++ L G+ I HIPG+ W Y+ + + ++ Q
Sbjct: 536 AILMDAKKAATLSSVLMLVFLLAGGYYI--QHIPGFIAWLKYVSFSHYCYKLLVGVQYTW 593
Query: 1378 VETKIIGPGFEGTVKEY 1394
E G G +V +Y
Sbjct: 594 DEVYECGSGLHCSVMDY 610
Score = 171 bits (434), Expect = 2e-42
Identities = 148/548 (27%), Positives = 251/548 (45%), Gaps = 88/548 (16%)
Query: 166 GIYRPKRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYN 225
G PK + L +L VSG++KPG + +LG G+GK++L+ ALAG+L L +G+++YN
Sbjct: 88 GSQEPKPNRL-VLKCVSGIVKPGELLAMLGPSGSGKTTLVTALAGRLQGKL--SGTVSYN 144
Query: 226 GHEADEFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERN 285
G E VKR +++Q D LTV ETL + A + K++ R E
Sbjct: 145 G-EPFTSSVKRKTGFVTQDDVLYPHLTVMETLTYTALLR--------LPKELTRKEKLEQ 195
Query: 286 IRPSPEIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTT 345
+ + ++ LGL C +++G ++RG+SGG+RKRV+
Sbjct: 196 V------------------------EMVVSDLGLTRCCNSVIGGGLIRGISGGERKRVSI 231
Query: 346 G-EMIVGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFD 404
G EM+V P L +DE ++GLDS+T +IV +++ TV+ + QP+ + +FD
Sbjct: 232 GQEMLVNP-SLLLLDEPTSGLDSTTAARIVATLRSLAR-GGRTVVTTIHQPSSRLYRMFD 289
Query: 405 DLVLLSEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQY 464
+++LSEG IY G V+E+F SIG++ + F+ D A +KQY
Sbjct: 290 KVLVLSEGCPIYSGDSGRVMEYFGSIGYQPG-----SSFVNPADFVLDLANGITSDTKQY 344
Query: 465 QFVPSG------EIAEAFRNSRFGSYVESL---------QTHPYDKSKCHPSALARTKYA 509
+ + E + + S SY ++L +T P D++ +A R K
Sbjct: 345 DQIETNGRLDRLEEQNSVKQSLISSYKKNLYPPLKEEVSRTFPQDQT----NARLRKKAI 400
Query: 510 VSRWEISKACFAREALLISRQRFLYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYV 569
+RW S + + ++L+ R ++ +R+ + G L+
Sbjct: 401 TNRWPTS--WWMQFSVLLKRG-------------------LLWWHSRVAHLQDQVGLLFF 439
Query: 570 SALFFGLVHMMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVI 629
++F+G F F+ + P+ K+R + Y ++ + V +P +I I
Sbjct: 440 FSIFWG----FFPLFNAIFTFPQERPMLIKERSSGIYRLSSYYIARTVGDLPMELILPTI 495
Query: 630 WTVIVYYTVGFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVI 689
+ I Y+ G PS F + I+ +A G+ + +I D A T S +LV
Sbjct: 496 FVTITYWMGGLKPSLTTFIMTLMIVLYNVLVAQGVGLALGAILMDAKKAATLSSVLMLVF 555
Query: 690 FLLGGFII 697
L GG+ I
Sbjct: 556 LLAGGYYI 563
>At1g31770 unknown protein
Length = 648
Score = 195 bits (495), Expect = 2e-49
Identities = 153/609 (25%), Positives = 294/609 (48%), Gaps = 40/609 (6%)
Query: 826 GMILPFQPLTMTFHNVSYFVDMPQEIRKQGIPETRLQ-LLSNVSGVFSPGVLTALVGSSG 884
G+ + P+T+ F V Y V + Q + G +++ + +L+ ++G+ PG A++G SG
Sbjct: 43 GLQMSMYPITLKFEEVVYKVKIEQTSQCMGSWKSKEKTILNGITGMVCPGEFLAMLGPSG 102
Query: 885 AGKTTLMDVLAGRKTGGYIEGDIKISGYPKEQRTFARISGYVEQNDIHSPQVTIEESLWF 944
+GKTTL+ L GR + + G + +G P R +G+V Q+D+ P +T+ E+L+F
Sbjct: 103 SGKTTLLSALGGRLSKTF-SGKVMYNGQPFSG-CIKRRTGFVAQDDVLYPHLTVWETLFF 160
Query: 945 SASLRLPKEISTDKKREFVEQVMKLVELDSLRNALVGMPGSSGLSTEQRKRLTIAVELVA 1004
+A LRLP ++ D+K E V++V+ + L+ N+++G P G+S ++KR++I E++
Sbjct: 161 TALLRLPSSLTRDEKAEHVDRVIAELGLNRCTNSMIGGPLFRGISGGEKKRVSIGQEMLI 220
Query: 1005 NPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDDLLLMKRG 1064
NPS++ +DEPTSGLD+ A ++ ++ GRTVV TIHQPS I+ FD ++L+ G
Sbjct: 221 NPSLLLLDEPTSGLDSTTAHRIVTTIKRLASGGRTVVTTIHQPSSRIYHMFDKVVLLSEG 280
Query: 1065 GRVIYGGKLGVQSQIMIDYFQGIRGIRPIPRGYNPATWVLEVTT---------------P 1109
+ YG + ++YF + + NPA +L++
Sbjct: 281 SPIYYGA-----ASSAVEYFSSLGFSTSLT--VNPADLLLDLANGIPPDTQKETSEQEQK 333
Query: 1110 SVEETIDADFAEIYNNSDQYRGVEASILEFEHPPAGSEPLKFDTIYSQSLLSQFYRCLWK 1169
+V+ET+ + + + + + A +E+ A ++ LK + + + QF L +
Sbjct: 334 TVKETLVSAYEKNISTKLKAELCNAESHSYEYTKAAAKNLKSEQ-WCTTWWYQF-TVLLQ 391
Query: 1170 QNLVYWRSPPYNAMRMYFTTISALVFGTVFWDIGSKRSSTQELYVVMGALYASCLFIGVN 1229
+ + R +N +R++ A + G ++W + +S Q+ ++ F +
Sbjct: 392 RGVRERRFESFNKLRIFQVISVAFLGGLLWWH--TPKSHIQDRTALLFFFSVFWGFYPLY 449
Query: 1230 NASTVQPIVSIERTVFYREKAAGMYSPIAYAVAQGLIEIPYIAVQAMVFGLITYFMINFE 1289
NA P E+ + +E+++GMY +Y +A+ + ++P F I Y+M +
Sbjct: 450 NAVFTFP---QEKRMLIKERSSGMYRLSSYFMARNVGDLPLELALPTAFVFIIYWMGGLK 506
Query: 1290 RTAGKFFLYLLFMFLTFTYFTFYGMMAVGLSPTQHLAAVISSAFYSLWNLLSGFLIPESH 1349
F L LL + + G+ L A ++S ++ + G+ + +
Sbjct: 507 PDPTTFILSLLVVLYSVLVAQGLGLAFGALLMNIKQATTLASVTTLVFLIAGGYYVQQ-- 564
Query: 1350 IPGWWIWFYYICPVQWTLRGVITSQLGDVETKIIGPGFEGTVKEYLSL------NLGYDP 1403
IP + +W Y+ + + ++ Q D + G V ++ ++ NL D
Sbjct: 565 IPPFIVWLKYLSYSYYCYKLLLGIQYTDDDYYECSKGVWCRVGDFPAIKSMGLNNLWIDV 624
Query: 1404 KIMGISTVG 1412
+MG+ VG
Sbjct: 625 FVMGVMLVG 633
Score = 168 bits (425), Expect = 2e-41
Identities = 152/625 (24%), Positives = 281/625 (44%), Gaps = 70/625 (11%)
Query: 171 KRHSLTILDNVSGVIKPGRMTLLLGAPGAGKSSLLLALAGKLDSNLKKTGSITYNGHEAD 230
K TIL+ ++G++ PG +LG G+GK++LL AL G+L +G + YNG
Sbjct: 75 KSKEKTILNGITGMVCPGEFLAMLGPSGSGKTTLLSALGGRLSKTF--SGKVMYNGQPFS 132
Query: 231 EFFVKRTCAYISQTDNHTAELTVRETLDFAARCQGAQEGFAAYTKDIGRLENERNIRPSP 290
+KR +++Q D LTV ETL F A +
Sbjct: 133 GC-IKRRTGFVAQDDVLYPHLTVWETLFFTALLR-------------------------- 165
Query: 291 EIDAFMKASSVGGKKHSVNTDYILKVLGLDICSETIVGSDMLRGVSGGQRKRVTTG-EMI 349
SS+ + + + D ++ LGL+ C+ +++G + RG+SGG++KRV+ G EM+
Sbjct: 166 ------LPSSLTRDEKAEHVDRVIAELGLNRCTNSMIGGPLFRGISGGEKKRVSIGQEML 219
Query: 350 VGPRKTLFMDEISTGLDSSTTFQIVKCIKNFVHLMDATVLMALLQPAPETFELFDDLVLL 409
+ P L +DE ++GLDS+T +IV IK TV+ + QP+ + +FD +VLL
Sbjct: 220 INP-SLLLLDEPTSGLDSTTAHRIVTTIKRLAS-GGRTVVTTIHQPSSRIYHMFDKVVLL 277
Query: 410 SEGHVIYEGPRENVLEFFESIGFKLPPRKGIADFLQEVSSRKDQAQYWADPSKQYQFVPS 469
SEG IY G + +E+F S+GF AD L ++++ + S+Q Q
Sbjct: 278 SEGSPIYYGAASSAVEYFSSLGFSTSLTVNPADLLLDLAN-GIPPDTQKETSEQEQKTVK 336
Query: 470 GEIAEAFR---NSRFGSYVESLQTHPYDKSKCHPSALARTKYAVSRWEISKACFAREALL 526
+ A+ +++ + + + ++H Y+ +K L ++ + W R
Sbjct: 337 ETLVSAYEKNISTKLKAELCNAESHSYEYTKAAAKNLKSEQWCTTWWYQFTVLLQRG--- 393
Query: 527 ISRQRF-----LYIFKTCQVAFVGFVTCTIFLRTRMHPTDEAYGNLYVSALFFG---LVH 578
+ +RF L IF+ VAF+G + ++ T + L+ ++F+G L +
Sbjct: 394 VRERRFESFNKLRIFQVISVAFLGGL---LWWHTPKSHIQDRTALLFFFSVFWGFYPLYN 450
Query: 579 MMFNGFSELSLMIARLPVFYKQRDNLFYPAWAWSLTNWVLRVPYSIIEAVIWTVIVYYTV 638
+F E ++I K+R + Y ++ + V +P + + I+Y+
Sbjct: 451 AVFTFPQEKRMLI-------KERSSGMYRLSSYFMARNVGDLPLELALPTAFVFIIYWMG 503
Query: 639 GFAPSAGRFFRYMFILFVMHQMAIGLFRMMASIARDMVLANTFGSAALLVIFLLGGFIIP 698
G P F + ++ +A GL ++ ++ A T S LV + GG+ +
Sbjct: 504 GLKPDPTTFILSLLVVLYSVLVAQGLGLAFGALLMNIKQATTLASVTTLVFLIAGGYYVQ 563
Query: 699 KGMIKPWWIWGYWLSPLTYGQRAITVNEFTASRWMKQSALGNNTIG-YNILHAQSLPSED 757
+ I P+ +W +LS Y + + ++T + + S +G + + + L +
Sbjct: 564 Q--IPPFIVWLKYLSYSYYCYKLLLGIQYTDDDYYECSKGVWCRVGDFPAIKSMGLNN-- 619
Query: 758 YWYWVSVAVLVTYAIIFNIMVTLAL 782
W+ V V+ + + +M +AL
Sbjct: 620 --LWIDVFVMGVMLVGYRLMAYMAL 642
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,299,444
Number of Sequences: 26719
Number of extensions: 1359919
Number of successful extensions: 4493
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 112
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 3568
Number of HSP's gapped (non-prelim): 541
length of query: 1441
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1329
effective length of database: 8,326,068
effective search space: 11065344372
effective search space used: 11065344372
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0346.8


