

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.5
(350 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
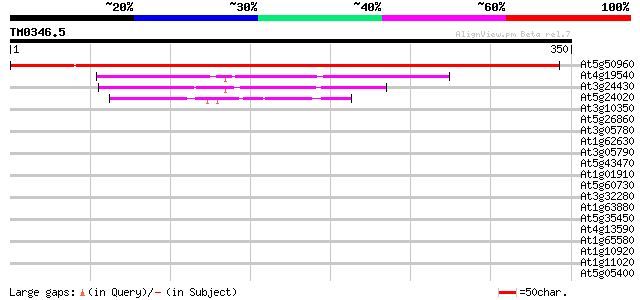
Score E
Sequences producing significant alignments: (bits) Value
At5g50960 nucleotide-binding protein 496 e-141
At4g19540 ATP binding protein - like 134 6e-32
At3g24430 mrp protein, putative 115 5e-26
At5g24020 septum site-determining MinD (dbj|BAA90261.1) 55 7e-08
At3g10350 putative ATPase 33 0.23
At5g26860 LON protease homolog (LON_ARA) 32 0.52
At3g05780 putative mitochondrial LON ATP-dependent protease 32 0.68
At1g62630 putative RPS-2 disease resistance protein (AA002294 32 0.68
At3g05790 putative mitochondrial LON ATP-dependent protease 31 1.2
At5g43470 disease resistance protein RPP8 30 1.5
At1g01910 arsA homolog (hASNA-I), putative 30 1.5
At5g60730 ATPase - like protein 30 2.0
At3g32280 hypothetical protein 30 2.6
At1g63880 disease resistance like protein 30 2.6
At5g35450 disease resistance protein 29 4.4
At4g13590 unknown protein 29 4.4
At1g65580 hypothetical protein 29 4.4
At1g10920 disease resistance protein RPM1 isolog 29 4.4
At1g11020 unknown protein 29 4.4
At5g05400 NBS/LRR disease resistance protein 28 5.7
>At5g50960 nucleotide-binding protein
Length = 350
Score = 496 bits (1277), Expect = e-141
Identities = 241/344 (70%), Positives = 292/344 (84%), Gaps = 2/344 (0%)
Query: 1 MEKGDIPENANDHCPGSQSDSAGKSDGCQGCPNQQTCATSSPKGPDSDLVAIKERMATVK 60
ME GDIPE+AN+HCPG QS+SAGKSD C GCPNQ+ CAT+ PKGPD DLVAI ERM+TVK
Sbjct: 1 MENGDIPEDANEHCPGPQSESAGKSDSCAGCPNQEACATA-PKGPDPDLVAIAERMSTVK 59
Query: 61 HKILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHELHRSNF 120
HKILVLSGKGGVGKSTFSAQL+FALA D +VGL+DIDICGPSIPKMLGLEG E+H+SN
Sbjct: 60 HKILVLSGKGGVGKSTFSAQLSFALAGMDHQVGLMDIDICGPSIPKMLGLEGQEIHQSNL 119
Query: 121 GCDPVYVQPNLGVMSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNELDFLIVDAPPG 180
G PVYV+ NLGVMSIGF+ + D+ +I RGPRK+ LI +FLKDVYW E+D+L+VDAPPG
Sbjct: 120 GWSPVYVEDNLGVMSIGFMLPNSDEAVIWRGPRKNGLIKQFLKDVYWGEIDYLVVDAPPG 179
Query: 181 TSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEVKVLGVVENMSGLCQ 240
TSDE ISIV+ L TG+DGAII+TTPQ+VSL DVRK V+FCK+ V VLGVVENMSGL Q
Sbjct: 180 TSDEHISIVQYLLPTGIDGAIIVTTPQEVSLIDVRKEVSFCKKVGVPVLGVVENMSGLSQ 239
Query: 241 PVMDFKFLKLAS-NGEQKDVTEWFLKFMREKAPEMVDMIACTEVFDSSGGGAVRMCKEMM 299
P+ D KF+KLA+ G +VTE + +R+ APE++D++AC+EVFDSSGGGA RMC+EM
Sbjct: 240 PLKDVKFMKLATETGSSINVTEDVIACLRKNAPELLDIVACSEVFDSSGGGAERMCREMG 299
Query: 300 IPFLGKVPLDPKLCKAAEEGRSCFDDKDCVASAFALKNIIEKLM 343
+PFLGKVP+DP+LCKAAE+G+SCF+D C+ SA ALK+II+K++
Sbjct: 300 VPFLGKVPMDPQLCKAAEQGKSCFEDNKCLISAPALKSIIQKVV 343
>At4g19540 ATP binding protein - like
Length = 313
Score = 134 bits (338), Expect = 6e-32
Identities = 87/223 (39%), Positives = 123/223 (55%), Gaps = 10/223 (4%)
Query: 55 RMATVKHKILVLSGKGGVGKSTFSAQLAFALAAK-DFEVGLLDIDICGPSIPKMLGLEGH 113
R+ VK I V SGKGGVGKS+ + LA ALA K + ++GLLD D+ GPS+P M+ +
Sbjct: 38 RLHGVKDIIAVASGKGGVGKSSTAVNLAVALANKCELKIGLLDADVYGPSVPIMMNINQK 97
Query: 114 ELHRSNFGCDPVYVQPNLGV--MSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNELD 171
+ PV N GV MS+G + + D P++ RGP + + K V W +LD
Sbjct: 98 PQVNQDMKMIPV---ENYGVKCMSMGLL-VEKDAPLVWRGPMVMSALAKMTKGVDWGDLD 153
Query: 172 FLIVDAPPGTSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEVKVLGV 231
L+VD PPGT D QISI + L L GA+I++TPQ V+L D +G++ + V +LG+
Sbjct: 154 ILVVDMPPGTGDAQISISQNL---KLSGAVIVSTPQDVALADANRGISMFDKVRVPILGL 210
Query: 232 VENMSGLCQPVMDFKFLKLASNGEQKDVTEWFLKFMREKAPEM 274
VENMS P + G ++ + LK + E EM
Sbjct: 211 VENMSCFVCPHCNEPSFIFGKEGARRTAAKKGLKLIGEIPLEM 253
>At3g24430 mrp protein, putative
Length = 532
Score = 115 bits (287), Expect = 5e-26
Identities = 69/182 (37%), Positives = 101/182 (54%), Gaps = 9/182 (4%)
Query: 56 MATVKHKILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHEL 115
++ + + I V S KGGVGKST + LA+ LA VG+ D D+ GPS+P M+ E L
Sbjct: 172 LSRISNIIAVSSCKGGVGKSTVAVNLAYTLAGMGARVGIFDADVYGPSLPTMVNPESRIL 231
Query: 116 HRSNFGCDPVYVQPNLGV--MSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWNELDFL 173
N + +GV +S GF I+RGP S +I + L W ELD+L
Sbjct: 232 -EMNPEKKTIIPTEYMGVKLVSFGFAG---QGRAIMRGPMVSGVINQLLTTTEWGELDYL 287
Query: 174 IVDAPPGTSDEQISIVKLLGATGLDGAIIITTPQQVSLTDVRKGVNFCKQAEVKVLGVVE 233
++D PPGT D Q+++ ++ L A+I+TTPQ+++ DV KGV + +V + VVE
Sbjct: 288 VIDMPPGTGDIQLTLCQV---APLTAAVIVTTPQKLAFIDVAKGVRMFSKLKVPCVAVVE 344
Query: 234 NM 235
NM
Sbjct: 345 NM 346
>At5g24020 septum site-determining MinD (dbj|BAA90261.1)
Length = 326
Score = 54.7 bits (130), Expect = 7e-08
Identities = 48/165 (29%), Positives = 77/165 (46%), Gaps = 26/165 (15%)
Query: 63 ILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHELHRSNFGC 122
+++ SGKGGVGK+T +A + +LA F V +D D+ ++ +LGLE +R N+ C
Sbjct: 61 VVITSGKGGVGKTTTTANVGLSLARYGFSVVAIDADLGLRNLDLLLGLE----NRVNYTC 116
Query: 123 ----------DPVYVQ----PNLGVMSIGFVNSDPDDPIILRGPRKSWLITEFLKDVYWN 168
D V+ N ++ I S P+ G WL+ + LK
Sbjct: 117 VEVINGDCRLDQALVRDKRWSNFELLCISKPRSKL--PMGFGGKALEWLV-DALKTRPEG 173
Query: 169 ELDFLIVDAPPGTSDEQISIVKLLGATGLDGAIIITTPQQVSLTD 213
DF+I+D P G I+ + T + A+++TTP +L D
Sbjct: 174 SPDFIIIDCPAGIDAGFITAI-----TPANEAVLVTTPDITALRD 213
>At3g10350 putative ATPase
Length = 386
Score = 33.1 bits (74), Expect = 0.23
Identities = 17/46 (36%), Positives = 24/46 (51%)
Query: 41 SPKGPDSDLVAIKERMATVKHKILVLSGKGGVGKSTFSAQLAFALA 86
S P + E ++ K K +L GKGGVGK++ +A LA A
Sbjct: 67 SVASPTETISEFDEMVSGTKRKYYMLGGKGGVGKTSCAASLAVRFA 112
>At5g26860 LON protease homolog (LON_ARA)
Length = 985
Score = 32.0 bits (71), Expect = 0.52
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 3/53 (5%)
Query: 48 DLVAIKERMATVKHKILVLSGKGGVGKSTFSAQLAFALAAKDFEV---GLLDI 97
+ +A+ T + KI+ LSG GVGK++ +A AL K F GL D+
Sbjct: 489 EFIAVGRLRGTSQGKIICLSGPPGVGKTSIGRSIARALNRKFFRFSVGGLADV 541
>At3g05780 putative mitochondrial LON ATP-dependent protease
Length = 924
Score = 31.6 bits (70), Expect = 0.68
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 3/53 (5%)
Query: 48 DLVAIKERMATVKHKILVLSGKGGVGKSTFSAQLAFALAAKDFEV---GLLDI 97
+ +A+ T + KI+ LSG GVGK++ +A AL K F GL D+
Sbjct: 427 EFIAVGRLRGTSQGKIICLSGPPGVGKTSIGRSIARALDRKFFRFSVGGLSDV 479
>At1g62630 putative RPS-2 disease resistance protein (AA002294
Length = 893
Score = 31.6 bits (70), Expect = 0.68
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 11/62 (17%)
Query: 63 ILVLSGKGGVGKSTFSAQLAFALAAKD---FEVGL-------LDIDICGPSIPKMLGLEG 112
I+ + G GGVGK+T QL F + KD F++G+ ++++ I + LGL G
Sbjct: 174 IMGMYGMGGVGKTTLLTQL-FNMFNKDKCGFDIGIWVVVSQEVNVEKIQDEIAQKLGLGG 232
Query: 113 HE 114
HE
Sbjct: 233 HE 234
>At3g05790 putative mitochondrial LON ATP-dependent protease
Length = 942
Score = 30.8 bits (68), Expect = 1.2
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 3/53 (5%)
Query: 48 DLVAIKERMATVKHKILVLSGKGGVGKSTFSAQLAFALAAKDFEV---GLLDI 97
+ +A+ T + KI+ LSG GVGK++ +A AL K F GL D+
Sbjct: 436 EFIAVGGLRGTSQGKIICLSGPTGVGKTSIGRSIARALDRKFFRFSVGGLSDV 488
>At5g43470 disease resistance protein RPP8
Length = 908
Score = 30.4 bits (67), Expect = 1.5
Identities = 14/56 (25%), Positives = 31/56 (55%), Gaps = 8/56 (14%)
Query: 34 QQTCATSSPKGPDSDLVAIKERMATVK--------HKILVLSGKGGVGKSTFSAQL 81
Q+ + P +SDLV +++ + + H+++ ++G GG+GK+T + Q+
Sbjct: 150 QREIRQTYPDSSESDLVGVEQSVKELVGHLVENDVHQVVSIAGMGGIGKTTLARQV 205
>At1g01910 arsA homolog (hASNA-I), putative
Length = 353
Score = 30.4 bits (67), Expect = 1.5
Identities = 16/37 (43%), Positives = 23/37 (61%)
Query: 62 KILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDID 98
K + + GKGGVGK+T S+ LA LA+ V ++ D
Sbjct: 20 KWVFVGGKGGVGKTTCSSILAICLASVRSSVLIISTD 56
>At5g60730 ATPase - like protein
Length = 393
Score = 30.0 bits (66), Expect = 2.0
Identities = 14/34 (41%), Positives = 22/34 (64%)
Query: 54 ERMATVKHKILVLSGKGGVGKSTFSAQLAFALAA 87
E ++ + K +L GKGGVGK++ +A LA A+
Sbjct: 62 EMVSVNQRKYYLLGGKGGVGKTSCAASLAVKFAS 95
>At3g32280 hypothetical protein
Length = 474
Score = 29.6 bits (65), Expect = 2.6
Identities = 18/63 (28%), Positives = 32/63 (50%), Gaps = 13/63 (20%)
Query: 108 LGLEGHELHRSNFGCDPVYVQ-PNLGVMSIGFVNSDPDDPIILRGPRKSWLITEFLKDVY 166
L L+ H + NF C+ + V+ PN +++ RGP K++L+ F+ D+Y
Sbjct: 62 LSLDPHHIEILNFLCNNLVVENPNGCILAQA------------RGPEKTYLMINFIYDLY 109
Query: 167 WNE 169
N+
Sbjct: 110 QND 112
>At1g63880 disease resistance like protein
Length = 1017
Score = 29.6 bits (65), Expect = 2.6
Identities = 22/91 (24%), Positives = 41/91 (44%), Gaps = 2/91 (2%)
Query: 59 VKHKILVLSGKGGVGKSTFSAQLAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHELHRS 118
V+ KI+ ++G G+GK+T A+ + L +K F++ +D S G +LH
Sbjct: 206 VEVKIVAIAGPAGIGKTTI-ARALYGLLSKRFQLSCF-VDNLRGSYHSGFDEYGFKLHLQ 263
Query: 119 NFGCDPVYVQPNLGVMSIGFVNSDPDDPIIL 149
V Q + + +G + + D +L
Sbjct: 264 EQFLSKVLNQSGMRICHLGAIKENLSDQRVL 294
>At5g35450 disease resistance protein
Length = 901
Score = 28.9 bits (63), Expect = 4.4
Identities = 14/56 (25%), Positives = 30/56 (53%), Gaps = 8/56 (14%)
Query: 34 QQTCATSSPKGPDSDLVAIKERMATV--------KHKILVLSGKGGVGKSTFSAQL 81
Q+ + P +SDLV +++ + + +++ +SG GG+GK+T + Q+
Sbjct: 148 QREIRQTFPNSSESDLVGVEQSVEELVGPMVEIDNIQVVSISGMGGIGKTTLARQI 203
>At4g13590 unknown protein
Length = 359
Score = 28.9 bits (63), Expect = 4.4
Identities = 19/46 (41%), Positives = 23/46 (49%), Gaps = 4/46 (8%)
Query: 4 GDIPENANDHCPGSQSDSAGKSDGCQGCPNQQTCATSS----PKGP 45
G A+D GS S S G+ DG Q Q+ ATSS P+GP
Sbjct: 61 GKFRVRASDAGVGSGSYSGGEEDGSQSSSLDQSPATSSESLKPRGP 106
>At1g65580 hypothetical protein
Length = 993
Score = 28.9 bits (63), Expect = 4.4
Identities = 25/99 (25%), Positives = 44/99 (44%), Gaps = 10/99 (10%)
Query: 81 LAFALAAKDFEVGLLDIDICGPSIPKMLGLEGHELHRSNFGCDPVYVQPNLGVMSIGFVN 140
L+ A++A E G L + ++ K L L+ E H + + Y+ P V + GF N
Sbjct: 231 LSIAISAYGSEGGALKVWPWDGALGKSLSLKMEERHMAALAVERSYIDPRNMVSANGFAN 290
Query: 141 SDPDDPIILRGPRKSWLITEFLKDVYW--NELDFLIVDA 177
+ D ++L+++ + W + L F I DA
Sbjct: 291 TLTSD--------VTFLVSDHTRARVWSASPLTFAIWDA 321
>At1g10920 disease resistance protein RPM1 isolog
Length = 821
Score = 28.9 bits (63), Expect = 4.4
Identities = 19/56 (33%), Positives = 33/56 (58%), Gaps = 12/56 (21%)
Query: 34 QQTCATSSPKGPDSDLVAIKERM-ATVKH-------KILVLSGKGGVGKSTFSAQL 81
+QT A SS +SDLV +++ + A H +++ +SG GG+GK+T + Q+
Sbjct: 129 RQTFANSS----ESDLVGVEQSVEALAGHLVENDNIQVVSISGMGGIGKTTLARQV 180
>At1g11020 unknown protein
Length = 321
Score = 28.9 bits (63), Expect = 4.4
Identities = 17/55 (30%), Positives = 24/55 (42%), Gaps = 2/55 (3%)
Query: 285 DSSGGGAVRMCKEMMIPFLGKVPLDPKLCKAAEE--GRSCFDDKDCVASAFALKN 337
D+S R+C E LG + P +CK ++ RSC D V FA +
Sbjct: 56 DASSAPCCRICLEDDSELLGDELISPCMCKGTQQFVHRSCLDHWRSVKEGFAFSH 110
>At5g05400 NBS/LRR disease resistance protein
Length = 874
Score = 28.5 bits (62), Expect = 5.7
Identities = 17/61 (27%), Positives = 31/61 (49%), Gaps = 2/61 (3%)
Query: 63 ILVLSGKGGVGKSTFSAQL--AFALAAKDFEVGLLDIDICGPSIPKMLGLEGHELHRSNF 120
+L + G GGVGK+T +Q+ F + DF++ + + P++ ++ G L N
Sbjct: 177 LLGIYGMGGVGKTTLLSQINNKFRTVSNDFDIAIWVVVSKNPTVKRIQEDIGKRLDLYNE 236
Query: 121 G 121
G
Sbjct: 237 G 237
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,337,134
Number of Sequences: 26719
Number of extensions: 362397
Number of successful extensions: 967
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 945
Number of HSP's gapped (non-prelim): 27
length of query: 350
length of database: 11,318,596
effective HSP length: 100
effective length of query: 250
effective length of database: 8,646,696
effective search space: 2161674000
effective search space used: 2161674000
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0346.5


