

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.15
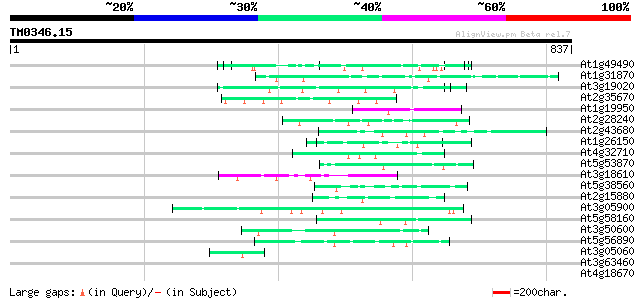
(837 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g49490 hypothetical protein 63 6e-10
At1g31870 unknown protein 53 8e-07
At3g19020 hypothetical protein 52 1e-06
At2g35670 fertilization-independent seed 2 protein (FIS2) 50 7e-06
At1g19950 unknown protein 50 7e-06
At2g28240 putative proline/hydroxyproline-rich glycoprotein 49 9e-06
At2g43680 SF16 protein {Helianthus annuus} like protein 49 1e-05
At1g26150 Pto kinase interactor, putative 49 2e-05
At4g32710 putative protein kinase 47 3e-05
At5g53870 predicted GPI-anchored protein 47 4e-05
At3g18610 unknown protein 47 4e-05
At5g38560 putative protein 46 8e-05
At2g15880 unknown protein 46 1e-04
At3g05900 unknown protein 45 1e-04
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 45 2e-04
At3g50600 unknown protein 44 3e-04
At5g56890 unknown protein 44 4e-04
At3g05060 putative SAR DNA-binding protein-1 43 6e-04
At3g63460 putative protein 42 0.001
At4g18670 extensin-like protein 42 0.001
>At1g49490 hypothetical protein
Length = 847
Score = 63.2 bits (152), Expect = 6e-10
Identities = 90/393 (22%), Positives = 147/393 (36%), Gaps = 29/393 (7%)
Query: 310 PPALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKR-KHEEPTKPDVPSD 368
PP E P + + ++ ++ S + PS K + + KP++ K EE KP+ P
Sbjct: 437 PPKHESPKPEEPENKHELPKQKESPKPQPS------KPEDSPKPEQPKPEESPKPEQPQI 490
Query: 369 GDDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDL 428
+ P + Q + P P + RR P +V + V S
Sbjct: 491 PEPTKPVSPPNEAQGPTPDDPYDASPVKN--RRSPPPPKVEDTRVPPPQPPMPSPSPPSP 548
Query: 429 NSLDALPISALLNRT-STPLTLILESQPAEQTISPPTSPRSSFFQPSPQE--APLWNMLQ 485
P+ + S+P + S P PP SP P P +P +
Sbjct: 549 IYSPPPPVHSPPPPVYSSPPPPHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFS 608
Query: 486 TPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRRTTDTESTTPCP 545
P PS SP H+ P + S P P S ++ P+ T T++ P P
Sbjct: 609 PPPPSPVYSPPPPS---HSPPPPVY--SPPPPTFSPPPTHNTNQPPMGAPTPTQA--PTP 661
Query: 546 SVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDH 605
S SS S+ S+ + + S S VP+P + L
Sbjct: 662 SSETTQVPTPSSESDQSQILSPVQAPTPVQSSTPSSEPTQVPTPSSSESYQAPNLSPVQA 721
Query: 606 PLPVFPLNAADPLAQPAQPAPENSESNNSS-------VRSPHPQ---VETSEPNLETHTS 655
P PV + +Q P+ E+++S + + V +P P V++ P+ E +S
Sbjct: 722 PTPVQAPTTSSETSQVPTPSSESNQSPSQAPTPILEPVHAPTPNSKPVQSPTPSSEPVSS 781
Query: 656 NDQTLDVGSPQGASEANSSNHPTSPETNLSIVP 688
+Q+ +V +P+ SS +SP T+ SI P
Sbjct: 782 PEQSEEVEAPEPTPVNPSSVPSSSPSTDTSIPP 814
Score = 60.5 bits (145), Expect = 4e-09
Identities = 88/384 (22%), Positives = 142/384 (36%), Gaps = 68/384 (17%)
Query: 319 PKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPD----VPSDGDDDAP 374
PK + K ++ E+ P + + + ++P+ KHE P + + PS +D
Sbjct: 414 PKPSDSSKPETPKTPEQPSPKPQPPKHESPKPEEPENKHELPKQKESPKPQPSKPEDSPK 473
Query: 375 PPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDLNSLDAL 434
P + K ++ E+P EP V S A + DD DA
Sbjct: 474 PEQPKPEESPKPEQPQIPEPTKPV----------------SPPNEAQGPTPDD--PYDAS 515
Query: 435 PISALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP--SEP 492
P+ NR S P + +++ + PP P S PSP +P + P P S P
Sbjct: 516 PVK---NRRSPPPPKVEDTR-----VPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSP 567
Query: 493 TSPITIQYNLHTSEPIILEQSEPEPRTSDHSVP-------------RASERPVRRTTDTE 539
P ++++ P + P P HS P PV
Sbjct: 568 PPP-----HVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPS 622
Query: 540 STTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPER 599
+ P P S P P++N+ + + SE VP+P + +
Sbjct: 623 HSPPPPVYSPPPPTFSPPPTHNTNQPPMGAPTPTQAPTPSSETTQ-VPTPSS-ESDQSQI 680
Query: 600 LVDPDHPLPVFPLNAADPLAQPAQ-PAPENSES----NNSSVRSPHPQVETSEPNLETHT 654
L P PV ++ P ++P Q P P +SES N S V++P P ++ T
Sbjct: 681 LSPVQAPTPV---QSSTPSSEPTQVPTPSSSESYQAPNLSPVQAPTP--------VQAPT 729
Query: 655 SNDQTLDVGSPQGASEANSSNHPT 678
++ +T V +P S + S PT
Sbjct: 730 TSSETSQVPTPSSESNQSPSQAPT 753
Score = 50.1 bits (118), Expect = 5e-06
Identities = 78/364 (21%), Positives = 119/364 (32%), Gaps = 94/364 (25%)
Query: 332 SSEESDPSDGVIQKKKKQADKPKRKHEEP---TKPDVPSDGDDDAPPPKKKKKQVRIVEK 388
S+ S PS + + + KP+ +P +KP+ P + +P P+ K + K
Sbjct: 388 SNGGSSPSPNPPRTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHE---SPK 444
Query: 389 PTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDLNSLDALPISALLNRTSTPLT 448
P E ++ ++ E+P
Sbjct: 445 PEEPENKHELPKQKESP------------------------------------------- 461
Query: 449 LILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPIT-----IQYNLH 503
+ QP++ P SP+ QP P+E+P Q P+P++P SP + +
Sbjct: 462 ---KPQPSK----PEDSPKPE--QPKPEESPKPEQPQIPEPTKPVSPPNEAQGPTPDDPY 512
Query: 504 TSEPIILEQSEPEPRTSDHSVPRASERPVRRTTDTESTTPCPSVSFPANVADSSPSNNSE 563
+ P+ +S P P+ D VP + + +P P V P SSP
Sbjct: 513 DASPVKNRRSPPPPKVEDTRVPPPQPPMPSPSPPSPIYSPPPPVHSPPPPVYSSPPP--- 569
Query: 564 SVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVF---PLNAADPLAQ 620
Y P P P P V P PVF P + P
Sbjct: 570 ----------------PHVYSPPPPVASPPPPSPPPPVHSPPPPPVFSPPPPVFSPPPPS 613
Query: 621 PAQPAPENSESNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGASEANSSNHPTSP 680
P P S S V SP P + P TH +N P GA + P+S
Sbjct: 614 PVYSPPPPSHSPPPPVYSPPPPTFSPPP---THNTNQ------PPMGAPTPTQAPTPSSE 664
Query: 681 ETNL 684
T +
Sbjct: 665 TTQV 668
Score = 49.7 bits (117), Expect = 7e-06
Identities = 50/192 (26%), Positives = 68/192 (35%), Gaps = 32/192 (16%)
Query: 463 PTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDH 522
P PR+S +PS E + + KP P +P EQ P+P+ H
Sbjct: 396 PNPPRTSEPKPSKPEPVMPKPSDSSKPETPKTP---------------EQPSPKPQPPKH 440
Query: 523 SVPRASERPVRRTTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEY 582
P+ E + + +P P S P + ES K ++ I E
Sbjct: 441 ESPKPEEPENKHELPKQKESPKPQPSKPEDSPKPEQPKPEES-------PKPEQPQIPEP 493
Query: 583 YHTVPSPRKYPGPRPERLVDPDHPLPVF------PLNAADPLAQPAQPAPENSESNNSSV 636
V P + GP P+ DP PV P D P QP P S S S +
Sbjct: 494 TKPVSPPNEAQGPTPD---DPYDASPVKNRRSPPPPKVEDTRVPPPQP-PMPSPSPPSPI 549
Query: 637 RSPHPQVETSEP 648
SP P V + P
Sbjct: 550 YSPPPPVHSPPP 561
Score = 31.2 bits (69), Expect = 2.5
Identities = 33/155 (21%), Positives = 53/155 (33%), Gaps = 20/155 (12%)
Query: 534 RTTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYP 593
RT++ + + P P + P++ + E Q K + E + P++
Sbjct: 400 RTSEPKPSKPEPVMPKPSDSSKPETPKTPEQPSPKPQPPKHESPKPEEPENKHELPKQKE 459
Query: 594 GPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETSEPNLETH 653
P+P+ D P +P QP PE S + P P S PN
Sbjct: 460 SPKPQPSKPEDSP-------------KPEQPKPEESPKPEQP-QIPEPTKPVSPPNEAQG 505
Query: 654 TSNDQTLDVGSPQGASEANSSNHPTSPETNLSIVP 688
+ D P AS + P P+ + VP
Sbjct: 506 PTPD------DPYDASPVKNRRSPPPPKVEDTRVP 534
>At1g31870 unknown protein
Length = 561
Score = 52.8 bits (125), Expect = 8e-07
Identities = 100/470 (21%), Positives = 182/470 (38%), Gaps = 61/470 (12%)
Query: 368 DGDDDAPPPKKKKKQVRIVEKPTRVEPAA------DVIRRIETPARVTRSTVRSSSKSAV 421
+ D KKKKKQ +KP++ EP D + + + + S+ ++ +
Sbjct: 15 ESSDVVEKKKKKKKQ----KKPSKPEPRGVLVVDEDPVWQKQVDPEEDENEDDSAEETPL 70
Query: 422 IDSDDDLNSLDALPI-------SALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPS 474
+D D ++ + L +A+ S +TL L + + ISPP R+ PS
Sbjct: 71 VDEDIEVKRMRRLEEIKARRAHNAIAEDGSGWVTLPLNREDTQSNISPPRRQRTRNDSPS 130
Query: 475 PQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRR 534
P+ P ++ ++N + EP + ++P SD S PR +R
Sbjct: 131 PEPGPRRSVADRVDTDMSPPRRRKRHNSPSPEP-NRKHTKPVSLDSDMSPPR------KR 183
Query: 535 TTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPG 594
+S +P P + + D SP V + + K S E + PR
Sbjct: 184 KARNDSPSPEPEAKYLSE--DLSPPRR-RHVHSPSRESSRKRSDSVELDDDLSPPR---- 236
Query: 595 PRPERLVDPDHPLPVFPL-NAADPLAQPAQ-----PAPENSESNNSSVRSPHPQVETSEP 648
R L H PV + ++ L+ P + P+PE + ++ S S + S P
Sbjct: 237 -RKRDL----HGSPVSDVKKKSNDLSPPRRRRYHSPSPEPARRSSKSFGS---NADLSPP 288
Query: 649 NLETHTSNDQTLDVGSPQGASEANSSNHPTSPETNLSIVPFTYLKPNNLLECISVFNHEA 708
+ + Q D+ + A++ S+ S +N P +P +
Sbjct: 289 GRNINMKDSQDSDLSPQRKAADLRRSSENVSRSSNFDSSP--PRRPR---------RDSS 337
Query: 709 SLMIRNVQGHTDLSEDPDSVAEEWNRLSDWLIAQVPSLMSRLTAERDQRIEAARQRFNRR 768
I Q T L D + E+ + + + ++ S LT + + + R + +
Sbjct: 338 PPQISKEQRKTGLISGKD-IGSEYRKKKEDEKLRFKNMDSELTGQNAEAV--FRDKITGK 394
Query: 769 VILHEQQLRSKLLEVVEEARRKKEQAEEAARQKAERIANAKLEAERLEAE 818
I E+ L+SK +V+E+ + K + + QK E A A+L+ LE +
Sbjct: 395 RISKEEYLKSKQKKVIEKPKEIKLEWGKGLAQKRE--AEARLQELELEKD 442
Score = 37.7 bits (86), Expect = 0.027
Identities = 42/173 (24%), Positives = 65/173 (37%), Gaps = 13/173 (7%)
Query: 310 PPALEFD-SPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPDVPSD 368
P +L+ D SPP+K+K R E S+ + +++ P R+ V +
Sbjct: 170 PVSLDSDMSPPRKRKARNDSPSPEPEAKYLSEDLSPPRRRHVHSPSRESSRKRSDSV--E 227
Query: 369 GDDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDL 428
DDD PP++K + P +DV ++ + R S S S
Sbjct: 228 LDDDLSPPRRK--------RDLHGSPVSDVKKKSNDLSPPRRRRYHSPSPEPARRSSKSF 279
Query: 429 NS-LDALPISALLN-RTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAP 479
S D P +N + S L + + A+ S RSS F SP P
Sbjct: 280 GSNADLSPPGRNINMKDSQDSDLSPQRKAADLRRSSENVSRSSNFDSSPPRRP 332
>At3g19020 hypothetical protein
Length = 951
Score = 52.0 bits (123), Expect = 1e-06
Identities = 90/390 (23%), Positives = 133/390 (34%), Gaps = 37/390 (9%)
Query: 310 PPALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPDVPSDG 369
PP E P+ K EES + P Q+ K + PK +P K + P
Sbjct: 522 PPKPEESPKPQPPKQETPKPEESPKPQPPK----QETPKPEESPK---PQPPKQETPKPE 574
Query: 370 DDDAPPPKKKKKQVRI---------VEKPTRVEPA-ADVI--RRIETPARVTRSTVRSSS 417
+ P P K+++ + +E P +P A I RR + P+ T T +S
Sbjct: 575 ESPKPQPPKQEQPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSP 634
Query: 418 KSAVIDSDDDLNSLDALP---ISALLNRTSTPLTLILESQPAEQTISPPT-SPRSSFFQP 473
+S + S + + P S S P + P PP SP P
Sbjct: 635 QSPPVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSP 694
Query: 474 SPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVR 533
P + +P P + P + H+ P + QS P P P P+
Sbjct: 695 PPPVHSPPPPVHSPPPPVHSPPPPV----HSPPPPV--QSPPPPPVFSPPPPA----PIY 744
Query: 534 RTTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYP 593
+P P V P SP S V + H+ P P
Sbjct: 745 SPPPPPVHSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPSPIY 804
Query: 594 GPRPERLVDPDHPLPVFPL-NAADPLAQPAQPAPENSESNNSSVRSPHPQVETSE-PNLE 651
P P P P PV PL A P+A P+ S ++ V++P P E E P+
Sbjct: 805 SPPPPVFSPP--PKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDS 862
Query: 652 THTSNDQTLDVGSPQGASEANSSNHPTSPE 681
H+ ++ SP E + + PE
Sbjct: 863 NHSPVFKSSPAPSPDSEPEVEAPVPSSEPE 892
Score = 50.1 bits (118), Expect = 5e-06
Identities = 80/340 (23%), Positives = 115/340 (33%), Gaps = 58/340 (17%)
Query: 310 PPALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPDVPSDG 369
PP LE S PK EES + PS KP+ EP+ P P
Sbjct: 421 PPNLEEPSKPKP--------EESPKPQQPS-----------PKPETPSHEPSNPKEPKPE 461
Query: 370 DDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDLN 429
PK ++ + + E P + P + + E P S + SSK ++
Sbjct: 462 SPKQESPKTEQPKPK-PESPKQESPKQEA-PKPEQPKPKPESPKQESSKQEPPKPEESPK 519
Query: 430 SLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP 489
P + P P ++T P SP+ QP QE P ++PKP
Sbjct: 520 PEPPKP-----EESPKP------QPPKQETPKPEESPKP---QPPKQETP--KPEESPKP 563
Query: 490 SEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRRTTDTESTTPCPSVSF 549
P T +P + P+P+ P +E P + ES P P+ +
Sbjct: 564 QPPKQ--------ETPKP----EESPKPQPPKQEQPPKTEAPKMGSPPLES--PVPNDPY 609
Query: 550 PAN-VADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLP 608
A+ + P S S + + S S H+ P P P P + P P+
Sbjct: 610 DASPIKKRRPQPPSPSTEE-----TKTTSPQSPPVHSPPPPPPVHSP-PPPVFSPPPPMH 663
Query: 609 VFPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETSEP 648
P P P P S V SP P V + P
Sbjct: 664 SPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPP 703
Score = 45.4 bits (106), Expect = 1e-04
Identities = 88/365 (24%), Positives = 131/365 (35%), Gaps = 34/365 (9%)
Query: 310 PPALEFDSPPKKKKTRKMIME--ESSEESDPSDGV-IQKKKKQADKPKRKHEEPTKPDVP 366
PP E P K + KM ES +DP D I+K++ Q P + + T P P
Sbjct: 581 PPKQE---QPPKTEAPKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTTSPQSP 637
Query: 367 SDGDDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDD 426
PPP P P V P S V
Sbjct: 638 PVHSPPPPPPVHSPPPPVFSPPPPMHSPPPPVYSP-PPPVHSPPPPPVHSPPPPVHSPPP 696
Query: 427 DLNSLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQT 486
++S P+ + +P + P Q SPP P S P+P +P + +
Sbjct: 697 PVHSPPP-PVHSPPPPVHSPPPPVHSPPPPVQ--SPPPPPVFSPPPPAPIYSPPPPPVHS 753
Query: 487 PKP---SEPTSPI-TIQYNLHTSEPIILEQSEP--EPRTSDHSVPRASE----RPVRRTT 536
P P S P P+ + +H+ P + P P HS P S P +
Sbjct: 754 PPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPVHSPPPPSPIYSPPPPVFSP 813
Query: 537 DTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIA---KEKESAISEYYHTVPSPRKYP 593
+ TP P + P A + S+ S + VQ E A S+ H+ P + P
Sbjct: 814 PPKPVTPLPPATSPMANAPTPSSSESGEISTPVQAPTPDSEDIEAPSDSNHS-PVFKSSP 872
Query: 594 GPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESN-NSSVRSPHPQVETSEPNLET 652
P P+ +P+ PV P ++P AP+ SE+ +SS S +P + + P E
Sbjct: 873 APSPDS--EPEVEAPV-------PSSEPEVEAPKQSEATPSSSPPSSNPSPDVTAPPSED 923
Query: 653 HTSND 657
+ D
Sbjct: 924 NDDGD 928
Score = 32.7 bits (73), Expect = 0.86
Identities = 23/114 (20%), Positives = 39/114 (34%), Gaps = 19/114 (16%)
Query: 587 PSPRKYPGPR---------PERLVDPDHPLP----------VFPLNAADPLAQPAQPAPE 627
PSP+ P P+ P L +P P P P + + P +P PE
Sbjct: 402 PSPKPTPTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPE 461
Query: 628 NSESNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGASEANSSNHPTSPE 681
+ + + P P+ E+ + + P+ + +S P PE
Sbjct: 462 SPKQESPKTEQPKPKPESPKQESPKQEAPKPEQPKPKPESPKQESSKQEPPKPE 515
>At2g35670 fertilization-independent seed 2 protein (FIS2)
Length = 488
Score = 49.7 bits (117), Expect = 7e-06
Identities = 67/303 (22%), Positives = 118/303 (38%), Gaps = 48/303 (15%)
Query: 317 SPPK-----KKKTRKMIMEESSEESDPSDGVIQKKKK------QADKPKRKHEEPTKPDV 365
SPP+ +K + ++ S P ++K + P + H
Sbjct: 169 SPPRAHSSAEKNESTHVNDDDDVSSPPRAHSLEKNESTHVNEDNISSPPKAHSSKKNEST 228
Query: 366 PSDGDDDAPPPK----KKKKQVRIVEKPTRVEPAADVIRRIET---------PARVTRST 412
+ +D + PP+ K+ + +P VEP+ +RR+ AR T+
Sbjct: 229 HMNDEDVSFPPRTRSSKETSDILTTTQPAIVEPSEPKVRRVSRRKQLYAKRYKARETQPA 288
Query: 413 VRSSSKSAVIDSDDDLNSLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQ 472
+ SS+ V+ +D+ ++ + P + L + S LT +QPA S P P +
Sbjct: 289 IAESSEPKVLHVNDE--NVSSPPEAHSLEKASDILTT---TQPAIAESSEPKVPHVNDEN 343
Query: 473 PSP---------QEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPR----T 519
S ++ N+ P P + S L T++P I E SEP+ R
Sbjct: 344 VSSTPRAHSSKKNKSTRKNVDNVPSPPKTRSSKKTSDILTTTQPTIAESSEPKVRHVNDD 403
Query: 520 SDHSVPRA-SERPVRRTTDTESTTPCP----SVSFPANV-ADSSPSNNSESVRKFVQIAK 573
+ S PRA S + + T + P P S +N+ A + P+ S K ++++
Sbjct: 404 NVSSTPRAHSSKKNKSTRKNDDNIPSPPKTRSSKKTSNILATTQPAKAEPSEPKVTRVSR 463
Query: 574 EKE 576
KE
Sbjct: 464 RKE 466
Score = 31.6 bits (70), Expect = 1.9
Identities = 26/81 (32%), Positives = 35/81 (43%), Gaps = 6/81 (7%)
Query: 618 LAQPAQPAPENSESNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGAS--EANSSN 675
LA P+ N+ +V SP ++E N TH ++D DV SP A E N S
Sbjct: 149 LAIAESSEPKVPHVNDGNVSSPPRAHSSAEKNESTHVNDDD--DVSSPPRAHSLEKNEST 206
Query: 676 HPTSPETNLSIVPFTYLKPNN 696
H E N+S P + N
Sbjct: 207 HVN--EDNISSPPKAHSSKKN 225
>At1g19950 unknown protein
Length = 315
Score = 49.7 bits (117), Expect = 7e-06
Identities = 45/174 (25%), Positives = 72/174 (40%), Gaps = 17/174 (9%)
Query: 512 QSEPEPRTSDHSVPRASERPVRRTTDTESTTPCPSVSFPANVADSSPSNNSE-------- 563
QS P+P+ + A E ++ D ++T+ S + + P ++
Sbjct: 145 QSTPKPKPKEKKQAAAPEEEEQKQPDLKATSQAASSNPQVRLQSKKPQLVTKEPISPKPL 204
Query: 564 -SVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPA 622
S RK Q+ E + A + T + PGP P P P P P AA A PA
Sbjct: 205 SSPRKQQQLQTETKEAKASVSQTKLTTLTPPGPPP-----PPPPPPPSPTTAAKRNADPA 259
Query: 623 QPAPENSESNNSSVRS-PHPQVETSEPNLETHTSNDQTLDV--GSPQGASEANS 673
QP+P +E + +V + P P E + T ++TL + GS + A A +
Sbjct: 260 QPSPTEAEEASQTVAALPEPASEIQRASSSKETIMEETLRITRGSLRKARSAGA 313
>At2g28240 putative proline/hydroxyproline-rich glycoprotein
Length = 660
Score = 49.3 bits (116), Expect = 9e-06
Identities = 71/300 (23%), Positives = 120/300 (39%), Gaps = 35/300 (11%)
Query: 407 RVTRSTVRSSSKSAVIDSDDDLNSL----DALPISALLNRTSTPLTLILESQPAEQTISP 462
R RS+ S+ + + ++ +NS ++ + +N + P + ++P T P
Sbjct: 289 RAPRSSQHSTQQQQAVQTNRHMNSTAPPRPSVTAAEPMNSAAPPRPSVTAAEPMNSTAPP 348
Query: 463 PTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLH---TSEPIILEQSEPEPRT 519
S ++ P APL + TP+PS + ++ N T+ P +E P
Sbjct: 349 RPSVTAAEATPPNLSAPLPHC-NTPQPSPISQQAAVESNTQMQSTALPRPSVTAEARPLH 407
Query: 520 SDHSVPRASERPVRR-------TTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIA 572
HS + RP+ + T T + P PS++ A + P +N+ R Q
Sbjct: 408 QPHS-NTSQPRPIPQQALAQSNTNITSTALPRPSITAEARLLHQ-PHSNTPQPRPIPQ-- 463
Query: 573 KEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPA--QPAPENSE 630
K A ++ T PRP LV + P P+ + P +P QPA ++
Sbjct: 464 KALVQANTDINSTAL-------PRP--LVTAEAP-PLHQSSCKAPQPKPISQQPAVQSKT 513
Query: 631 SNNSSVRSPHPQVET-SEPNLETHTSNDQTLDVGSP---QGASEANSSNHPTSPETNLSI 686
+S P P V T + P + + Q V P Q +E NS+ HP T+ +I
Sbjct: 514 DIINSTALPRPSVTTEARPLHQPRSKTPQPKPVSQPPAKQSNTEINSTPHPRPSVTSKAI 573
>At2g43680 SF16 protein {Helianthus annuus} like protein
Length = 668
Score = 48.9 bits (115), Expect = 1e-05
Identities = 78/366 (21%), Positives = 142/366 (38%), Gaps = 65/366 (17%)
Query: 462 PPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSD 521
PPT R + + SP P + +P+P+ P + P L P PR
Sbjct: 78 PPTPDRPNPYSASPPPRPASPRVASPRPTSPRVASPRVPSPRAEVPRTLSPKPPSPRA-- 135
Query: 522 HSVPRA-SERPVRRTTDTESTTPCPSVSFPANVAD-----SSPSNNSESVRKFVQIAKEK 575
VPR+ S +P P P P +++ S PS+ S + ++ A +
Sbjct: 136 -EVPRSLSPKP-----------PSPRADLPRSLSPKPFDRSKPSSASANAPPTLRPASTR 183
Query: 576 ESAISEYYHTVPSPR--------------KYPGPR--PERLVDPDHPLPVFPLNAADP-- 617
+ H+VPSPR K P PR P L P P P ADP
Sbjct: 184 VPSQRITPHSVPSPRPSSPRGASPQAISSKPPSPRAEPPTLDTPRPPSPRAASLRADPPR 243
Query: 618 --LAQPAQPAPENSESNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGASEANSSN 675
A+P P P + ++ + +P P + P + S+ LD P ++
Sbjct: 244 LDAARPTTPRPPSPLADAPRLDAPRP----TTPKPPSPRSDPPRLDAPRP-------TTP 292
Query: 676 HPTSPETNLSIVPFTYLKPNNLLECISVFNHEASLMIRNVQGHTDLSEDPDSVAEEWNRL 735
P SP + + P + V+ E +L ++ H ++ + R
Sbjct: 293 KPPSPRS---------VSPRAVQRREIVYRPEPTLPVQ----HASATKIQGAFRGYMARK 339
Query: 736 SDWLIAQVPSLMSRLTAERDQRIEAARQRFNRRVILHEQQLRSKLLEVVE-EARRKKEQA 794
S + + L + +R ++ ++V+ + Q++S+ ++++E +A+ +K++A
Sbjct: 340 SFRALKGLVRLQGVVRGYSVKRQTINAMKYMQQVVRVQSQIQSRRIKMLENQAQVEKDEA 399
Query: 795 EEAARQ 800
+ AA +
Sbjct: 400 KWAASE 405
Score = 34.3 bits (77), Expect = 0.30
Identities = 50/188 (26%), Positives = 72/188 (37%), Gaps = 31/188 (16%)
Query: 349 QADKPKRKHEEPTKPDVPSDGDDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARV 408
+AD P+ +P PS +APP + TRV + +P
Sbjct: 149 RADLPRSLSPKPFDRSKPSSASANAPPTLRPAS--------TRVPSQRITPHSVPSP--- 197
Query: 409 TRSTVRSSSKSAVIDSDDDLNSLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRS 468
S+ R +S A+ S A P + R +P L + P + PT+PR
Sbjct: 198 RPSSPRGASPQAISSKPP---SPRAEPPTLDTPRPPSPRAASLRADPPRLDAARPTTPR- 253
Query: 469 SFFQPSP-QEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRA 527
PSP +AP L P+P+ P P S+P L+ P P T PR+
Sbjct: 254 ---PPSPLADAP---RLDAPRPTTPKPPSP------RSDPPRLD--APRPTTPKPPSPRS 299
Query: 528 -SERPVRR 534
S R V+R
Sbjct: 300 VSPRAVQR 307
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 48.5 bits (114), Expect = 2e-05
Identities = 59/229 (25%), Positives = 79/229 (33%), Gaps = 33/229 (14%)
Query: 444 STPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP-SEPTSPITIQYNL 502
S P L E+ P +S P P SS P P EAP + +P P + P P +L
Sbjct: 99 SPPPPLPTEAPPPANPVSSPP-PESSPPPPPPTEAPPTTPITSPSPPTNPPPPPESPPSL 157
Query: 503 HTSEPIILEQSEPEPRTSDHSVPR---------------------ASERPVRRTTDTEST 541
+P P+ HS PR ASERP +D+E
Sbjct: 158 PAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPASERPSTPPSDSEHP 217
Query: 542 TPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKE----SAISEYYHTVPSPRKYPGPRP 597
+P P P P + + K S S T+P P+ P P P
Sbjct: 218 SP-PPPGHPKRREQPPPPGSKRPTPSPPSPSDSKRPVHPSPPSPPEETLPPPKPSPDPLP 276
Query: 598 ERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETS 646
P LP P + P P+ P S +N S +P P + S
Sbjct: 277 SNSSSPPTLLP--PSSVVSP---PSPPRKSVSGPDNPSPNNPTPVTDNS 320
Score = 48.5 bits (114), Expect = 2e-05
Identities = 62/238 (26%), Positives = 79/238 (33%), Gaps = 37/238 (15%)
Query: 458 QTISPPTSPRSSFFQPSPQEAPLWNMLQTPKPSE-PTSPITIQYNLHTSEPIILEQSEPE 516
Q+ PP +P SS P P+ +P L P P+ P SP S P L P
Sbjct: 59 QSSPPPETPLSS---PPPEPSPPSPSLTGPPPTTIPVSPPP-----EPSPPPPLPTEAPP 110
Query: 517 PRTSDHSVPRASERPVRRTTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKE 576
P S P S P T+ TTP S S P N P + S+
Sbjct: 111 PANPVSSPPPESSPPPPPPTEAPPTTPITSPSPPTN--PPPPPESPPSL----------- 157
Query: 577 SAISEYYHTVPSPRKYPGPRPERLVDPDHP----LPVFPLNAADPLAQ--PAQPAPENSE 630
P P P P P +LV P H LP P + P + P+ PA E
Sbjct: 158 --------PAPDPPSNPLP-PPKLVPPSHSPPRHLPSPPASEIPPPPRHLPSPPASERPS 208
Query: 631 SNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGASEANSSNHPTSPETNLSIVP 688
+ S P P E SP S++ HP+ P +P
Sbjct: 209 TPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSDSKRPVHPSPPSPPEETLP 266
Score = 40.0 bits (92), Expect = 0.005
Identities = 58/262 (22%), Positives = 89/262 (33%), Gaps = 32/262 (12%)
Query: 310 PPALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPDVPSDG 369
PP SPP + + E+ ++P + P PT P
Sbjct: 86 PPTTIPVSPPPEPSPPPPLPTEAPPPANPVSS--PPPESSPPPPPPTEAPPTTPITSPSP 143
Query: 370 DDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDLN 429
+ PPP + + + P+ P ++ +P R S S
Sbjct: 144 PTNPPPPPESPPSLPAPDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIPPPP-------- 195
Query: 430 SLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP 489
LP R STP S + PP P+ P P + P P
Sbjct: 196 --RHLPSPPASERPSTP-----PSDSEHPSPPPPGHPKRREQPPPPGS-------KRPTP 241
Query: 490 SEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVR----RTTDTESTTPCP 545
S P SP + +H S P E++ P P+ S +P S P + + + P
Sbjct: 242 S-PPSPSDSKRPVHPSPPSPPEETLPPPKPSPDPLPSNSSSPPTLLPPSSVVSPPSPPRK 300
Query: 546 SVSFPANVADSSP---SNNSES 564
SVS P N + ++P ++NS S
Sbjct: 301 SVSGPDNPSPNNPTPVTDNSSS 322
>At4g32710 putative protein kinase
Length = 731
Score = 47.4 bits (111), Expect = 3e-05
Identities = 67/253 (26%), Positives = 96/253 (37%), Gaps = 34/253 (13%)
Query: 423 DSDDDLNSLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWN 482
DS D +S A P+S L S+P L + T SPP P S P P E+P
Sbjct: 24 DSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVESPPSP-PIESPPPP 82
Query: 483 MLQTPKPSEPTSPITIQYNLHT---SEPIILEQSEPEPRTS---DHSVPRASE-RPVRRT 535
+L++P P SP ++ S P+ ++P P S +VP + P R+
Sbjct: 83 LLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISPPPRS 142
Query: 536 TDTESTTPC------------------PSVSFPANVADSSPSNNSESVRKFVQIAKEKES 577
+EST P PS P N + +PS N+ S+ + K S
Sbjct: 143 LPSESTPPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNTPSLPETSPPPKPPLS 202
Query: 578 AISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPA--PENSESNNSS 635
+ P P+K P P LP +A P P P+ S + + S
Sbjct: 203 TTPFPSSSTPPPKKSPAAVTLPFFGPAGQLP------DGTVAPPIGPVIEPKTSPAESIS 256
Query: 636 VRSPHPQVETSEP 648
+P P V S P
Sbjct: 257 PGTPQPLVPKSLP 269
Score = 40.4 bits (93), Expect = 0.004
Identities = 53/207 (25%), Positives = 76/207 (36%), Gaps = 16/207 (7%)
Query: 453 SQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP-------SEPT-SPITIQYNLHT 504
+ P ++ P S + P+P +PL L +P P S PT SP +
Sbjct: 13 TSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVESPP 72
Query: 505 SEPIILEQSEPEPRTSDHSVPRASERPVRRTTDTESTTPCPSVSF-PANVADSSPSNNSE 563
S PI P P + P E P + + + P + F PA + S SE
Sbjct: 73 SPPI----ESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSE 128
Query: 564 SVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQ 623
+V I+ S SE V + P P R P P P+N++ P P
Sbjct: 129 TVPPGNTISPPPRSLPSESTPPVNTASPPPPSPPRRRSGPKPSFPP-PINSSPPNPSPNT 187
Query: 624 PA-PENSESNNSSV-RSPHPQVETSEP 648
P+ PE S + +P P T P
Sbjct: 188 PSLPETSPPPKPPLSTTPFPSSSTPPP 214
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 47.0 bits (110), Expect = 4e-05
Identities = 60/244 (24%), Positives = 88/244 (35%), Gaps = 24/244 (9%)
Query: 463 PTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDH 522
P++P S P P +P Q PK P SP+ S+P S +P S
Sbjct: 134 PSAPAHS---PVPSVSPT----QPPKSHSPVSPVAPASAPSKSQPPRSSVSPAQPPKSSS 186
Query: 523 SVPRASERPVRRTTDTESTTPCPSVSFPANVADS---SPSNN-SESVRKFVQIAKEKESA 578
+ T TP PS P+ V+ S SP++ S S + +
Sbjct: 187 PISHTPALSPSHATSHSPATPSPSPKSPSPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPS 246
Query: 579 ISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESNNSSVRS 638
S + SP P P P P ++ P PA P+P +S S V S
Sbjct: 247 HSPAHAPSHSPAHAPSHSPAHSPSHSPATPKSPSPSSSPAQSPATPSPMTPQS-PSPVSS 305
Query: 639 PHPQVETS----------EPNLETHTSNDQTLDVGSPQGASEANSSNHPTSPETNLSIVP 688
P P + P+ T T+++ T SP+ + + S TS + L
Sbjct: 306 PSPDQSAAPSDQSTPLAPSPSETTPTADNITAPAPSPR--TNSASGLAVTSVMSTLFSAT 363
Query: 689 FTYL 692
FT+L
Sbjct: 364 FTFL 367
Score = 39.3 bits (90), Expect = 0.009
Identities = 46/201 (22%), Positives = 78/201 (37%), Gaps = 16/201 (7%)
Query: 446 PLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTS 505
P + +SQP ++SP P+SS P L +P + SP T + +
Sbjct: 162 PASAPSKSQPPRSSVSPAQPPKSS--SPISHTPAL-----SPSHATSHSPATPSPSPKSP 214
Query: 506 EPIILEQSEPEPRTSDHSVPRA-SERPVRRTTDTESTTPCPSVSF-PANVADSSPSNNSE 563
P+ S T HS S P + + + P S + P++ SPS++
Sbjct: 215 SPVSHSPSHSPAHTPSHSPAHTPSHSPAHAPSHSPAHAPSHSPAHAPSHSPAHSPSHSPA 274
Query: 564 SVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDP-DHPLPVFP-----LNAADP 617
+ + + +S + T SP P P++ P D P+ P AD
Sbjct: 275 TPKSPSPSSSPAQSPATPSPMTPQSPSPVSSPSPDQSAAPSDQSTPLAPSPSETTPTADN 334
Query: 618 LAQPAQPAPENSESNNSSVRS 638
+ PA P+P + ++ +V S
Sbjct: 335 ITAPA-PSPRTNSASGLAVTS 354
>At3g18610 unknown protein
Length = 636
Score = 47.0 bits (110), Expect = 4e-05
Identities = 63/282 (22%), Positives = 114/282 (40%), Gaps = 65/282 (23%)
Query: 312 ALEFDSPPKKKKTRKMIMEESSEESDP---SDGVIQKKKKQADKPKRKHEEPTKPDVPSD 368
A + D K K+T + +ESS E + SD I KK+ + K+ E + D S
Sbjct: 69 ASDSDEEEKTKETPSKLKDESSSEEEDDSSSDEEIAPAKKRPEPIKKAKVESSSSDDDST 128
Query: 369 GDDDAPPPKKKKKQVRIVEKPTRVEPAA---------DVIRRIETPARVTRSTVRSSSKS 419
D++ P KKQ ++EK +VE ++ + + + PA + ++ + SSS
Sbjct: 129 SDEETAP---VKKQPAVLEK-AKVESSSSDDDSSSDEETVPVKKQPAVLEKAKIESSS-- 182
Query: 420 AVIDSDDDLNSLDALPISALLNRTSTPL---TLILESQPAEQTISPPTSPRSSFFQPSPQ 476
SDDD +S + + P+ T +LE AE + S S SS +P+P
Sbjct: 183 ----SDDDSSS----------DEETVPMKKQTAVLEKAKAESSSSDDGS--SSDEEPTPA 226
Query: 477 EAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRRTT 536
+ EPI++++ + +SD P ++P
Sbjct: 227 K---------------------------KEPIVVKKDSSDESSSDEETPVVKKKPTTVVK 259
Query: 537 DTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESA 578
D ++ + S ++ + +P+ V+ AK+ S+
Sbjct: 260 DAKAES-SSSEEESSSDDEPTPAKKPTVVKNAKPAAKDSSSS 300
Score = 36.2 bits (82), Expect = 0.078
Identities = 71/373 (19%), Positives = 129/373 (34%), Gaps = 57/373 (15%)
Query: 320 KKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPDVPSDGDDDAPPPKKK 379
K+ + M+ + ++ V+QK+K + PK K E + SD ++ K
Sbjct: 26 KRDAEEDLDMQVTKKQKKELIDVVQKEKAEKTVPK-KVESSSSDASDSDEEEKTKETPSK 84
Query: 380 KKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDLNSLDALPISAL 439
K E+ ++ + P + ++ V SSS SDDD
Sbjct: 85 LKDESSSEEEDDSSSDEEIAPAKKRPEPIKKAKVESSS------SDDD------------ 126
Query: 440 LNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQ 499
TS T ++ QPA + S S S +E T P+ Q
Sbjct: 127 --STSDEETAPVKKQPAVLEKAKVESSSSDDDSSSDEE---------------TVPVKKQ 169
Query: 500 YNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRRTTDTESTT----------PCPSVSF 549
+ I S+ + + + +VP + V ES++ P P+
Sbjct: 170 PAVLEKAKIESSSSDDDSSSDEETVPMKKQTAVLEKAKAESSSSDDGSSSDEEPTPAKKE 229
Query: 550 PANV-ADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLP 608
P V DSS ++S+ + + K+K + + V + E D P P
Sbjct: 230 PIVVKKDSSDESSSD---EETPVVKKKPTTV------VKDAKAESSSSEEESSSDDEPTP 280
Query: 609 VFPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGA 668
A+PA +SE ++ S + T + + + TS ++ S +
Sbjct: 281 AKKPTVVKN-AKPAAKDSSSSEEDSDEEESDDEKPPTKKAKVSSKTSKQESSSDESSDES 339
Query: 669 SEANSSNHPTSPE 681
+ S + +P+
Sbjct: 340 DKEESKDEKVTPK 352
>At5g38560 putative protein
Length = 681
Score = 46.2 bits (108), Expect = 8e-05
Identities = 65/233 (27%), Positives = 82/233 (34%), Gaps = 51/233 (21%)
Query: 455 PAEQTISPPTSPRSSFFQPSPQEAPLWNMLQ---TPKPSEPTSPITIQYNLHTSEPIILE 511
P +SPP+S S+ P Q P TP PS P SP P ++
Sbjct: 5 PPLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSP-----------PPVVS 53
Query: 512 QSEPEPRTSDHSVPRASERPVRRTTDTESTTPCPSV--SFPANVADSSPSNNSESVRKFV 569
S P P S S P +S P P P V S P VA S P V
Sbjct: 54 SSPPPPVVS--SPPPSSSPP-----------PSPPVITSPPPTVASSPPPP--------V 92
Query: 570 QIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPENS 629
IA S + T P+P + P P P P P +P +P+ P +
Sbjct: 93 VIASPPPSTPAT---TPPAPPQTVSPPPPPDASPSPPAP----TTTNPPPKPSPSPPGET 145
Query: 630 ESNNSSVRSPHPQVETSEPNLETHTSNDQTLDVGSPQGASEANSSNHPTSPET 682
S SP P+ S P T TS P S + S++PT P T
Sbjct: 146 PSPPGETPSP-PKPSPSTPTPTTTTSPP------PPPATSASPPSSNPTDPST 191
Score = 40.0 bits (92), Expect = 0.005
Identities = 68/285 (23%), Positives = 94/285 (32%), Gaps = 87/285 (30%)
Query: 361 TKPDVPSDGDDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSS---S 417
T+P PS APPP + P+ + V+ P V+ SS S
Sbjct: 27 TQPTTPS-----APPP--------VTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPS 73
Query: 418 KSAVIDSDDDLNSLDALPISALLNRTSTPLTLILESQPAE-QTISPPTSPRSSFFQPSPQ 476
+ + S P+ STP T + PA QT+SPP P +S PSP
Sbjct: 74 PPVITSPPPTVASSPPPPVVIASPPPSTPAT----TPPAPPQTVSPPPPPDAS---PSPP 126
Query: 477 EAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRRTT 536
N P PS P P P S P+ S T
Sbjct: 127 APTTTNPPPKPSPSPPGET-------------------PSPPGETPSPPKPSP---STPT 164
Query: 537 DTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPR 596
T +T+P P + A+ S+P++
Sbjct: 165 PTTTTSPPPPPATSASPPSSNPTD------------------------------------ 188
Query: 597 PERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESNNSSVRSPHP 641
P L P PLPV P P+A+P PA S + N+++ S P
Sbjct: 189 PSTLAPPPTPLPVVP--REKPIAKPTGPA---SNNGNNTLPSSSP 228
>At2g15880 unknown protein
Length = 727
Score = 45.8 bits (107), Expect = 1e-04
Identities = 51/202 (25%), Positives = 74/202 (36%), Gaps = 42/202 (20%)
Query: 453 SQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQ 512
S A + SP P +P P + ++P+P++P + P+ +
Sbjct: 389 SSQATPSKSPSPVPTRPVHKPQPPK-------ESPQPNDP----------YNQSPVKFRR 431
Query: 513 SEPEPRTSDHSV----PRASERPVRRTTDTESTTPCPSVSFPANVADSSPSNN--SESVR 566
S P P+ H V P AS P T+ +TP P V P +S N+ +S
Sbjct: 432 SPPPPQQPHHHVVHSPPPASSPP---TSPPVHSTPSP-VHKPQPPKESPQPNDPYDQSPV 487
Query: 567 KFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPAP 626
KF + S H+ P P P P + P P PV+ P+ P P P
Sbjct: 488 KF------RRSPPPPPVHSPPPPSPIHSPPPPPVYSPPPPPPVYSPPPPPPVYSPPPPPP 541
Query: 627 ENSESNNSSVRSPHPQVETSEP 648
V SP P V + P
Sbjct: 542 ---------VHSPPPPVHSPPP 554
Score = 34.7 bits (78), Expect = 0.23
Identities = 76/362 (20%), Positives = 105/362 (28%), Gaps = 44/362 (12%)
Query: 333 SEESDPSDGVIQKKKKQADKPKRKHEEPTKPDV----PSDGDDDAPPPKKKKKQVRIVEK 388
S ++ PS + KP+ E P D P PPP++ V
Sbjct: 389 SSQATPSKSPSPVPTRPVHKPQPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPP 448
Query: 389 PTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSDDDLNSLDALPISALLNRTSTPLT 448
P P + + +P + S + D P S P
Sbjct: 449 PASSPPTSPPVHSTPSPVHKPQPPKESPQPNDPYDQSPVKFRRSPPPPPV----HSPPPP 504
Query: 449 LILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPI 508
+ S P SPP P + P P P+++ P P P+ H+ P
Sbjct: 505 SPIHSPPPPPVYSPPPPP--PVYSPPPPP-PVYSPPPPPPVHSPPPPV------HSPPPP 555
Query: 509 ILEQSEPEPRTSDHSVPRASERPVRRTTDTESTTPCPSVSFPANVADSSPSNNSESVRKF 568
+ S P P S + PV + P P V P S P
Sbjct: 556 V--HSPPPPVHSPPPPVHSPPPPVHSPPPPVYSPPPPPVHSPPPPVHSPPPP-------- 605
Query: 569 VQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPEN 628
H+ P P Y P P + P P+ P P P P
Sbjct: 606 --------------VHSPPPP-VYSPPPPPPVHSPPPPVFSPPPPVHSPPPPVYSPPPPV 650
Query: 629 SESNNSSVRSPHPQVETSEPNLETHTSNDQT-LDVGSPQGASEANSSNHPTSPETNLSIV 687
V+SP P S P L S+ T V SP + + + P P I
Sbjct: 651 YSPPPPPVKSPPPPPVYSPPLLPPKMSSPPTQTPVNSPPPRTPSQTVEAP-PPSEEFIIP 709
Query: 688 PF 689
PF
Sbjct: 710 PF 711
>At3g05900 unknown protein
Length = 660
Score = 45.4 bits (106), Expect = 1e-04
Identities = 92/495 (18%), Positives = 177/495 (35%), Gaps = 64/495 (12%)
Query: 243 LKKMGLIKTKVAPAHEDTSDEVRKRRVPLVDYPLWSQADAPEAILCYIDDLRQQGYEIDV 302
LK+ L+K VA + +T D + V+ L E I D+ + E +V
Sbjct: 92 LKEPILVKETVAEVNVETVDTEKAEEKQTVENVLIEDHKDQEETK--IVDVSESTDEAEV 149
Query: 303 DEFFR-NLPPALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPT 361
+ ++ P + + +K ++ EE+ + + ++ + DK + +
Sbjct: 150 QQVEPVDVQPVKDAEKAEEKPTVESVVEEETKDREETK--IVDVSESAGDKQVESVDVQS 207
Query: 362 KPDVPSDGDDDA----------PPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRS 411
DV ++ ++ P P+ +K +EK +E +V++ ET ++
Sbjct: 208 VRDVSAEIAEEKVKDVEALEVEPKPETSEKVETQLEKARELETEVEVVKAEETAEATEQA 267
Query: 412 TVRSSSK---SAVIDSDDDLNSLDAL-----------------------PISALLNRTST 445
V K V + D ++NS D PI S
Sbjct: 268 KVELEGKLEDVIVEEKDSEINSKDEKTSESGSALCSEEILSTIQESNTDPIKETEGDASY 327
Query: 446 PLTLILESQPAEQ-TISPPTS---PRSSFFQPSPQEAPLWNMLQTPKPSEPT-----SPI 496
P+ +I ++ E+ + P + P S SP++ N KP E T +P
Sbjct: 328 PIDVIEKAITEEKHVVDEPANEEKPSESSAALSPEKVVPINQDSDTKPKEETEGDAAAPA 387
Query: 497 TIQYNLHTSEPIILEQSEPEPRTSDHSVPRASER--PVRRTTDTESTTPC-PSVSFPANV 553
+ T E ++++ + TS+ E+ P + DTE VS PA++
Sbjct: 388 DVIEKAITEEKYVVDEPSKDETTSESGSALCPEKAVPTNQDLDTEPKKETEEDVSSPADI 447
Query: 554 ADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLN 613
+ + + V + + K ES + V + P++ + D P P +
Sbjct: 448 IEKAITEEKHVVEEPSKDEKTSESGSALSPEKVVPTNQDSDTEPKKETEGDVPSPADVIE 507
Query: 614 AADPLAQPAQPAPENSESNN-SSVRSPHPQVETSEPNLETHT-------SNDQTL---DV 662
A + P E N S + ++ + N++ T +++TL D
Sbjct: 508 KAITDEKHVVEEPLKDEQENVSEAKDVVTKLAAEDENIKKDTDTPVAEGKSEETLKETDT 567
Query: 663 GSPQGASEANSSNHP 677
S + + AN P
Sbjct: 568 ESVEKEAAANKQEEP 582
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 45.1 bits (105), Expect = 2e-04
Identities = 59/261 (22%), Positives = 93/261 (35%), Gaps = 31/261 (11%)
Query: 458 QTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP-SEPTSPITIQYNLHTSEPIILEQSEPE 516
Q +PP P P E +++Q +P S+ S +++ + + P
Sbjct: 544 QAGAPPPPPPLPAAASKPSEQLQHSVVQATEPLSQGNSWMSLAGSTFQTVPNEKNLITLP 603
Query: 517 PRTSDHSVPRASERPVRRTTDTESTTPCPSVSFPAN-------------------VADSS 557
P S AS P +TT++ +P S + P N +D+
Sbjct: 604 PTPPLASTSHASPEPSSKTTNSLLLSPQASPATPTNPSKTVSVDFFGAATSPHLGASDNV 663
Query: 558 PSNNSESVRKFVQIAK-EKESAISEYYHTVPSP-------RKYPGPRPERLVDPDHPLPV 609
SN + R I+ +K+ A+ P P K P P P P P+ V
Sbjct: 664 ASNLGQPARSPPPISNSDKKPALPRPPPPPPPPPMQHSTVTKVPPPPPPAPPAPPTPI-V 722
Query: 610 FPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETSEPNLETHTSN--DQTLDVGSPQG 667
+ P P PAP +SN S P + P L TH+++ T P G
Sbjct: 723 HTSSPPPPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPPRLPTHSASPPPPTAPPPPPLG 782
Query: 668 ASEANSSNHPTSPETNLSIVP 688
+ A S+ P P+ + P
Sbjct: 783 QTRAPSAPPPPPPKLGTKLSP 803
>At3g50600 unknown protein
Length = 852
Score = 44.3 bits (103), Expect = 3e-04
Identities = 62/290 (21%), Positives = 104/290 (35%), Gaps = 34/290 (11%)
Query: 346 KKKQADKPKRKHEEPTKPDVPSDGD----DDAPPPKKKKKQVRIVEKPTRVEPAADVIRR 401
K+ AD+ + EE K + GD D+ K KK Q+RI EKPT + ++
Sbjct: 352 KQNVADEQAKAAEEFKKTMYGATGDGSSSDEEGVTKPKKLQIRIREKPTSTTVDVNKLKE 411
Query: 402 IETPARVTRSTVRSSSKSAVIDSDDDLNSLDALPISALLNRTSTPLTLILESQPAEQTIS 461
++ + S++ I N S L +L SQP+ T++
Sbjct: 412 AAKTFKLGDGLGLTMSRTKSI------------------NAGSQDLGQML-SQPSSSTVA 452
Query: 462 PPTSPRSSFFQPSPQEAPLWNM----LQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEP 517
T+P S+ P W + P P +PI + +T + + ++ P P
Sbjct: 453 TTTAPSSASAPVDPFAMSSWTQQPQPVSQPAPPGVAAPIPEDFFQNTIPSVEVAKTLPPP 512
Query: 518 RTSDHSVPRASERPV--RRTTDTESTTPCPSVSFP-ANVADSSPSNNSESVRKFVQIAKE 574
T + +A+ + + + + TP P + P V P S+ Q
Sbjct: 513 GTYLSKMDQAARAAIAAQGGPNQANNTPLPDIGLPDGGVPQQYPQQTSQQPGAPFQTVGL 572
Query: 575 KESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQP 624
+ + + Y P + P P D + P N D P QP
Sbjct: 573 PDGGVRQQY---PGQNQVPSQVPVSTQPLDLSVLGVP-NTGDSGKPPGQP 618
>At5g56890 unknown protein
Length = 1113
Score = 43.9 bits (102), Expect = 4e-04
Identities = 71/306 (23%), Positives = 108/306 (35%), Gaps = 36/306 (11%)
Query: 366 PSDGDDDAPPPKKKKKQVRIVEKPTRVEPAADVIRRIETPARVTRSTVRSSSKSAVIDSD 425
P D PP K Q + P + PA V V ++ A I
Sbjct: 53 PESHKSDNVPPSKASSQPSLPPLADLAAPPPSDSVGGKAPAGVPVVFVPNAPAPATIPVK 112
Query: 426 DDLNSLDALPI-SALLNRTSTPLTLILESQPAEQTISPPTSPRSSFFQPSPQEAPLWNM- 483
D LP+ S + + TP+ P + PP S R + P+P +P+ ++
Sbjct: 113 D-------LPVASPPVLQPITPIASPPRFIPGDAPKEPPFSGRVT---PAPVSSPVSDIP 162
Query: 484 ----LQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASERPVRRTTD-- 537
+ P P+ P N H PI + P S ++ PV
Sbjct: 163 PIPSVALPPPTPSNVPPRNASNNHKPPPI---EKSIAPVASPPTISIDIAPPVHPVIPKL 219
Query: 538 TESTTPCPSVSFPANVADSSPSNNSESVRKFVQI---AKEKESAISEYYHTVPSPR---- 590
T S++P P+ + SP N + I A + A + H PS
Sbjct: 220 TPSSSPVPT----STPTKGSPRRNPPTTHPVFPIESPAVSPDHAANPVKHPPPSDNGDDS 275
Query: 591 KYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETSEPNL 650
K PG P + PLPVFP A+ P P+ P + S +P P +++ N+
Sbjct: 276 KSPGAAPAN--ETAKPLPVFPHKASPPSIAPSAPKFNRHSHHTSPSTTPPP--DSTPSNV 331
Query: 651 ETHTSN 656
H S+
Sbjct: 332 HHHPSS 337
>At3g05060 putative SAR DNA-binding protein-1
Length = 533
Score = 43.1 bits (100), Expect = 6e-04
Identities = 29/97 (29%), Positives = 39/97 (39%), Gaps = 14/97 (14%)
Query: 298 YEIDVDEFFRNLPPALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKK----------- 346
Y D E S K KK +K + EE EE +PS+ +KK
Sbjct: 436 YNTAADSLLGETSAKSEEPSKKKDKKKKKKVEEEKPEEEEPSEKKKKKKAEAETEAVVEV 495
Query: 347 ---KKQADKPKRKHEEPTKPDVPSDGDDDAPPPKKKK 380
+K+ +K KRKHEE + P+ D KK K
Sbjct: 496 AKEEKKKNKKKRKHEEEETTETPAKKKDKKEKKKKSK 532
Score = 31.2 bits (69), Expect = 2.5
Identities = 19/87 (21%), Positives = 42/87 (47%), Gaps = 1/87 (1%)
Query: 311 PALEFDSPPKKKKTRKMIMEESSEESDPSDGVIQKKKKQADKPKRKHEEPTKPDVPSDGD 370
P +E + KK + +I + + +D ++ + ++++P +K ++ K V +
Sbjct: 413 PKIEVYNKDKKMGSGGLITPAKTYNT-AADSLLGETSAKSEEPSKKKDKKKKKKVEEEKP 471
Query: 371 DDAPPPKKKKKQVRIVEKPTRVEPAAD 397
++ P +KKKK+ E VE A +
Sbjct: 472 EEEEPSEKKKKKKAEAETEAVVEVAKE 498
>At3g63460 putative protein
Length = 1097
Score = 42.4 bits (98), Expect = 0.001
Identities = 62/251 (24%), Positives = 108/251 (42%), Gaps = 40/251 (15%)
Query: 428 LNSLDALPISALLNRTSTPLTLILESQPAEQTISPPTSPRSSF-FQPSPQEAPLWNMLQT 486
L LD+ +S L+ ++L E + T S T P+S+ + P +A N+L
Sbjct: 754 LKVLDSGGLSPELSILRDRISLSAEPE-TNTTASGNTQPQSTMPYNQEPTQAQP-NVLAN 811
Query: 487 PKPSEPTSPITIQYNL-HTSEPIILE-------QSEPEPRTSDHSVPRASERPVRRTTDT 538
P ++ P T Y + S P + + Q++P P+ S P ++ +P RTT
Sbjct: 812 PYDNQYQQPYTDSYYVPQVSHPPMQQPTMFMPHQAQPAPQPSFTPAPTSNAQPSMRTTFV 871
Query: 539 ESTTPC---------PSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSP 589
ST P P++S + + + PSNN+ V +Y + PS
Sbjct: 872 PSTPPALKNADQYQQPTMS---SHSFTGPSNNAYPV----------PPGPGQYAPSGPSQ 918
Query: 590 -RKYPGPRPERLVDP-DHPLPVFPL---NAADPLAQPAQPAPENSESNNS-SVRSPHPQV 643
+YP P+ ++V P P+ P+ A QPA P + + + + + +P P V
Sbjct: 919 LGQYPNPKMPQVVAPAAGPIGFTPMATPGVAPRSVQPASPPTQQAAAQAAPAPATPPPTV 978
Query: 644 ETSE-PNLETH 653
+T++ N+ H
Sbjct: 979 QTADTSNVPAH 989
>At4g18670 extensin-like protein
Length = 839
Score = 42.0 bits (97), Expect = 0.001
Identities = 56/211 (26%), Positives = 75/211 (35%), Gaps = 42/211 (19%)
Query: 444 STPLTLILESQPAEQTI--SPPTSPRSSFFQPSPQEAPLWNMLQTPKPSEPTSPIT-IQY 500
S P I+ S P+ SPPTSP + SP +P TP S P+SP T
Sbjct: 448 SPPSPSIVPSPPSTTPSPGSPPTSPTTPTPGGSPPSSP---TTPTPGGSPPSSPTTPTPG 504
Query: 501 NLHTSEPIILEQ--SEPEPRTSDH---SVPRASERPVRRTTDTESTTPCPSVSFPANVAD 555
S P S P P S +VP P + ++P PS P+
Sbjct: 505 GSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPSPPTP 564
Query: 556 S------SPSNNSESVRKFVQIAKEKESAISEYYHTVPSPRKYPGPRPERLVDPDHPLPV 609
S SP NS + +PSP + GP P P PLP
Sbjct: 565 STPPTPISPGQNSPPI--------------------IPSP-PFTGPSPPS--SPSPPLP- 600
Query: 610 FPLNAADPLAQPAQPAPENSESNNSSVRSPH 640
P+ + P+ P +P S ++ PH
Sbjct: 601 -PVIPSPPIVGPTPSSPPPSTPTPGTLLHPH 630
Score = 38.9 bits (89), Expect = 0.012
Identities = 72/288 (25%), Positives = 93/288 (32%), Gaps = 59/288 (20%)
Query: 414 RSSSKSAVIDSDDDLNSLDALPISALLNRTSTPLTLI---LESQPAEQTISPPTSPRSSF 470
RS+ V+ S S P S ++ S P+T+ P SP P
Sbjct: 402 RSTRPPVVVPSPPTTPSPGGSPPSPSIS-PSPPITVPSPPTTPSPGGSPPSPSIVPSPPS 460
Query: 471 FQPSPQEAPLWNMLQTPKPSEPTSPITIQYNLHTSEPIILEQSEPEPRTSDHSVPRASER 530
PSP P TP S P+SP T P P S S P
Sbjct: 461 TTPSPGSPPTSPTTPTPGGSPPSSPTT-----------------PTPGGSPPSSP----- 498
Query: 531 PVRRTTDTESTTPCPSVSFPANVADSSPSNNSESVRKFVQIAKEKESAISEYYHTVPSPR 590
TTP P S P++ SP + S S TVPSP
Sbjct: 499 ----------TTPTPGGSPPSSPTTPSPGGSPPS-----------PSISPSPPITVPSPP 537
Query: 591 KYPGPRPERLVDPDHPLPVFPLNAADPLAQPAQPAPENSESNNSSVRSPHPQVETSEPNL 650
P P P P P P+ + P+ P S NS P P + P+
Sbjct: 538 STP-TSPGSPPSPSSPTPSSPIPSP---PTPSTPPTPISPGQNSPPIIPSPPF--TGPSP 591
Query: 651 ETHTSNDQTLDVGSPQGASEANSSNHPTSPETNLSIVPFTYLKPNNLL 698
+ S + SP SS P++P P T L P++LL
Sbjct: 592 PSSPSPPLPPVIPSPPIVGPTPSSPPPSTP------TPGTLLHPHHLL 633
Score = 30.0 bits (66), Expect = 5.6
Identities = 22/76 (28%), Positives = 27/76 (34%), Gaps = 6/76 (7%)
Query: 455 PAEQTISPPTSPRSSFFQPSPQEAPLWNMLQTPKP-----SEPTSPITIQYNLHTSEPII 509
P ISPP P + P PQ P P P + P P Y+ S P+
Sbjct: 728 PTYHYISPPPPP-TPIHSPPPQSHPPCIEYSPPPPPTVHYNPPPPPSPAHYSPPPSPPVY 786
Query: 510 LEQSEPEPRTSDHSVP 525
S P P +S P
Sbjct: 787 YYNSPPPPPAVHYSPP 802
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.129 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,810,447
Number of Sequences: 26719
Number of extensions: 964463
Number of successful extensions: 5553
Number of sequences better than 10.0: 351
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 291
Number of HSP's that attempted gapping in prelim test: 4367
Number of HSP's gapped (non-prelim): 973
length of query: 837
length of database: 11,318,596
effective HSP length: 108
effective length of query: 729
effective length of database: 8,432,944
effective search space: 6147616176
effective search space used: 6147616176
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0346.15


