

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.14
(1622 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
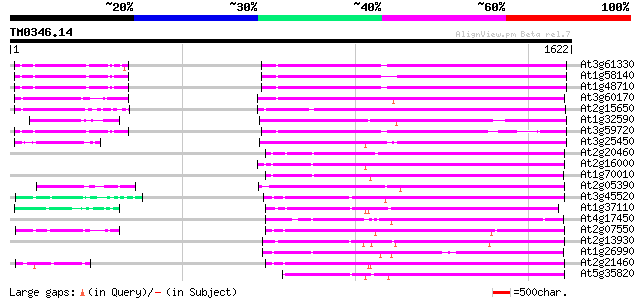
Score E
Sequences producing significant alignments: (bits) Value
At3g61330 copia-type polyprotein 671 0.0
At1g58140 hypothetical protein 669 0.0
At1g48710 hypothetical protein 667 0.0
At3g60170 putative protein 661 0.0
At2g15650 putative retroelement pol polyprotein 628 e-180
At1g32590 hypothetical protein, 5' partial 586 e-167
At3g59720 copia-type reverse transcriptase-like protein 577 e-164
At3g25450 hypothetical protein 566 e-161
At2g20460 putative retroelement pol polyprotein 551 e-156
At2g16000 putative retroelement pol polyprotein 544 e-154
At1g70010 hypothetical protein 538 e-152
At2g05390 putative retroelement pol polyprotein 516 e-146
At3g45520 copia-like polyprotein 499 e-141
At1g37110 499 e-141
At4g17450 retrotransposon like protein 495 e-140
At2g07550 putative retroelement pol polyprotein 484 e-136
At2g13930 putative retroelement pol polyprotein 483 e-136
At1g26990 polyprotein, putative 480 e-135
At2g21460 putative retroelement pol polyprotein 477 e-134
At5g35820 copia-like retrotransposable element 465 e-131
>At3g61330 copia-type polyprotein
Length = 1352
Score = 671 bits (1731), Expect = 0.0
Identities = 363/890 (40%), Positives = 512/890 (56%), Gaps = 23/890 (2%)
Query: 727 KCL-LSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKV 785
+CL + EE W+ H R GH + + LS+ +VRGLP + + +CE C GK K+
Sbjct: 455 QCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKM 513
Query: 786 PFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHS 845
F ++ +PLEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL K E
Sbjct: 514 SFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFE 573
Query: 846 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVER 905
+F F A V+ E I +RSD GGEF + +F + GI + PR+PQQNGVVER
Sbjct: 574 IFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVVER 633
Query: 906 KNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISY 965
KNRT+ EMAR+ML+ + K WAE V A Y+ NR + + KTP E W KP +S+
Sbjct: 634 KNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSH 693
Query: 966 FHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 1025
FG + + ++ K D KS K + +GY + SKG++ YN D K S ++ FD++
Sbjct: 694 LRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEE 753
Query: 1026 LDSDQSKLVEKFADLSISVSDKGKALEEAEP-EEDDPEEAGPSDSQPRKKSRIIASHPKE 1084
+ D + E + D+ + E P EE P+ SQ + S
Sbjct: 754 GEWDWNSNEEDYNFFPHFEEDEPEPTREEPPSEEPTTPPTSPTSSQIEESS--------- 804
Query: 1085 LILGNKDEPVRTRSAFRPSEETL----LSMKGLVSLIEPKSIDEALQDKDWILAMEEEVN 1140
+ R RS E T L++ L + EP +A++ K W AM+EE+
Sbjct: 805 -----SERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQKAIEKKTWRNAMDEEIK 859
Query: 1141 EFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETF 1200
KND W L P IG KWV++ K N KG+V R KARLVAKGYSQ+ GIDY E F
Sbjct: 860 SIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRVGIDYDEVF 919
Query: 1201 VPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFK 1260
PVARLE +RL+IS + + +HQMDVKSAFLNG + EEVY+ QP G+ + D V +
Sbjct: 920 APVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLR 979
Query: 1261 LKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGS 1320
LKK LYGLKQAPRAW R+ + E +F++ + L+ K K+DILI +YVD +IF
Sbjct: 980 LKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTG 1039
Query: 1321 ANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTES 1380
N S+ +EF K M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKKF + +S
Sbjct: 1040 NNPSIFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKIDDS 1099
Query: 1381 TIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRE 1440
TPM L K++ V ++ ++GSL YLT +RPD L++V + +R+ P
Sbjct: 1100 NPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTT 1159
Query: 1441 THLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNL 1500
TH A KRILRY+KGT N GL Y TS+YKL GY D D+ GD RKST G ++G
Sbjct: 1160 THFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTA 1219
Query: 1501 VSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESNIPIYCDNTAAISL 1559
+W SK+Q + LST +AEY++A C +W+++ L++ + E I+ DN +AI+L
Sbjct: 1220 FTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIAL 1279
Query: 1560 SKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KALSR 1608
+KNP+ H R+KHI+ +YH+IR+ V K + L++V T + AD + K L R
Sbjct: 1280 AKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADFFTKPLKR 1329
Score = 93.2 bits (230), Expect = 1e-18
Identities = 75/337 (22%), Positives = 158/337 (46%), Gaps = 24/337 (7%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKLYAQ 73
P+ ++ W R+++ LG D+W+I+ G+ P + + + + +K +
Sbjct: 11 PVLTKSNYDNWSLRMKAI-LGAH-DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---R 65
Query: 74 HHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIM 133
KA ++ + + ++K+ + A +E L+ S++G +VK+ + +L ++E+ M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 134 EPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVERM 193
+ E + + FSR + ++ + + +V+RSL + +VT IE T+D+E M
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 194 SLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDE 253
++E+L+ L+ +E K+ + +D+ ++ + + + K E ++ Q
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIAEQVLNM----QITKEENGQSYQRRGGGQVRGRGRGGY 241
Query: 254 DELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYKSDC- 312
++ + ++ RG GK KS KSS+K C+ C + GHY S+C
Sbjct: 242 GNGRGWRPHEDNTNQRGENSSRGRGKGHPKSR---YDKSSVK---CYNCGKFGHYASECK 295
Query: 313 -PKLKKDKRPKKHFKTKK------SLMVTFDESESED 342
P KK + K H+ +K LM ++ + E ++
Sbjct: 296 APSNKKFEE-KAHYVEEKIQEEDMLLMASYKKDEQKE 331
>At1g58140 hypothetical protein
Length = 1320
Score = 669 bits (1726), Expect = 0.0
Identities = 359/886 (40%), Positives = 502/886 (56%), Gaps = 47/886 (5%)
Query: 727 KCL-LSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKV 785
+CL + EE W+ H R GH + + LS+ +VRGLP + + +CE C GK K+
Sbjct: 455 QCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKM 513
Query: 786 PFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHS 845
F ++ +PLEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL K E
Sbjct: 514 SFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFE 573
Query: 846 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVER 905
+F F A V+ E I +RSD GGEF + +F + GI + PR+PQQNGV ER
Sbjct: 574 IFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAER 633
Query: 906 KNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISY 965
KNRT+ EMAR+ML+ + K WAE V A Y+ NR + + KTP E W KP +S+
Sbjct: 634 KNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSH 693
Query: 966 FHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 1025
FG + + ++ K D KS K + +GY + SKG++ YN D K S ++ FD++
Sbjct: 694 LRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEE 753
Query: 1026 LDSDQSKLVEKFADLSISVSDKGKALEEAEP-EEDDPEEAGPSDSQPRKKSRIIASHPKE 1084
+ D + E + DK + E P EE P+ SQ +K
Sbjct: 754 GEWDWNSNEEDYNFFPHFEEDKPEPTREEPPSEEPTTPPTSPTSSQIEEKC--------- 804
Query: 1085 LILGNKDEPVRTRSAFRPSEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNEFSK 1144
EP EA++ K W AM+EE+ K
Sbjct: 805 ---------------------------------EPMDFQEAIEKKTWRNAMDEEIKSIQK 831
Query: 1145 NDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVA 1204
ND W L P IG KWV++ K N KG+V R KARLVAKGYSQ+ GIDY E F PVA
Sbjct: 832 NDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRAGIDYDEVFAPVA 891
Query: 1205 RLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFKLKKS 1264
RLE +RL+IS + + +HQMDVKSAFLNG + EEVY+ QP G+ + D V +LKK+
Sbjct: 892 RLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKA 951
Query: 1265 LYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGSANQS 1324
LYGLKQAPRAW R+ + E +F++ + L+ K K+DILI +YVD +IF N S
Sbjct: 952 LYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPS 1011
Query: 1325 LCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAK 1384
+ +EF K M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKKF M +S
Sbjct: 1012 MFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVC 1071
Query: 1385 TPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLT 1444
TPM L K++ V ++ ++GSL YLT +RPD L++V + +R+ P TH
Sbjct: 1072 TPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFK 1131
Query: 1445 AVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWA 1504
A KRILRY+KGT N GL Y TS+YKL GY D D+ GD RKST G ++G +W
Sbjct: 1132 AAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWM 1191
Query: 1505 SKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESNIPIYCDNTAAISLSKNP 1563
SK+Q + LST +AEY++A C +W+++ L++ + E I+ DN +AI+L+KNP
Sbjct: 1192 SKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNP 1251
Query: 1564 ILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KALSR 1608
+ H R+KHI+ +YH+IR+ V K + L++V T + ADI+ K L R
Sbjct: 1252 VFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKR 1297
Score = 93.2 bits (230), Expect = 1e-18
Identities = 74/336 (22%), Positives = 156/336 (46%), Gaps = 22/336 (6%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKLYAQ 73
P+ ++ W R+++ LG D+W+I+ G+ P + + + + +K +
Sbjct: 11 PVLTKSNYDNWSLRMKAI-LGAH-DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---R 65
Query: 74 HHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIM 133
KA ++ + + ++K+ + A +E L+ S++G +VK+ + +L ++E+ M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 134 EPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVERM 193
+ E + + FSR + ++ + + +V+RSL + +VT IE T+D+E M
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 194 SLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDE 253
++E+L+ L+ +E K+ + +D+ ++ + + + K E ++ Q
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIVEQVLNM----QITKEENGQSYQRRGGGQVRGRGRGGY 241
Query: 254 DELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYKSDC- 312
++ + ++ RG GK KS KSS+K C+ C + GHY S+C
Sbjct: 242 GNGRGWRPHEDNTNQRGENSSRGRGKGHPKSR---YDKSSVK---CYNCGKFGHYASECK 295
Query: 313 -PKLKKDKRPKKHFKTK-----KSLMVTFDESESED 342
P KK + + + K LM ++ + E E+
Sbjct: 296 APSNKKFEEKANYVEEKIQEEDMLLMASYKKDEQEE 331
>At1g48710 hypothetical protein
Length = 1352
Score = 667 bits (1720), Expect = 0.0
Identities = 362/890 (40%), Positives = 510/890 (56%), Gaps = 23/890 (2%)
Query: 727 KCL-LSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKV 785
+CL + EE W+ H R GH + + LS+ +VRGLP + + +CE C GK K+
Sbjct: 455 QCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKM 513
Query: 786 PFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHS 845
F ++ + LEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL K E
Sbjct: 514 SFPKESSSRAQKSLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFE 573
Query: 846 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVER 905
+F F A V+ E I +RSD GGEF + +F + GI + PR+PQQNGV ER
Sbjct: 574 IFKKFKAHVEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAER 633
Query: 906 KNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISY 965
KNRT+ EMAR+ML+ + K WAE V A Y+ NR + + KTP E W K +S+
Sbjct: 634 KNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKSGVSH 693
Query: 966 FHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 1025
FG + + ++ K D KS K + +GY + SKG++ YN D K S ++ FD++
Sbjct: 694 LRVFGSIAHAHVPDEKRSKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEE 753
Query: 1026 LDSDQSKLVEKFADLSISVSDKGKALEEAEP-EEDDPEEAGPSDSQPRKKSRIIASHPKE 1084
+ D + E + D+ + E P EE P+ SQ + S
Sbjct: 754 GEWDWNSNEEDYNFFPHFEEDEPEPTREEPPSEEPTTPPTSPTSSQIEESS--------- 804
Query: 1085 LILGNKDEPVRTRSAFRPSEETL----LSMKGLVSLIEPKSIDEALQDKDWILAMEEEVN 1140
+ R RS E T L++ L + EP EA++ K W AM+EE+
Sbjct: 805 -----SERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQEAIEKKTWRNAMDEEIK 859
Query: 1141 EFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETF 1200
KND W L P IG KWV++ K N KG+V R KARLVAKGY Q+ GIDY E F
Sbjct: 860 SIQKNDTWELTSLPNGHKTIGVKWVYKAKKNSKGEVERYKARLVAKGYIQRAGIDYDEVF 919
Query: 1201 VPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFK 1260
PVARLE +RL+IS + + +HQMDVKSAFLNG + EEVY+ QP G+ + D V +
Sbjct: 920 APVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLR 979
Query: 1261 LKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGS 1320
LKK+LYGLKQAPRAW R+ + E +F++ + L+ K K+DILI +YVD +IF
Sbjct: 980 LKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTG 1039
Query: 1321 ANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTES 1380
N S+ +EF K M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKKF M +S
Sbjct: 1040 NNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDS 1099
Query: 1381 TIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRE 1440
TPM L K++ V ++ ++GSL YLT +RPD L++V + +R+ P
Sbjct: 1100 NPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTT 1159
Query: 1441 THLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNL 1500
TH A KRILRY+KGT N GL Y TS+YKL GY D D+ GD RKST G ++G
Sbjct: 1160 THFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTA 1219
Query: 1501 VSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESNIPIYCDNTAAISL 1559
+W SK+Q + LST +AEY++A C +W+++ L++ + E I+ DN +AI+L
Sbjct: 1220 FTWMSKKQPIVVLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIAL 1279
Query: 1560 SKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KALSR 1608
+KNP+ H R+KHI+ +YH+IR+ V K + L++V T + ADI+ K L R
Sbjct: 1280 AKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKR 1329
Score = 93.2 bits (230), Expect = 1e-18
Identities = 74/336 (22%), Positives = 156/336 (46%), Gaps = 22/336 (6%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKLYAQ 73
P+ ++ W R+++ LG D+W+I+ G+ P + + + + +K +
Sbjct: 11 PVLTKSNYDNWSLRMKAI-LGAH-DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---R 65
Query: 74 HHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIM 133
KA ++ + + ++K+ + A +E L+ S++G +VK+ + +L ++E+ M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 134 EPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVERM 193
+ E + + FSR + ++ + + +V+RSL + +VT IE T+D+E M
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 194 SLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDE 253
++E+L+ L+ +E K+ + +D+ ++ + + + K E ++ Q
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIIEQVLNM----QITKEENGQSYQRRGGGQVRGRGRGGY 241
Query: 254 DELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYKSDC- 312
++ + ++ RG GK KS KSS+K C+ C + GHY S+C
Sbjct: 242 GNGRGWRPHEDNTNQRGENSSRGRGKGHPKSR---YDKSSVK---CYNCGKFGHYASECK 295
Query: 313 -PKLKKDKRPKKHFKTK-----KSLMVTFDESESED 342
P KK + + + K LM ++ + E E+
Sbjct: 296 APSNKKFEEKANYVEEKIQEEDMLLMASYKKDEQEE 331
>At3g60170 putative protein
Length = 1339
Score = 661 bits (1705), Expect = 0.0
Identities = 371/905 (40%), Positives = 525/905 (57%), Gaps = 21/905 (2%)
Query: 718 LSELEAQNVKCLLS---VNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALC 774
L+ +N CL + +++E + H R GH + + L+ +V GLP LK + + +C
Sbjct: 412 LASKPQKNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILKATKE-IC 470
Query: 775 EACQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWV 834
C GK + K +S L+L+H D+ GP+ S GKRY + +DD++R TWV
Sbjct: 471 AICLTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWV 530
Query: 835 KFLSRKDESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCP 894
FL K E+ + F F A V+ E + +R+D GGEF +++F S+GI+ +
Sbjct: 531 YFLHEKSEAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGISRQLTAA 590
Query: 895 RTPQQNGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYE 954
TPQQNGV ERKNRT+ R+ML E + K FW+E + +IQNR + TP E
Sbjct: 591 FTPQQNGVAERKNRTIMNAVRSMLSERQVPKMFWSEATKWSVHIQNRSPTAAVEGMTPEE 650
Query: 955 LWKNIKPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTI 1014
W KP + YF FGC+ YV + K D KS KC+ LG S+ SK +R Y+ K I
Sbjct: 651 AWSGRKPVVEYFRVFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLYDPVMKKI 710
Query: 1015 EESIHVRFDD--KLDSDQSKLVEKFADLSISVSDKGKALEEAEP-EEDDPEEAGPSDSQP 1071
S V FD+ D DQ+ + K L D K E EP P G SD+
Sbjct: 711 VISKDVVFDEDKSWDWDQADVEAKEVTLECGDEDDEKNSEVVEPIAVASPNHVG-SDNNV 769
Query: 1072 RKKSRIIASHPKELILGNKDEPVRTRSAFRPSEET--------LLSMKGLVSLIE--PKS 1121
+ S P + K R + ET LS+ L+ + E P
Sbjct: 770 SSSPILAPSSPAPSPVAAKVTRERRPPGWMADYETGEGEEIEENLSVMLLMMMTEADPIQ 829
Query: 1122 IDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKA 1181
D+A++DK W AME E+ KN+ W L P+ IG KWV++ KLNE G+V + KA
Sbjct: 830 FDDAVKDKIWREAMEHEIESIVKNNTWELTTLPKGFTPIGVKWVYKTKLNEDGEVDKYKA 889
Query: 1182 RLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEV 1241
RLVAKGY+Q GIDYTE F PVARL+ +R +++ S N + Q+DVKSAFL+G + EEV
Sbjct: 890 RLVAKGYAQCYGIDYTEVFAPVARLDTVRTILAISSQFNWEIFQLDVKSAFLHGELKEEV 949
Query: 1242 YVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKT 1301
YV QP GF E + V+KL+K+LYGLKQAPRAWY R+ ++ L+ EF R + TLF KT
Sbjct: 950 YVRQPEGFIREGEEEKVYKLRKALYGLKQAPRAWYSRIEAYFLKEEFERCPSEHTLFTKT 1009
Query: 1302 YKDDILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTY 1361
+ILIV +YVD +IF +++++C EF K M EFEMS +G++K+FLGI+V Q+ G +
Sbjct: 1010 RVGNILIVSLYVDDLIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIEVKQSDGGIF 1069
Query: 1362 IHQSKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASR 1421
I Q +Y +E+L +F M ES K P+ P L K++ KV + +++ ++GSL+YLT +R
Sbjct: 1070 ICQRRYAREVLARFGMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVGSLMYLTVTR 1129
Query: 1422 PDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMY--KKTSEYKLSGYCDVDY 1479
PD ++ V L +RF S+PR +H A KRILRYLKGT LG+ Y +K KL + D DY
Sbjct: 1130 PDLMYGVCLISRFMSNPRMSHWLAAKRILRYLKGTVELGIFYRRRKNRSLKLMAFTDSDY 1189
Query: 1480 AGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLED 1539
AGD R+ST G + S + WASK+Q + LST +AEYI+AA C+ Q +W++ LE
Sbjct: 1190 AGDLNDRRSTSGFVFLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQCVWLRKVLEK 1249
Query: 1540 YQILE-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHK 1598
E S I CDN++ I LSK+P+LH ++KHIEV++H++RD V V+ L++ T+ +
Sbjct: 1250 LGAEEKSATVINCDNSSTIQLSKHPVLHGKSKHIEVRFHYLRDLVNGDVVKLEYCPTEDQ 1309
Query: 1599 WADIY 1603
ADI+
Sbjct: 1310 VADIF 1314
Score = 85.9 bits (211), Expect = 2e-16
Identities = 72/331 (21%), Positives = 142/331 (42%), Gaps = 46/331 (13%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKLYAQ 73
P FDG +++W +E+F +LW ++ +G V E+ KL +
Sbjct: 13 PRFDGY-YDFWSMTMENFLRS--RELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKL--K 67
Query: 74 HHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIM 133
K + L AI E + I D + I+ES+K ++G+ KVK ++ +L +++E M
Sbjct: 68 DLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAM 127
Query: 134 EPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVERM 193
+ E I+ R +V ++ + V +++RSL + +V SIE + D+ +
Sbjct: 128 KEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTL 187
Query: 194 SLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDE 253
S++EL L HE + ++ ALK +E+ S+
Sbjct: 188 SIDELHGSLLVHE---QRLNGHVQEEQALKVTHEERPSQ--------------------- 223
Query: 254 DELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYKSDCP 313
+ +RGS +G+ G+ ++ V C++C GH++ +CP
Sbjct: 224 --------------GRGRGVFRGS---RGRGRGRGRSGTNRAIVECYKCHNLGHFQYECP 266
Query: 314 KLKKDKRPKKHFKTKKSLMVTFDESESEDVD 344
+ +K+ + + ++ L++ + E + D
Sbjct: 267 EWEKNANYAELEEEEELLLMAYVEQNQANRD 297
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 628 bits (1619), Expect = e-180
Identities = 349/908 (38%), Positives = 523/908 (57%), Gaps = 30/908 (3%)
Query: 716 IRLSELEAQNVKCLLSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCE 775
I+LS +E + + + EE W H+RLGH S +++ Q+ LV GLP K + + C+
Sbjct: 436 IKLSSVEEEAMTANVQT-EETW--HKRLGHVSNKRLQQMQDKELVNGLPRFKVTKET-CK 491
Query: 776 ACQKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVK 835
AC GK ++ F ++ T LE++H D+ GP++ +SI G RY ++ +DDY+ WV
Sbjct: 492 ACNLGKQSRKSFPKESQTKTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVY 551
Query: 836 FLSRKDESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPR 895
FL +K E+ + F F A V+ + C I +R E + GI + P
Sbjct: 552 FLKQKSETFATFKKFKALVEKQSNCSIKTLRP----------MEVFCEDEGINRQVTLPY 601
Query: 896 TPQQNGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNK-TPYE 954
+PQQNG ERKNR+L EMAR+ML E + WAE V T+ Y+QNR+ + I + TP E
Sbjct: 602 SPQQNGAAERKNRSLVEMARSMLVEQDLPLKLWAEAVYTSAYLQNRLPSKAIEDDVTPME 661
Query: 955 LWKNIKPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTI 1014
W KPN+S+ FG +CYV + K DAK+ +L+GYS+++KG+R + + + +
Sbjct: 662 KWCGHKPNVSHLRIFGSICYVHIPDQKRRKLDAKAKCGILIGYSNQTKGYRVFLLEDEKV 721
Query: 1015 EESIHVRF--DDKLDSDQSKLVEKFADLSISVSDKGKALEEAEPEE----DDPEEAGPSD 1068
E S V F D K D D+ + V+K +SI+ + + +E + DD G +
Sbjct: 722 EVSRDVVFQEDKKWDWDKQEEVKKTFVMSINDIQESRDQQETSSHDLSQIDDHANNGEGE 781
Query: 1069 SQPRKKSRIIASHPKELILGNKD----EPVRTRSAFRPSEETLLSMKG-LVSLIEPKSID 1123
+ S++ +E K + + ++ ++E ++ LV+ EP++ D
Sbjct: 782 TSSHVLSQVNDQEERETSESPKKYKSMKEILEKAPRMENDEAAQGIEACLVANEEPQTYD 841
Query: 1124 EALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARL 1183
EA DK+W AM EE+ KN W LV KP+ +VI KW+++ K + G+ V++KARL
Sbjct: 842 EARGDKEWEEAMNEEIKVIEKNRTWKLVDKPEKKNVISVKWIYKIKTDASGNHVKHKARL 901
Query: 1184 VAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYV 1243
VA+G+SQ+ GIDY ETF PV+R + IR L++++ L+QMDVKSAFLNG + EEVYV
Sbjct: 902 VARGFSQEYGIDYLETFAPVSRYDTIRALLAYAAQMKWRLYQMDVKSAFLNGELEEEVYV 961
Query: 1244 HQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYK 1303
QPPGF E + V +L K+LYGLKQAPRAWYER+ S+ ++N F R D L+ K
Sbjct: 962 TQPPGFVIEGKEEKVLRLYKALYGLKQAPRAWYERIDSYFIQNGFARSMNDAALYSKKKG 1021
Query: 1304 DDILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIH 1363
+D+LIV +YVD +I N L F K M+ EFEM+ +G L YFLG++V+Q G ++
Sbjct: 1022 EDVLIVSLYVDDLIITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGMEVNQDDSGIFLS 1081
Query: 1364 QSKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGK--VCQKLYRCMIGSLLYLTASR 1421
Q KY +L+ KF M ES TP+ P + + K YR ++G LLYL ASR
Sbjct: 1082 QEKYANKLIDKFGMKESKSVSTPLTPQGKRKGVEGDDKEFADPTKYRRIVGGLLYLCASR 1141
Query: 1422 PDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAG 1481
PD +++ +R+ S P H KR+LRY+KGT+N G+++ +L GY D D+ G
Sbjct: 1142 PDVMYASSYLSRYMSSPSIQHYQEAKRVLRYVKGTSNFGVLFTSKETPRLVGYSDSDWGG 1201
Query: 1482 DRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQ 1541
+KST G LG + W S +Q T+ STA+AEYI+ + Q +W++ ED+
Sbjct: 1202 SLEDKKSTTGYVFTLGLAMFCWQSCKQQTVAQSTAEAEYIAVCAATNQAIWLQRLFEDFG 1261
Query: 1542 I-LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWA 1600
+ + IPI CDN +AI++ +NP+ H R KHIE+KYHF+R+ KG++ L++ + + A
Sbjct: 1262 LKFKEGIPILCDNKSAIAIGRNPVQHRRTKHIEIKYHFVREAEHKGLIQLEYCKGEDQLA 1321
Query: 1601 DIY-KALS 1607
D+ KALS
Sbjct: 1322 DVLTKALS 1329
Score = 95.9 bits (237), Expect = 2e-19
Identities = 78/335 (23%), Positives = 157/335 (46%), Gaps = 39/335 (11%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYE-RPVDAEGKKIPRSEMTAEQKKLYA 72
P+FDG+++++W ++ + F LW ++ +G PV AE T ++ +
Sbjct: 10 PIFDGEKYDFWSIKMATIFR--TRKLWSVVEEGVPVEPVQAEETPETARAKTLREEAV-T 66
Query: 73 QHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFI 132
A IL +A++ + + +I + ++ LK ++G+ +V+ K SL ++YE+
Sbjct: 67 NDTMALQILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYENLK 126
Query: 133 MEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVER 192
M N++I+ + +L + + T + +++ SLP + +V+ +E TRD++
Sbjct: 127 MYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRDLDA 186
Query: 193 MSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSD 252
+++ EL+ ILK E + + ++ K+ + Y ++K +S Q D+
Sbjct: 187 LTMSELLGILKAQEARVTAREESTKEG----AFYVRSKGRESGFKQ-----------DNT 231
Query: 253 EDELTLISKRLNHIWKHKQSKY-RGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYKSD 311
+ + K HK SK+ + K K++ G+ K S + C++C + GHY ++
Sbjct: 232 NNRVNQDKKWCGF---HKSSKHTEEECREKPKNDDHGKNKRS--NIKCYKCGKIGHYANE 286
Query: 312 CPKLKKDKRPKKHFKTKKSLMVTFDESESEDVDSD 346
C K K+ VT +E EDV+ D
Sbjct: 287 C-----------RSKNKERAHVTLEE---EDVNED 307
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 586 bits (1511), Expect = e-167
Identities = 324/903 (35%), Positives = 501/903 (54%), Gaps = 51/903 (5%)
Query: 722 EAQNVKCLLSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSD-ALCEACQKG 780
E + +CL + + + H+R GH + + + L++ +V+GLP + A+C+ C KG
Sbjct: 379 ETEETRCLQVIGKANNMWHKRFGHLNHQGLRSLAEKEMVKGLPKFDLGEEEAVCDICLKG 438
Query: 781 KFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRK 840
K + ++ +++ L+L+H D+ GP+ S GKRY + +DD+SR W LS K
Sbjct: 439 KQIRESIPKESAWKSTQVLQLVHTDICGPINPASTSGKRYILNFIDDFSRKCWTYLLSEK 498
Query: 841 DESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQN 900
E+ F F A+V+ E ++V +RSD GGE+ + +F+ +GI + TPQQN
Sbjct: 499 SETFQFFKEFKAEVERESGKKLVCLRSDRGGEYNSREFDEYCKEFGIKRQLTAAYTPQQN 558
Query: 901 GVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIK 960
GV ERKNR++ M R ML E+ + + FW E V A YI NR + + + TP E W + K
Sbjct: 559 GVAERKNRSVMNMTRCMLMEMSVPRKFWPEAVQYAVYILNRSPSKALNDITPEEKWSSWK 618
Query: 961 PNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHV 1020
P++ + FG + Y L + K D KS KC++ G S SK +R Y+ I S V
Sbjct: 619 PSVEHLRIFGSLAYALVPYQKRIKLDEKSIKCVMFGVSKESKAYRLYDPATGKILISRDV 678
Query: 1021 RFDDKLDSD-QSKLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIA 1079
+FD++ + + K +E+ +L SD A EE PE + + +++ +++
Sbjct: 679 QFDEERGWEWEDKSLEE--ELVWDNSDHEPAGEEG-PEINHNGQQDQEETEEEEETVAET 735
Query: 1080 SHPKELILGN-----KDEPVRTRSAFRPSEETLLSMKGLVSLI-------EPKSIDEALQ 1127
H +G + +PV + + L++ ++ +P +EA Q
Sbjct: 736 VHQNLPAVGTGGVRQRQQPVWMKDYVVGNARVLITQDEEDEVLALFIGPDDPVCFEEAAQ 795
Query: 1128 DKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKG 1187
+ W AME E+ +N+ W LV+ P+ VIG KW+F+ K NEKG+V + KARLVAKG
Sbjct: 796 LEVWRKAMEAEITSIEENNTWELVELPEEAKVIGLKWIFKTKFNEKGEVDKFKARLVAKG 855
Query: 1188 YSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPP 1247
Y Q+ G+D+ E F PVA+ + IRL++ + + Q+DVKSAFL+G + E+V+V QP
Sbjct: 856 YHQRYGVDFYEVFAPVAKWDTIRLILGLAAEKGWSVFQLDVKSAFLHGDLKEDVFVEQPK 915
Query: 1248 GFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDIL 1307
GFE E+ V+KLKK+LYGLKQAPRAWY R+ F + F + + TLF K + D L
Sbjct: 916 GFEVEEESSKVYKLKKALYGLKQAPRAWYSRIEEFFGKEGFEKCYCEHTLFVKKERSDFL 975
Query: 1308 IVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKY 1367
+V +YVD +I+ ++ + + F M EF M+ +G++KYFLG++V Q G +I+Q KY
Sbjct: 976 VVSVYVDDLIYTGSSMEMIEGFKNSMMEEFAMTDLGKMKYFLGVEVIQDERGIFINQRKY 1035
Query: 1368 TKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFS 1427
E++KK+ M K P+ P L K A
Sbjct: 1036 AAEIIKKYGMEGCNSVKNPIVPGQKLTKAGA----------------------------- 1066
Query: 1428 VHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RK 1487
+R+ P E HL AVKRILRY++GT +LG+ Y++ +L G+ D DYAGD RK
Sbjct: 1067 ---VSRYMESPNEQHLLAVKRILRYVQGTLDLGIQYERGGATELVGFVDSDYAGDVDDRK 1123
Query: 1488 STFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESN 1546
ST G LG ++WASK+Q + LST +AE++SA+ + Q +W+++ LE+ E
Sbjct: 1124 STSGYVFMLGGGAIAWASKKQPIVTLSTTEAEFVSASYGACQAVWLRNVLEEIGCRQEGG 1183
Query: 1547 IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KA 1605
++CDN++ I LSKNP+LH R+KHI V+YHF+R+ V++G + L + T + ADI KA
Sbjct: 1184 TLVFCDNSSTIKLSKNPVLHGRSKHIHVRYHFLRELVKEGTIRLDYCTTTDQVADIMTKA 1243
Query: 1606 LSR 1608
+ R
Sbjct: 1244 VKR 1246
Score = 78.2 bits (191), Expect = 4e-14
Identities = 63/273 (23%), Positives = 122/273 (44%), Gaps = 54/273 (19%)
Query: 58 IPRSE----MTAEQKKLYAQH----HKARAILLSAISYEEYQKITDLEFANGIFESLKMS 109
IPR E +T Q+ A+ HK + L ++I + I E + ++ES+K
Sbjct: 1 IPRPERNVILTGAQRTELAEKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRK 60
Query: 110 HEGNKKVKESKTLSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRV 169
++GN +V+ ++ L + +E M+ E+I FSR + +R L + V ++
Sbjct: 61 YQGNDRVQSAQLQRLRRSFEVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKI 120
Query: 170 IRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKA 229
+R+L E + +V +IE + +++ ++++ L S L HE Q+L + +
Sbjct: 121 LRTLVEKFTYVVCAIEESNNIKELTVDGLQSSLMVHE------QNLSRHDV--------- 165
Query: 230 KSEKSKALQAEAEESEEASEDSDEDELTLISKRLNHIWKHKQSKYRGS----GKAKGKSE 285
+ + L+AE + W+ + RG G+ +G +
Sbjct: 166 ---EERVLKAETQ------------------------WRPDGGRGRGGSPSRGRGRGGYQ 198
Query: 286 SSGQKKSSIKEVTCFECKESGHYKSDCPKLKKD 318
G+ + V CF+C + GHYK++CP +K+
Sbjct: 199 GRGRGYVNRDTVECFKCHKMGHYKAECPSWEKE 231
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 577 bits (1488), Expect = e-164
Identities = 335/890 (37%), Positives = 470/890 (52%), Gaps = 103/890 (11%)
Query: 727 KCL-LSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKV 785
+CL + EE W+ H R GH + + LS+ +VRGLP + + +CE C G K+
Sbjct: 455 QCLKMCYKEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQ-VCEGCLLGNQFKM 513
Query: 786 PFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHS 845
F ++ +PLEL+H D+ GP+K +S+G Y ++ +DD+SR TWV FL K E
Sbjct: 514 SFPKESSSRAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFE 573
Query: 846 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVER 905
+F F A V+ E I +RSD GGEF + +F + GI + PR+PQQNGV ER
Sbjct: 574 IFKKFKAHVEKESGLVIKTMRSDSGGEFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVAER 633
Query: 906 KNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISY 965
KNRT+ EMAR+ML+ + K WAE V A Y+ NR + + KTP E W KP +S+
Sbjct: 634 KNRTILEMARSMLKSKRLPKELWAEAVACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSH 693
Query: 966 FHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 1025
FG + + ++ +K D KS K + +GY + SKG++ YN D K S ++ FD++
Sbjct: 694 LRVFGSIAHAHVPDEKRNKLDDKSEKYIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEE 753
Query: 1026 LDSDQSKLVEKFADLSISVSDKGKALEEAEP-EEDDPEEAGPSDSQPRKKSRIIASHPKE 1084
+ D + E + DK + E P EE P+ SQ + S
Sbjct: 754 GEWDWNSNEEDYNFFPHFEEDKPEPTREEPPSEEPTTPPTSPTSSQIEESS--------- 804
Query: 1085 LILGNKDEPVRTRSAFRPSEETL----LSMKGLVSLIEPKSIDEALQDKDWILAMEEEVN 1140
+ R RS E T L++ L + EP EA++ K W AM+EE+
Sbjct: 805 -----SERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQEAIEKKTWRNAMDEEIK 859
Query: 1141 EFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETF 1200
KND W L P IG KWV++ K N KG+V R KARLVAKGYSQ+ GIDY E F
Sbjct: 860 SIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRAGIDYDEIF 919
Query: 1201 VPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKNPDHVFK 1260
PVARLE +RL+IS + + +HQMDVKSAFLNG + EEVY+ QP G+ + D V +
Sbjct: 920 APVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLR 979
Query: 1261 LKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGS 1320
LKK LYGLKQAPRAW R+ + E +F++ + L+ K K+DILI +YVD +IF
Sbjct: 980 LKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTG 1039
Query: 1321 ANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTES 1380
N S+ +EF K M EFEM+ +G + Y+LGI+V Q G +I Q Y KE+LKKF M +S
Sbjct: 1040 NNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDS 1099
Query: 1381 TIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRE 1440
+ ++GSL YLT +RPD L++V + +R+ P
Sbjct: 1100 NPS--------------------------LVGSLRYLTCTRPDILYAVGVVSRYMEHPTT 1133
Query: 1441 THLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNL 1500
TH A KRILRY+KGT N GL Y TS DY
Sbjct: 1134 THFKAAKRILRYIKGTVNFGLHYSTTS----------DY--------------------- 1162
Query: 1501 VSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESNIPIYCDNTAAISL 1559
+C +W+++ L++ + E I+ DN +AI+L
Sbjct: 1163 ---------------------KLVVCHA--IWLRNLLKELSLPQEEPTKIFVDNKSAIAL 1199
Query: 1560 SKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADIY-KALSR 1608
+KNP+ H R+KHI+ +YH+IR+ V K + L++V T + ADI+ K L R
Sbjct: 1200 AKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKR 1249
Score = 94.7 bits (234), Expect = 4e-19
Identities = 75/336 (22%), Positives = 156/336 (46%), Gaps = 22/336 (6%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKLYAQ 73
P+ ++ W R+++ LG D+W+I+ G+ P + + + + +K +
Sbjct: 11 PVLTKSNYDNWSLRMKAI-LGAH-DVWEIVEKGFIEPENEGSLSQTQKDGLRDSRK---R 65
Query: 74 HHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIM 133
KA ++ + + ++K+ + A +E L+ S++G +VK+ + +L ++E+ M
Sbjct: 66 DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQM 125
Query: 134 EPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVERM 193
+ E + + FSR + ++ + + +V+RSL + +VT IE T+D+E M
Sbjct: 126 KEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAM 185
Query: 194 SLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDE 253
++E+L+ L+ +E K+ + +D+ ++ + + + K E ++ Q
Sbjct: 186 TIEQLLGSLQAYEEKKKKKEDIVEQVLNM----QITKEENGQSYQRRGGGQVRGRGRGGY 241
Query: 254 DELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYKSDC- 312
++ + ++ RG GK KS KSS+K C+ C + GHY S+C
Sbjct: 242 GNGRGWRPHEDNTNQRGENSSRGRGKGHPKSR---YDKSSVK---CYNCGKFGHYASECK 295
Query: 313 -PKLKKDKRPKKHFKTK-----KSLMVTFDESESED 342
P KK K + + K LM ++ + E E+
Sbjct: 296 APSNKKFKEKANYVEEKIQEEDMLLMASYKKDEQEE 331
>At3g25450 hypothetical protein
Length = 1343
Score = 567 bits (1460), Expect = e-161
Identities = 329/922 (35%), Positives = 504/922 (53%), Gaps = 50/922 (5%)
Query: 721 LEAQNVKCL-LSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQK 779
LE +N KCL L+ E + H RLGH S I + K LV G+ + C +C
Sbjct: 406 LEVENSKCLQLTTTNESTIWHARLGHISFETIKAMIKKELVIGISSSVPQEKETCGSCLF 465
Query: 780 GKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSR 839
GK + F ++ LEL+H DL GP+ + KRY V++DD+SR+ W L
Sbjct: 466 GKQARHSFPKATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLKE 525
Query: 840 KDESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQ 899
K E+ F F A V+ E I R+D GGEF + +F+ GI + P TPQQ
Sbjct: 526 KSEAFGKFKEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGINRHLTAPYTPQQ 585
Query: 900 NGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNI 959
NGVVER+NRTL M R++L+ + M + W E V + Y+ NR+ R + N+TPYE++K+
Sbjct: 586 NGVVERRNRTLLGMTRSILKHMNMPNYLWGEAVRHSTYLINRVGTRSLSNQTPYEVFKHK 645
Query: 960 KPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIH 1019
KPN+ + FGCV Y L K D +S + LG SK +R + + I S
Sbjct: 646 KPNVEHLRVFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRIFVSRD 705
Query: 1020 VRFDDK-------------------------------LDSDQSKLVEKFADLSISVSDKG 1048
V FD+ ++D S E+ + I+ D+
Sbjct: 706 VVFDENRSWMWQESSSETDKESGTFTITLSEFGNNGVTENDISTEPEETEEAEINGEDE- 764
Query: 1049 KALEEAEPEEDDPEEAGPSDSQPRKKSRIIASHPKELILGNKDEPVRTRSAFRPSEETLL 1108
+EEAE EE D + P + ++ I ++ K+ +L + E +E LL
Sbjct: 765 NIIEEAETEEHDQSQEEPQPVRRSQRQVIRPNYLKDYVLCAEIE----------AEHLLL 814
Query: 1109 SMKGLVSLIEPKSIDEALQDKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRN 1168
++ EP EA + K+W A +EE+ KN WSLV P IG KWVF+
Sbjct: 815 AVND-----EPWDFKEANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAIGVKWVFKL 869
Query: 1169 KLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDV 1228
K N G + + KARLVAKGY Q+ G+D+ E F PVAR+E +RL+I+ + ++ +H +DV
Sbjct: 870 KHNSDGSINKYKARLVAKGYVQRHGVDFEEVFAPVARIETVRLIIALAASNGWEIHHLDV 929
Query: 1229 KSAFLNGYISEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEF 1288
K+AFL+G + E+VYV QP GF ++++ + V+KL K+LYGL+QAPRAW +L+ L E +F
Sbjct: 930 KTAFLHGELREDVYVSQPEGFTNKESKEKVYKLHKALYGLRQAPRAWNTKLNEILKELKF 989
Query: 1289 VRGKVDTTLFCKTYKDDILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYF 1348
+ + +L+ K ++IL+V +YVD ++ +N + F K M +FEMS +G+L Y+
Sbjct: 990 EKCHKEPSLYRKQEGENILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYY 1049
Query: 1349 LGIQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYR 1408
LGI+V Q+ +G + Q +Y K++L++ M++ TPM + L K ++ + YR
Sbjct: 1050 LGIEVLQSKDGITLKQERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYR 1109
Query: 1409 CMIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSE 1468
IG L YL +RPD ++V + +R+ +PRE+H A+K+ILRYL+GTT+ GL +KK
Sbjct: 1110 RNIGCLRYLLHTRPDLSYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHGLYFKKGEN 1169
Query: 1469 YKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICST 1528
L GY D + D KST G+ +L ++W S++Q + LS+ +AE+++A +
Sbjct: 1170 AGLIGYSDSSHNVDLDDGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAK 1229
Query: 1529 QMLWMKHQLEDYQILE-SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGV 1587
Q +W++ L + E + I DN +AI+L+KNP+ H R+KHI +YHFIR+ V+ G
Sbjct: 1230 QAIWLQELLAEVIGTECEKVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFIRECVENGQ 1289
Query: 1588 LLLKFVDTDHKWADIY-KALSR 1608
+ ++ V + ADI KAL +
Sbjct: 1290 IEVEHVPGVRQKADILTKALGK 1311
Score = 54.3 bits (129), Expect = 6e-07
Identities = 50/251 (19%), Positives = 111/251 (43%), Gaps = 41/251 (16%)
Query: 14 PMFDGQRFEYWKDRLESFFLGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKLYAQ 73
PM + + W R+E+ LW I G +A+++K
Sbjct: 23 PMLNSVNYTVWTMRMEAVLRVHK--LWGTIEPG-----------------SADEEK---- 59
Query: 74 HHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIM 133
+ ARA+L +I ++ + ++ ++E++K + G ++VKE++ +L+ +++ M
Sbjct: 60 NDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAEFDKLKM 119
Query: 134 EPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLP-ESWMPLVTSIELTRDVER 192
+ +E+I++ R + L + V + ++SLP + ++ +V ++E D++
Sbjct: 120 KDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQVLDLKT 179
Query: 193 MSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSD 252
+ E++ +K +E + + D E L E E EE +D +
Sbjct: 180 TTFEDIAGRIKTYEDRVWDDDD---------------SHEDQGKLMTEVE--EEVVDDLE 222
Query: 253 EDELTLISKRL 263
E+E +I+K +
Sbjct: 223 EEEEEVINKEI 233
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 551 bits (1420), Expect = e-156
Identities = 308/877 (35%), Positives = 482/877 (54%), Gaps = 26/877 (2%)
Query: 740 HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPFKAKNVVSTSRPL 799
H+RLGH S ++ LS+ V G K A C C K K+ F + N + S
Sbjct: 570 HKRLGHPSFSRLDSLSE---VLGTTRHKNKKSAYCHVCHLAKQKKLSFPSANNICNST-F 625
Query: 800 ELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQNEKA 859
ELLHID++GP E++ G +Y + IVDD+SR TW+ L K + +VF FI V+N+
Sbjct: 626 ELLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWIYLLKSKSDVLTVFPAFIDLVENQYD 685
Query: 860 CRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQ 919
R+ VRSD+ E F + + GI SCP TP+QN VVERK++ + +AR ++
Sbjct: 686 TRVKSVRSDNAKELA---FTEFYKAKGIVSFHSCPETPEQNSVVERKHQHILNVARALMF 742
Query: 920 EIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVLNTK 979
+ M+ +W + V TA ++ NR + NKTP+E+ P+ S +FGC+CY +
Sbjct: 743 QSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLPDYSQLKTFGCLCYSSTSS 802
Query: 980 DRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL----DSDQSKLVE 1035
+ HKF +S C+ LGY KG++ + ++ + S +V F ++L S QS
Sbjct: 803 KQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEFHEELFPLASSQQSATTA 862
Query: 1036 KFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIASHPKEL--ILGNKDE- 1092
+ G ++ P + PS +++ +H ++ NKD+
Sbjct: 863 SDVFTPMDPLSSGNSITSHLPSP----QISPSTQISKRRITKFPAHLQDYHCYFVNKDDS 918
Query: 1093 -PVRTRSAFRP-SEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNEFSKNDVWSL 1150
P+ + ++ S +L + + + P+S EA K+W A+++E+ + D W +
Sbjct: 919 HPISSSLSYSQISPSHMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEI 978
Query: 1151 VKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFVPVARLEAIR 1210
P +G KWVF K + G + R KAR+VAKGY+Q+EG+DYTETF PVA++ ++
Sbjct: 979 TSLPPGKKAVGCKWVFTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVK 1038
Query: 1211 LLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFEDEKN----PDHVFKLKKSLY 1266
LL+ S + L+Q+D+ +AFLNG + E +Y+ P G+ D K P+ V +LKKS+Y
Sbjct: 1039 LLLKVSASKKWYLNQLDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIY 1098
Query: 1267 GLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKIIFGSANQSLC 1326
GLKQA R W+ + S+ LL F + D TLF + + +++ +YVD I+ S +
Sbjct: 1099 GLKQASRQWFLKFSNSLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAA 1158
Query: 1327 KEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNMTESTIAKTP 1386
+ ++ ++A F++ +G LKYFLG++V +T EG + Q KY ELL +M + + P
Sbjct: 1159 QSLTEALKASFKLRELGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIP 1218
Query: 1387 MHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSDPRETHLTAV 1446
M P L K D +++YR ++G L+YLT +RPD F+V+ +F S PR HL AV
Sbjct: 1219 MTPNIRLSKNDGLLLEDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAV 1278
Query: 1447 KRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASK 1506
++L+Y+KGT GL Y + L GY D D+ R+ST G F+GS+L+SW SK
Sbjct: 1279 YKVLQYIKGTVGQGLFYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSK 1338
Query: 1507 RQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILESNIPI-YCDNTAAISLSKNPIL 1565
+Q T+ S+A+AEY + A+ S +M W+ L ++ S +PI Y D+TAA+ ++ NP+
Sbjct: 1339 KQPTVSRSSAEAEYRALALASCEMAWLSTLLLALRV-HSGVPILYSDSTAAVYIATNPVF 1397
Query: 1566 HSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADI 1602
H R KHIE+ H +R+ + G L L V T + ADI
Sbjct: 1398 HERTKHIEIDCHTVREKLDNGQLKLLHVKTKDQVADI 1434
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 544 bits (1402), Expect = e-154
Identities = 307/898 (34%), Positives = 483/898 (53%), Gaps = 22/898 (2%)
Query: 718 LSELEAQNVKCLLSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEAC 777
L ++ Q++ V+ W HRRLGHAS++++ +S G K C C
Sbjct: 539 LLDVGDQSISVNAVVDISMW--HRRLGHASLQRLDAISDS---LGTTRHKNKGSDFCHVC 593
Query: 778 QKGKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFL 837
K K+ F N V +LLHID++GP E++ G +Y + IVDD+SR TW+ L
Sbjct: 594 HLAKQRKLSFPTSNKVC-KEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLL 652
Query: 838 SRKDESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTP 897
K E +VF FI QV+N+ ++ VRSD+ E KF S + GI SCP TP
Sbjct: 653 KTKSEVLTVFPAFIQQVENQYKVKVKAVRSDNAPEL---KFTSFYAEKGIVSFHSCPETP 709
Query: 898 QQNGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWK 957
+QN VVERK++ + +AR ++ + + W + V TA ++ NR + ++NKTPYE+
Sbjct: 710 EQNSVVERKHQHILNVARALMFQSQVPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILT 769
Query: 958 NIKPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEES 1017
P +FGC+CY + + HKF +S CL LGY KG++ + ++ T+ S
Sbjct: 770 GTAPVYEQLRTFGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFIS 829
Query: 1018 IHVRFDDKL--------DSDQSKLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDS 1069
+V+F +++ KL +S + + P + S
Sbjct: 830 RNVQFHEEVFPLAKNPGSESSLKLFTPMVPVSSGIISDTTHSPSSLPSQISDLPPQISSQ 889
Query: 1070 QPRKKSRIIASHPKELILGNKDEPVRTRSAF-RPSEETLLSMKGLVSLIEPKSIDEALQD 1128
+ RK + + + + P+ + ++ + S + + + + P + EA
Sbjct: 890 RVRKPPAHLNDYHCNTMQSDHKYPISSTISYSKISPSHMCYINNITKIPIPTNYAEAQDT 949
Query: 1129 KDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGY 1188
K+W A++ E+ K + W + P+ +G KWVF K G++ R KARLVAKGY
Sbjct: 950 KEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGY 1009
Query: 1189 SQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPG 1248
+Q+EG+DYT+TF PVA++ I+LL+ S + L Q+DV +AFLNG + EE+++ P G
Sbjct: 1010 TQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEG 1069
Query: 1249 FEDEKN----PDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD 1304
+ + K + V +LK+S+YGLKQA R W+++ SS LL F + D TLF K Y
Sbjct: 1070 YAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDG 1129
Query: 1305 DILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQ 1364
+ +IV +YVD I+ S +++ + ++ + F++ +G+LKYFLG++V +T G I Q
Sbjct: 1130 EFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQ 1189
Query: 1365 SKYTKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDN 1424
KY ELL+ M PM P + K+D + YR ++G L+YLT +RPD
Sbjct: 1190 RKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDI 1249
Query: 1425 LFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT 1484
F+V+ +F S PR THLTA R+L+Y+KGT GL Y +S+ L G+ D D+A +
Sbjct: 1250 TFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQD 1309
Query: 1485 *RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILE 1544
R+ST F+G +L+SW SK+Q T+ S+A+AEY + A+ + +M+W+ L Q
Sbjct: 1310 SRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASP 1369
Query: 1545 SNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADI 1602
+Y D+TAAI ++ NP+ H R KHI++ H +R+ + G L L V T+ + ADI
Sbjct: 1370 PVPILYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADI 1427
>At1g70010 hypothetical protein
Length = 1315
Score = 538 bits (1386), Expect = e-152
Identities = 311/888 (35%), Positives = 480/888 (54%), Gaps = 33/888 (3%)
Query: 740 HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPFKAKNVVSTSRPL 799
H+RLGH S++K+ +S L P K ++D C C K +PF + N S SRP
Sbjct: 408 HKRLGHPSVQKLQPMSSL---LSFPKQKNNTDFHCRVCHISKQKHLPFVSHNNKS-SRPF 463
Query: 800 ELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQNEKA 859
+L+HID +GP ++ G RY + IVDDYSR TWV L K + +V TF+ V+N+
Sbjct: 464 DLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLRNKSDVLTVIPTFVTMVENQFE 523
Query: 860 CRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTMLQ 919
I VRSD+ E F + S GI SCP TPQQN VVERK++ + +AR++
Sbjct: 524 TTIKGVRSDNAPELN---FTQFYHSKGIVPYHSCPETPQQNSVVERKHQHILNVARSLFF 580
Query: 920 EIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVLNTK 979
+ + +W + + TA Y+ NR+ + +K P+E+ P + FGC+CY +
Sbjct: 581 QSHIPISYWGDCILTAVYLINRLPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSP 640
Query: 980 DRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL----DSDQSKLVE 1035
HKF ++ C +GY KG++ + + +I S HV F ++L SD S+ +
Sbjct: 641 KDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVFHEELFPFLGSDLSQEEQ 700
Query: 1036 KF-ADLS------------ISVSDKGKALE---EAEPEEDDPEEAGPSDSQPRKKSRIIA 1079
F DL+ ++ SD ++E A P + PE + + + KK +
Sbjct: 701 NFFPDLNPTPPMQRQSSDHVNPSDSSSSVEILPSANPTNNVPEPSVQTSHRKAKKPAYLQ 760
Query: 1080 SHPKELILGNKDEPVRTRSAF-RPSEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEE 1138
+ ++ + +R ++ R ++ L + L EP + EA + + W AM E
Sbjct: 761 DYYCHSVVSSTPHEIRKFLSYDRINDPYLTFLACLDKTKEPSNYTEAEKLQVWRDAMGAE 820
Query: 1139 VNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTE 1198
+ W + P IG +W+F+ K N G V R KARLVA+GY+Q+EGIDY E
Sbjct: 821 FDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSDGSVERYKARLVAQGYTQKEGIDYNE 880
Query: 1199 TFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFE----DEKN 1254
TF PVA+L +++LL+ + + L Q+D+ +AFLNG + EE+Y+ P G+ D
Sbjct: 881 TFSPVAKLNSVKLLLGVAARFKLSLTQLDISNAFLNGDLDEEIYMRLPQGYASRQGDSLP 940
Query: 1255 PDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVD 1314
P+ V +LKKSLYGLKQA R WY + SS LL F++ D T F K L V +Y+D
Sbjct: 941 PNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLGFIQSYCDHTCFLKISDGIFLCVLVYID 1000
Query: 1315 KIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKK 1374
II S N + M++ F++ +GELKYFLG+++ ++ +G +I Q KY +LL +
Sbjct: 1001 DIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFLGLEIVRSDKGIHISQRKYALDLLDE 1060
Query: 1375 FNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARF 1434
+ PM P+ + + V YR +IG L+YL +RPD F+V+ A+F
Sbjct: 1061 TGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMYLNITRPDITFAVNKLAQF 1120
Query: 1435 QSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQ 1494
PR+ HL AV +IL+Y+KGT GL Y TSE +L Y + DY R R+ST G C
Sbjct: 1121 SMAPRKAHLQAVYKILQYIKGTIGQGLFYSATSELQLKVYANADYNSCRDSRRSTSGYCM 1180
Query: 1495 FLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESNIPIYCDN 1553
FLG +L+ W S++Q + S+A+AEY S ++ + +++W+ + L++ Q+ L ++CDN
Sbjct: 1181 FLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDELVWLTNFLKELQVPLSKPTLLFCDN 1240
Query: 1554 TAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWAD 1601
AAI ++ N + H R KHIE H +R+ + KG+ L ++T+ + AD
Sbjct: 1241 EAAIHIANNHVFHERTKHIESDCHSVRERLLKGLFELYHINTELQIAD 1288
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 516 bits (1328), Expect = e-146
Identities = 304/896 (33%), Positives = 475/896 (52%), Gaps = 36/896 (4%)
Query: 720 ELEAQNVKCLLSVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQK 779
+L +NVKCL + + + + LV G+ + + C +C
Sbjct: 393 DLNVENVKCL------------------QLEAATMVRKELVIGISNIPKEKET-CGSCLL 433
Query: 780 GKFTKVPFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSR 839
GK + PF S+ LEL+H DL GP+ + KRY +V++DD++R+ W L
Sbjct: 434 GKQARQPFPKATTYRASQVLELVHGDLCGPITQSTTAKKRYILVLIDDHTRYMWSMLLKE 493
Query: 840 KDESHSVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQ 899
K E+ F F +V+ E +I R+D GGEF + +F+ GI + P TPQQ
Sbjct: 494 KSEAFEKFRDFKTKVEQESGVKIKTFRTDKGGEFVSQEFQDFCAKEGINRHLTAPYTPQQ 553
Query: 900 NGVVERKNRTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNI 959
NGVVER+NRTL M R++L+ + M + W E V + YI NR+ R + N+TPYE++K
Sbjct: 554 NGVVERRNRTLLGMTRSILKHMKMPNYLWGEAVRHSTYIINRVGTRSLQNQTPYEVFKQR 613
Query: 960 KPNISYFHSFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYN-TDAKTIEESI 1018
KPN+ + FGC+ Y L K D +S + LG SK +R + T+ K I+ +
Sbjct: 614 KPNVEHLRVFGCIGYAKIEGPHLRKLDDRSKMLVYLGTEPGSKAYRLLDPTNRKIIKWNN 673
Query: 1019 HVRFDDKLDSDQSKLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRII 1078
+ S + +F + I SD + + E E+ EE G ++ +++
Sbjct: 674 SDSETRDISGTFSLTLGEFGNNGIQESDDIETEKNGEESENSHEEEGENEHNEQEQIDAE 733
Query: 1079 ASHPKELILGNKDEPVRTRSAFRPSEETLLSMKGLVSLIEPKSIDEALQD---------- 1128
+ P P RS + + L L++ IE + + A+ D
Sbjct: 734 ETQPSHAT----PLPTLRRSTRQVGKPNYLDDYVLMAEIEGEQVLLAINDEPWDFKEANK 789
Query: 1129 -KDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKG 1187
K+W A +EE+ KN WSL+ P VIG KWVF+ K N G + + KARLVAKG
Sbjct: 790 LKEWRDACKEEILSIEKNKTWSLIDLPVRRKVIGLKWVFKIKRNSDGSINKYKARLVAKG 849
Query: 1188 YSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPP 1247
Y Q+ GIDY E F VAR+E IR++I+ + ++ +H +DVK+AFL+G + E+VYV QP
Sbjct: 850 YVQRHGIDYDEVFAHVARIETIRVIIALAASNGWEVHHLDVKTAFLHGELREDVYVTQPE 909
Query: 1248 GFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDIL 1307
GF ++ N V+KL K+LYGLKQAPRAW +L+ L E FV+ + +++ + + +L
Sbjct: 910 GFTNKDNEGKVYKLHKALYGLKQAPRAWNTKLNKILQELNFVKCSKEPSVYRRQEEKKLL 969
Query: 1308 IVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKY 1367
IV IYVD ++ ++ L F K M +FEMS +G+L Y+LGI+V G + Q +Y
Sbjct: 970 IVAIYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQLTYYLGIEVLHRKNGIILRQERY 1029
Query: 1368 TKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFS 1427
+++++ M+ PM L K + ++ YR MIG L Y+ +RPD +
Sbjct: 1030 AMKIIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITERDYRRMIGCLRYIVHTRPDLSYC 1089
Query: 1428 VHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RK 1487
V + +R+ PRE+H A+K++LRYLKGT + GL K+ + L GY D ++ D K
Sbjct: 1090 VGVLSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKRGFKSGLVGYSDSSHSADLDDGK 1149
Query: 1488 STFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQL-EDYQILESN 1546
ST G+ +L ++W S++Q + LS+ +AE+++A + Q +W++ E
Sbjct: 1150 STAGHIFYLHQCPITWCSQKQQVVALSSCEAEFMAATEAAKQAIWLQDLFAEVCGTTSEK 1209
Query: 1547 IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWADI 1602
+ I DN +AI+L+KN + H R+KHI +YHFIR+ V+ ++ + V + ADI
Sbjct: 1210 VMIRVDNKSAIALTKNLVFHGRSKHIHRRYHFIRECVENNLVEVDHVPGVEQRADI 1265
Score = 70.5 bits (171), Expect = 8e-12
Identities = 59/289 (20%), Positives = 124/289 (42%), Gaps = 43/289 (14%)
Query: 77 ARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIMEPN 136
ARA+L ++ ++ + + ++E++K + G ++VKE+K +L+ +++ M+ N
Sbjct: 27 ARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRLNMKDN 86
Query: 137 ESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLP-ESWMPLVTSIELTRDVERMSL 195
E+I+E R + L + V + ++SLP + ++ ++ ++E D+
Sbjct: 87 ETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDLNTTGF 146
Query: 196 EELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDEDE 255
E+++ +K +E + + D + E+ K + A +E S +
Sbjct: 147 EDIVGRMKTYEDRVCDEDD--------------SPEEQGKLMYANSESSYDT-------- 184
Query: 256 LTLISKRLNHIWKHKQSKYRGSGKAKGKSESS-GQKKSSIKEVTCFECKESGHYKSDCPK 314
+ + RG G++ G+ G ++ +V C+ C ++GHY S+C
Sbjct: 185 --------------RGGRGRGRGRSSGRGRGGYGYQQRDKSKVICYRCDKTGHYASECL- 229
Query: 315 LKKDKRPKKHFKTKKSLMVTFDESESEDVDSDGEVQGLMTIVKDKEAES 363
R K K ++ D+ E E + V VK KE E+
Sbjct: 230 ----DRLLKLIKAQEQQQNNEDDDEIESLMMHEVVYLNERSVKPKEFEA 274
>At3g45520 copia-like polyprotein
Length = 1363
Score = 499 bits (1286), Expect = e-141
Identities = 304/911 (33%), Positives = 476/911 (51%), Gaps = 50/911 (5%)
Query: 733 NEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPFKAKNV 792
N++ + HRRL H S + +S L K L K S CE C G+ K+ F
Sbjct: 442 NDDTVLWHRRLCHMSQKNMSLLIKKGF---LDKKKVSMLDTCEDCIYGRAKKIGFNLAQH 498
Query: 793 VSTSRPLELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFI 851
T + LE +H DL+G P S+G +Y + +DDY+R WV FL KDE+ F ++I
Sbjct: 499 -DTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYTRKVWVYFLKTKDEAFEKFVSWI 557
Query: 852 AQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQ 911
+ V+N+ R+ +R+D+G EF N F+ + G +C TPQQNGVVER NRT+
Sbjct: 558 SLVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKGFQRHRTCAYTPQQNGVVERMNRTIM 617
Query: 912 EMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGC 971
E R+ML + G+ K FWAE +TA + N+ I + P + W P SY +GC
Sbjct: 618 EKVRSMLCDSGLPKRFWAEATHTAVLLINKTPCSAINFEFPDKRWSGKAPIYSYLRRYGC 677
Query: 972 VCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQS 1031
V +V +L+ ++ K +L+GY KG++ + + K S +V F + ++
Sbjct: 678 VTFVHTDGGKLN---LRAKKGVLIGYPSGVKGYKVWLIEEKKCVVSRNVSFQE--NAVYK 732
Query: 1032 KLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQ----PRKKSRIIASHPK---- 1083
L+++ +S D + + + E D +G SQ P + + ++ P+
Sbjct: 733 DLMQRKEQVSCEEDDHAGSYIDLDLEADKDNSSGGEQSQAQVTPATRGAVTSTPPRYETD 792
Query: 1084 --------------ELILGNKDEPVRTRSAFRPSE---ETLLSMKGLVSLIEPKSIDEAL 1126
L+ + +R F + E L + + +EP EA+
Sbjct: 793 DIEETDVHQSPLSYHLVRDRERREIRAPRRFDDEDYYAEALYTTED-GDAVEPADYKEAV 851
Query: 1127 QDKDWI---LAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKG-DVVRNKAR 1182
+D++W LAM EE+ KND W+ V +P+ +IG++W+++ K G + R KAR
Sbjct: 852 RDENWDKWRLAMNEEIESQLKNDTWTTVTRPEKQRIIGSRWIYKYKQGIPGVEEPRFKAR 911
Query: 1183 LVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVY 1242
LVAKGY+Q+EG+DY E F PV + +IR+L+S N+ L Q+DVK+AFL+G + E++Y
Sbjct: 912 LVAKGYAQREGVDYHEIFAPVVKHVSIRILLSIVAQENLELEQLDVKTAFLHGELKEKIY 971
Query: 1243 VHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTY 1302
+ P G E + V L KSLYGLKQAPR W E+ + ++ E F R D+ + K
Sbjct: 972 MMPPEGCESLFKENEVCLLNKSLYGLKQAPRQWNEKFNHYMTEIGFKRSDYDSCAYTKKL 1031
Query: 1303 KDD-ILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQV--DQTPEG 1359
DD + + YVD ++ + N K + +FEM +G K LGI++ D+
Sbjct: 1032 SDDSTMYLLFYVDDMLVAANNMQAIDALKKELSIKFEMKDLGAAKKILGIEIIIDREAGV 1091
Query: 1360 TYIHQSKYTKELLKKFNMTESTIAKTPMHP-----TCILEKEDASGKVCQKL-YRCMIGS 1413
++ Q Y ++LK FNM ES A TP+ + EK + + Y +GS
Sbjct: 1092 LWLSQESYLNKVLKTFNMLESKPALTPLGAHLKMKSATEEKLSTEEEYMNSVPYSSAVGS 1151
Query: 1414 LLY-LTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLS 1472
++Y + +RPD + V + +RF S P + H VK +LRY+KGT + L YK+ S++ +
Sbjct: 1152 IMYAMIGTRPDLAYPVGVVSRFMSQPAKEHWLGVKWVLRYIKGTVDTRLCYKRNSDFSIC 1211
Query: 1473 GYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLW 1532
GYCD DYA D R+S G LG N +SW S Q + S+ + EY+S + +W
Sbjct: 1212 GYCDADYAADLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSSTECEYMSLTEAVKEAIW 1271
Query: 1533 MKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKF 1592
+K L+D+ + N+ I+CD+ +AI+LSKN + H R KHI+VK+HFIR+ + G + +
Sbjct: 1272 LKGLLKDFGYEQKNVEIFCDSQSAIALSKNNVHHERTKHIDVKFHFIREIIADGKVEVSK 1331
Query: 1593 VDTDHKWADIY 1603
+ T+ ADI+
Sbjct: 1332 ISTEKNPADIF 1342
Score = 63.2 bits (152), Expect = 1e-09
Identities = 94/374 (25%), Positives = 150/374 (39%), Gaps = 76/374 (20%)
Query: 16 FDGQR-FEYWKDRLESFF--LGFDADLWDIIVD-GYERPVDA--EGKKIPRSEMTAEQKK 69
FDG+ + WK++L + LG A L + G ER + E +K R +M A ++K
Sbjct: 11 FDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEEREKMEAFEEK 70
Query: 70 LYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQKYE 129
KAR+ ++ ++S +KI A + E+L + + L QK
Sbjct: 71 ----KRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNR---IYLKQKLY 123
Query: 130 SFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTSIELTRD 189
SF M N SIE F +VA + LN + +D I ++ SLP+ + L +++ +
Sbjct: 124 SFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSSG 183
Query: 190 VERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEESEEASE 249
+SL+E+ + + EL+ + KKSI K + + E+ SE
Sbjct: 184 KTVLSLDEVAAAIYSRELEFGSV----KKSI---------KGQAEGLYVKDKAENRGRSE 230
Query: 250 DSDEDELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECKESGHYK 309
D K KGK S K+ C+ C E GH K
Sbjct: 231 QKD--------------------------KGKGKRSKSKSKRG------CWICGEDGHLK 258
Query: 310 SDCPKLKKDKRPKKHFKTKKSLMVTFDESESEDVDSDGE-VQGLMTIVKDKEAESKEVPD 368
S CP + K FK + S ++ E G V+G + V ES +
Sbjct: 259 STCP-----NKNKPQFKNQGS-------NKGESSGGKGNLVEGSVNFV-----ESAGMFV 301
Query: 369 SDSASEEDPNSDDE 382
S++ S D + +DE
Sbjct: 302 SEALSSTDIHLEDE 315
>At1g37110
Length = 1356
Score = 499 bits (1284), Expect = e-141
Identities = 298/890 (33%), Positives = 476/890 (53%), Gaps = 56/890 (6%)
Query: 740 HRRLGHASMRKISQLSKLNLV--RGLPTLKFSSDALCEACQKGKFTKVPFKAKNVVSTSR 797
H RLGH SM + L+ L+ + + L+F CE C GK KV F S
Sbjct: 441 HSRLGHMSMNNLKVLAGKGLIDRKEINELEF-----CEHCVMGKSKKVSFNVGKHTSEDA 495
Query: 798 PLELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQN 856
L +H DL+G P T SI GK+Y + I+DD +R W+ FL KDE+ F + + V+N
Sbjct: 496 -LSYVHADLWGSPNVTPSISGKQYFLSIIDDKTRKVWLYFLKSKDETFDKFCEWKSLVEN 554
Query: 857 EKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMART 916
+ ++ +R+D+G EF N +F+S +GI +C TPQQNGV ER NRT+ E R
Sbjct: 555 QVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHRTCTYTPQQNGVAERMNRTIMEKVRC 614
Query: 917 MLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVL 976
+L + G+ + FWAE TA Y+ NR I + P E+W N KP + FG + YV
Sbjct: 615 LLNKSGVEEVFWAEAAATAAYLINRSPASAINHNVPEEMWLNRKPGYKHLRKFGSIAYVH 674
Query: 977 NTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL---------- 1026
+ +L ++ K LGY +KG++ + + + S +V F + +
Sbjct: 675 QDQGKLKP---RALKGFFLGYPAGTKGYKVWLLEEEKCVISRNVVFQESVVYRDLKVKED 731
Query: 1027 DSDQSKLVE---------KFADLS-----ISVSDKGKALEEAEPEEDDPEEAGPSDSQ-- 1070
D+D E KFA+ S I + + + E E D EE S+
Sbjct: 732 DTDNLNQKETTSSEVEQNKFAEASGSGGVIQLQSDSEPITEGEQSSDSEEEVEYSEKTQE 791
Query: 1071 -PRKKSRIIASHPKELILGNKDEPVRTRSAFRPSEETLLSMKGLVSLI--EPKSIDEALQ 1127
P++ ++ + N + P R F ++ + + I EP+S EA++
Sbjct: 792 TPKRTGLTTYKLARDRVRRNINPPTR----FTEESSVTFALVVVENCIVQEPQSYQEAME 847
Query: 1128 DKD---WILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKG-DVVRNKARL 1183
+D W +A +E++ KN W LV KP+ +IG +W+F+ K G + R KARL
Sbjct: 848 SQDCEKWDMATHDEMDSLMKNGTWDLVDKPKDRKIIGCRWLFKLKSGIPGVEPTRFKARL 907
Query: 1184 VAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYV 1243
VAKGY+Q+EG+DY E F PV + +IR+L+S V+ ++ L QMDVK+ FL+G + EE+Y+
Sbjct: 908 VAKGYTQREGVDYQEIFAPVVKHVSIRILMSLVVDKDLELEQMDVKTTFLHGDLEEELYM 967
Query: 1244 HQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYK 1303
QP GF + + + V +LKKSLYGLKQ+PR W +R F+ +F+R + D ++ K
Sbjct: 968 EQPEGFVSDSSENKVCRLKKSLYGLKQSPRQWNKRFDRFMSSQQFIRSEHDACVYVKHVS 1027
Query: 1304 D-DILIVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYI 1362
+ D + + +YVD ++ A+++ + + EFEM MG LGI + + +G +
Sbjct: 1028 EHDFIYLLLYVDDMLIAGASKAEINRVKEQLSTEFEMKDMGGASRILGIDIYRDRKGGVL 1087
Query: 1363 HQSK--YTKELLKKFNMTESTIAKTPM---HPTCILEKEDASGKVCQKLYRCMIGSLLY- 1416
S+ Y +++L +FNM+ + + P+ + +ED Y +GS++Y
Sbjct: 1088 KLSQEIYIRKVLDRFNMSGAKMTNAPVGAHFKLAAVREEDECVDTDVVPYSSAVGSIMYA 1147
Query: 1417 LTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCD 1476
+ +RPD +++ L +R+ S P H AVK ++RYLKG +L L++ K ++ ++GYCD
Sbjct: 1148 MLGTRPDLAYAICLISRYMSKPGSMHWEAVKWVMRYLKGAQDLNLVFTKEKDFTVTGYCD 1207
Query: 1477 VDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQ 1536
+YA D R+S G +G N VSW + Q + +ST +AEYI+ A + + +W+K
Sbjct: 1208 SNYAADLDRRRSISGYVFTIGGNTVSWKASLQPVVAMSTTEAEYIALAEAAKEAMWIKGL 1267
Query: 1537 LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKG 1586
L+D + + + I+CD+ +AI LSKN + H R KHI+V++++IRD V+ G
Sbjct: 1268 LQDMGMQQDKVKIWCDSQSAICLSKNSVYHERTKHIDVRFNYIRDVVESG 1317
Score = 47.0 bits (110), Expect = 9e-05
Identities = 71/314 (22%), Positives = 121/314 (37%), Gaps = 62/314 (19%)
Query: 15 MFDGQR-FEYWKDRLESFF--LGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKKL- 70
+F+G R F WK R+++ LG L D + +E K+ ++ K++
Sbjct: 12 VFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTKEVP 71
Query: 71 ----YAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLIQ 126
Q +A+ I+++ IS K+ ++ +L NKK E+ + I
Sbjct: 72 DPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATL------NKKYMETSLPNRIY 125
Query: 127 ---KYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTS 183
K SF M +I++ F +VA + L + I ++ SLP S + L
Sbjct: 126 TQLKLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLK-- 183
Query: 184 IELTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEE 243
H LK K++ ++ AKS E E
Sbjct: 184 ---------------------HTLKYGN------KTLTVQDVTSSAKS-------LEREL 209
Query: 244 SEEASEDSDEDELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECK 303
+E D + + ++R + ++ Q G+ KG+S S+ + K V C+ CK
Sbjct: 210 AEAVDLDKGQAAVLYTTERGRPLVRNNQK----GGQGKGRSRSNSKTK-----VPCWYCK 260
Query: 304 ESGHYKSDCPKLKK 317
+ GH K DC KK
Sbjct: 261 KEGHVKKDCYSRKK 274
>At4g17450 retrotransposon like protein
Length = 1433
Score = 495 bits (1275), Expect = e-140
Identities = 295/887 (33%), Positives = 463/887 (51%), Gaps = 61/887 (6%)
Query: 740 HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDA--LCEACQKGKFTKVPFKAKNVVSTSR 797
H+RLGH + KI LS + ++ K S +C C K + F+++ + S
Sbjct: 554 HKRLGHPAYSKIDLLSDVLNLKVKKINKEHSPVCHVCHVCHLSKQKHLSFQSRQNMC-SA 612
Query: 798 PLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQNE 857
+L+HID +GP + TW+ L K + VF FI V +
Sbjct: 613 AFDLVHIDTWGPFSVPTNDA--------------TWIYLLKNKSDVLHVFPAFINMVHTQ 658
Query: 858 KACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTM 917
++ VRSD+ E KF LF ++GI SCP TP+QN VVERK++ + +AR +
Sbjct: 659 YQTKLKSVRSDNAHEL---KFTDLFAAHGIVAYHSCPETPEQNSVVERKHQHILNVARAL 715
Query: 918 LQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVLN 977
L + + FW + V TA ++ NR+ + NK+PYE KNI P +FGC+CY
Sbjct: 716 LFQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAYESLKTFGCLCYSST 775
Query: 978 TKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKF 1037
+ + HKF+ ++ C+ LGY KG++ + + + S HV F + + F
Sbjct: 776 SPKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFHEDI----------F 825
Query: 1038 ADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIASHPKELILGNKD----EP 1093
+S ++ D +++ P P +D P +++ II +HP + + +K +P
Sbjct: 826 PFISSTIKDD---IKDFFPLLQFPAR---TDDLPLEQTSIIDTHPHQDVSSSKALVPFDP 879
Query: 1094 VRTRSAFRP------------SEETLLSMKGLVSLIEPKSIDEALQDKDWILAMEEEVNE 1141
+ R P +E + + + + P+ EA K W AM+EE+
Sbjct: 880 LSKRQKKPPKHLQDFHCYNNTTEPFHAFINNITNAVIPQRYSEAKDFKAWCDAMKEEIGA 939
Query: 1142 FSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKGYSQQEGIDYTETFV 1201
+ + WS+V P + IG KWVF K N G + R KARLVAKGY+Q+EG+DY ETF
Sbjct: 940 MVRTNTWSVVSLPPNKKAIGCKWVFTIKHNADGSIERYKARLVAKGYTQEEGLDYEETFS 999
Query: 1202 PVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFED---EKNPDH- 1257
PVA+L ++R+++ + +HQ+D+ +AFLNG + EE+Y+ PPG+ D E P H
Sbjct: 1000 PVAKLTSVRMMLLLAAKMKWSVHQLDISNAFLNGDLDEEIYMKIPPGYADLVGEALPPHA 1059
Query: 1258 VFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDKII 1317
+ +L KS+YGLKQA R WY +LS+ L F + D TLF K ++ V +YVD I+
Sbjct: 1060 ICRLHKSIYGLKQASRQWYLKLSNTLKGMGFQKSNADHTLFIKYANGVLMGVLVYVDDIM 1119
Query: 1318 FGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKYTKELLKKFNM 1377
S + +F+ +++ F++ +G KYFLGI++ ++ +G I Q KY ELL
Sbjct: 1120 IVSNSDDAVAQFTAELKSYFKLRDLGAAKYFLGIEIARSEKGISICQRKYILELLSTTGF 1179
Query: 1378 TESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFSVHLCARFQSD 1437
S + P+ P+ L KED YR ++G L+YL +RPD ++V+ +F
Sbjct: 1180 LGSKPSSIPLDPSVKLNKEDGVPLTDSTSYRKLVGKLMYLQITRPDIAYAVNTLCQFSHA 1239
Query: 1438 PRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RKSTFGNCQFLG 1497
P HL+AV ++LRYLKGT GL Y ++ L GY D D+ R+ C F+G
Sbjct: 1240 PTSVHLSAVHKVLRYLKGTVGQGLFYSADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIG 1299
Query: 1498 SNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQILESNIP---IYCDNT 1554
LVSW SK+Q T+ +STA+AE+ + + + +M+W+ +D+++ IP +YCDNT
Sbjct: 1300 DYLVSWKSKKQDTVSMSTAEAEFRAMSQGTKEMIWLSRLFDDFKV--PFIPPAYLYCDNT 1357
Query: 1555 AAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWAD 1601
AA+ + N + H R K +E+ + R+ V+ G L FV+T + AD
Sbjct: 1358 AALHIVNNSVFHERTKFVELDCYKTREAVESGFLKTMFVETGEQVAD 1404
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 484 bits (1247), Expect = e-136
Identities = 305/907 (33%), Positives = 486/907 (52%), Gaps = 56/907 (6%)
Query: 740 HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPFKAKNVVSTSRPL 799
H+RL H S + + L + L K SS +CE C GK + F + T L
Sbjct: 442 HQRLCHMSQKNMEILVRKGF---LDKKKVSSLDVCEDCIYGKAKRKSFSLAHH-DTKEKL 497
Query: 800 ELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQNEK 858
E +H DL+G P S+G +Y M I+DD++R WV F+ KDE+ F ++ V+N+
Sbjct: 498 EYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYFMKTKDEAFEKFVEWVNLVENQT 557
Query: 859 ACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTML 918
R+ +R+D+G EF N F+ +S GI +C TPQQNGV ER NRT+ E R+ML
Sbjct: 558 DRRVKTLRTDNGLEFCNKLFDGFCESIGIHRHRTCAYTPQQNGVAERMNRTIMEKVRSML 617
Query: 919 QEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVLNT 978
+ G+ K FWAE +T + N+ + + P + W P SY +GCV +V +T
Sbjct: 618 SDSGLPKRFWAEATHTTVLLINKTPSSALNFEIPDKKWSGNPPVYSYLRRYGCVAFV-HT 676
Query: 979 KDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLVEKFA 1038
D K + ++ K +L+GY KG++ + D + S ++ F + ++ L+++
Sbjct: 677 DDG--KLEPRAKKGVLIGYPVGVKGYKVWILDERKCVVSRNIIFQE--NAVYKDLMQRQE 732
Query: 1039 DLSISVSDK-GKALE---EAE---------------PEEDDPEEAGPS--DSQPRKKSRI 1077
++S D+ G LE EAE P + P + P+ D+ + S +
Sbjct: 733 NVSTEEDDQTGSYLEFDLEAERDVISGGDQEMVNTIPAPESPVVSTPTTQDTNDDEDSDV 792
Query: 1078 IASHPKELILGNKDE-PVRTRSAFRPSE---ETLLSMKGLVSLIEPKSIDEALQDKD--- 1130
S ++ ++D+ +R F + E L + + +EP++ +A D +
Sbjct: 793 NQSPLSYHLVRDRDKREIRAPRRFDDEDYYAEALYTTED-GEAVEPENYRKAKLDANFDK 851
Query: 1131 WILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKG-DVVRNKARLVAKGYS 1189
W LAM+EE++ KN+ W++V +P++ +IG +W+F+ KL G + R KARLVAKGY+
Sbjct: 852 WKLAMDEEIDSQEKNNTWTIVTRPENQRIIGCRWIFKYKLGILGVEEPRFKARLVAKGYA 911
Query: 1190 QQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGF 1249
Q+EGIDY E F PV + +IR+L+S ++ L Q+DVK+AFL+G + E++Y+ P G+
Sbjct: 912 QKEGIDYHEIFAPVVKHVSIRVLLSIVAQEDLELEQLDVKTAFLHGELKEKIYMSPPEGY 971
Query: 1250 EDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKD-DILI 1308
E + V L K+LYGLKQAP+ W E+ +F+ E FV+ D+ + K D ++
Sbjct: 972 ESMFKANEVCLLNKALYGLKQAPKQWNEKFDNFMKEICFVKSAYDSCAYTKVLPDGSVMY 1031
Query: 1309 VQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQV--DQTPEGTYIHQSK 1366
+ IYVD I+ S N+ + FEM +G K LG+++ D+T ++ Q
Sbjct: 1032 LLIYVDDILVASKNKEAITALKANLGMRFEMKDLGAAKKILGMEIIRDRTLGVLWLSQEG 1091
Query: 1367 YTKELLKKFNMTESTIAKTPMHPTC---------ILEKEDASGKVCQKLYRCMIGSLLY- 1416
Y ++L+ +NM E+ A TP+ ++ ED V Y +GS++Y
Sbjct: 1092 YLNKILETYNMAEAKPAMTPLGAHFKFQAATEQKLIRDEDFMKSVP---YSSAVGSIMYA 1148
Query: 1417 LTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCD 1476
+ +RPD + V + +RF S P + H VK +LRY+KGT L YKK+S + + GYCD
Sbjct: 1149 MLGTRPDLAYPVGIISRFMSQPIKEHWLGVKWVLRYIKGTLKTRLCYKKSSSFSIVGYCD 1208
Query: 1477 VDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQ 1536
DYA D R+S G LG N +SW S Q + ST ++EY+S + +W+K
Sbjct: 1209 ADYAADLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSTTESEYMSLTEAVKEAIWLKGL 1268
Query: 1537 LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTD 1596
L+D+ + ++ I+CD+ +AI+LSKN + H R KHI+VKYHFIR+ + G + + + T+
Sbjct: 1269 LKDFGYEQKSVEIFCDSQSAIALSKNNVHHERTKHIDVKYHFIREIISDGTVEVLKISTE 1328
Query: 1597 HKWADIY 1603
ADI+
Sbjct: 1329 KNPADIF 1335
Score = 61.6 bits (148), Expect = 4e-09
Identities = 73/316 (23%), Positives = 134/316 (42%), Gaps = 74/316 (23%)
Query: 16 FDGQR-FEYWKDRLESFF--LGFDADLWDIIVDGYERPVDAEG-----KKIPRSEMTAEQ 67
FDG+ + WK++L + LG + L + G ++ V E +K+ + E E+
Sbjct: 11 FDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEALEEK 70
Query: 68 KKLYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLSLI-- 125
KK KAR+ ++ +++ +KI A + +L K+ SK L
Sbjct: 71 KK------KARSAIVLSVTDRVLRKIKKESTAAAMLLALD-------KLYMSKALPNRIY 117
Query: 126 --QKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTS 183
QK SF M N S+E F ++ + +N + +D I ++ +LP+++ L +
Sbjct: 118 PKQKLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDT 177
Query: 184 IELTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSK--YEKAKSEKSKALQAEA 241
++ + ++L+E+ + + EL+ + KKSI ++++ Y K K+E
Sbjct: 178 LKYSSGKSILTLDEVAAAIYSKELELGSV----KKSIKVQAEGLYVKDKNEN-------- 225
Query: 242 EESEEASEDSDEDELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFE 301
+G G+ KGK + G+K S K+ C+
Sbjct: 226 ---------------------------------KGKGEQKGKGK--GKKGKSKKKPGCWT 250
Query: 302 CKESGHYKSDCPKLKK 317
C E GH++S CP K
Sbjct: 251 CGEEGHFRSSCPNQNK 266
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 483 bits (1242), Expect = e-136
Identities = 310/916 (33%), Positives = 481/916 (51%), Gaps = 55/916 (6%)
Query: 732 VNEEQWV*HRRLGHASMRKISQLSKLNLVRG--LPTLKFSSDALCEACQKGKFTKVPFKA 789
V +E + H RLGH S + + L K +R + L+F CE C GK +V F
Sbjct: 410 VKDETALWHSRLGHMSQKGMEILVKKGCLRREVIKELEF-----CEDCVYGKQHRVSFAP 464
Query: 790 KNVVSTSRPLELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFS 848
V T L +H DL+G P S+G +Y + VDDYSR W+ FL +KDE+ F
Sbjct: 465 AQHV-TKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRKVWIYFLRKKDEAFEKFV 523
Query: 849 TFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNR 908
+ V+N+ ++ ++R+D+G E+ N FE GI +C TPQQNG+ ER NR
Sbjct: 524 EWKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVRHKTCAYTPQQNGIAERLNR 583
Query: 909 TLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHS 968
T+ + R+ML GM K FWAE +TA Y+ NR I P E W P++S
Sbjct: 584 TIMDKVRSMLSRSGMEKKFWAEAASTAVYLINRSPSTAINFDLPEEKWTGALPDLSSLRK 643
Query: 969 FGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRF------ 1022
FGC+ Y+ + +L + +S K + Y + KG++ + + K S +V F
Sbjct: 644 FGCLAYIHADQGKL---NPRSKKGIFTSYPEGVKGYKVWVLEDKKCVISRNVIFREQVMF 700
Query: 1023 -DDKLDSDQSKLVEKFADLSIS-------VSDKGKALEE-AEPEE----DDPEEAGPSDS 1069
D K DS + DL ++ +D+G A ++ + P E +P P+ S
Sbjct: 701 KDLKGDSQNTISESDLEDLRVNPDMNDQEFTDQGGATQDNSNPSEATTSHNPVLNSPTHS 760
Query: 1070 QPRKKSRIIASHPKELILGN--KDEPVRTRSAFRPSEETLLSMKGLV------SLIEPKS 1121
Q + + ++L +D RT A E+ +M G EPKS
Sbjct: 761 QDEESEEEDSDAVEDLSTYQLVRDRVRRTIKANPKYNES--NMVGFAYYSEDDGKPEPKS 818
Query: 1122 IDEALQDKD---WILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKG-DVV 1177
EAL D D W AM+EE+ SKN W LV KP+ V +IG +WVF K G +
Sbjct: 819 YQEALLDPDWEKWNAAMKEEMVSMSKNHTWDLVTKPEKVKLIGCRWVFTRKAGIPGVEAP 878
Query: 1178 RNKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYI 1237
R ARLVAKG++Q+EG+DY E F PV + +IR L+S V++N+ L QMDVK+AFL+G++
Sbjct: 879 RFIARLVAKGFTQKEGVDYNEIFSPVVKHVSIRYLLSMVVHYNMELQQMDVKTAFLHGFL 938
Query: 1238 SEEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTL 1297
EE+Y+ QP GFE ++ + V LK+SLYGLKQ+PR W R F+ ++ R D+ +
Sbjct: 939 EEEIYMAQPEGFEIKRGSNKVCLLKRSLYGLKQSPRQWNLRFDEFMRGIKYTRSAYDSCV 998
Query: 1298 FCKTYKDDILI-VQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQT 1356
+ K D I + +YVD ++ SAN+S E +++ EFEM +G+ K LG+++ +
Sbjct: 999 YFKKCNGDTYIYLLLYVDDMLIASANKSEVNELKQLLSREFEMKDLGDAKKILGMEISRD 1058
Query: 1357 PEG--TYIHQSKYTKELLKKFNMTESTIAKTP------MHPTCILEKEDASGKVCQKLYR 1408
+ + Q Y K++L+ F M + TP + E E+ ++ Y
Sbjct: 1059 RDAGLLTLSQEGYVKKVLRSFQMDNAKPVSTPLGIHFKLKAATDKEYEEQFERMKIVPYA 1118
Query: 1409 CMIGSLLY-LTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTS 1467
IGS++Y + +RPD +S+ + +RF S P + H AVK +LRY++GT L ++K
Sbjct: 1119 NTIGSIMYSMIGTRPDLAYSLGVISRFMSKPLKDHWQAVKWVLRYMRGTEKKKLCFRKQE 1178
Query: 1468 EYKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICS 1527
++ L GYCD DY + R+S G +G N +SW SK Q + +S+ +AEY++
Sbjct: 1179 DFLLRGYCDSDYGSNFDTRRSITGYVFTVGGNTISWKSKLQKVVAISSTEAEYMALTEAV 1238
Query: 1528 TQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGV 1587
+ LW+K + + + ++ D+ +AI+L+KN + H R KHI+++ HFIRD + G+
Sbjct: 1239 KEALWLKGFAAELGHSQDYVEVHSDSQSAITLAKNSVHHERTKHIDIRLHFIRDIICAGL 1298
Query: 1588 LLLKFVDTDHKWADIY 1603
+ + + T+ A+I+
Sbjct: 1299 IKVVKIATECNPANIF 1314
Score = 43.1 bits (100), Expect = 0.001
Identities = 48/262 (18%), Positives = 112/262 (42%), Gaps = 13/262 (4%)
Query: 39 LWDIIVDGYERPVDAEGKKIP--RSEMTAEQKKLYAQHHKARAILLSAISYEEYQKITDL 96
L D+++ +P+ E ++ P R + A++ + KA+ ++ ++ + +KI
Sbjct: 9 LKDVVLGTTPKPLTEEEEEDPEKRKKRDADEVARLERCDKAKNVIFLNVADKVLRKIELC 68
Query: 97 EFANGIFESLKMSHEGNKKVKESKTLSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPL 156
+ A +E+L T + +F M+ N+ I+E F +VA + L
Sbjct: 69 KTAAEAWETLDRLFMIRSLPHRVYTQL---SFYTFKMQENKKIDENIDDFLKIVADLNHL 125
Query: 157 NKSYTTKDHVIRVIRSLPESWMPLVTSIELTRDVERMSLEELISILKCHELKRSEMQDLR 216
T + I ++ SLP + LV +++ + E++ L++++ + E + S+
Sbjct: 126 QIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVMVAARDKERELSQ----- 180
Query: 217 KKSIALKSKYEKAKSEKSKALQAEAEESEEASEDSDEDELTLISKRLNHIWKHKQSKYRG 276
++ + + + + Q ++ S+ +D + I + H K+ Y+
Sbjct: 181 NNRPVVEGHFARGRPDGKNNNQGNKGKNRSRSKSADGKRVCWICGKEGHF---KKQCYKW 237
Query: 277 SGKAKGKSESSGQKKSSIKEVT 298
+ K K + S +SS+ + T
Sbjct: 238 IERNKSKQQGSDNGESSLAKST 259
>At1g26990 polyprotein, putative
Length = 1436
Score = 480 bits (1235), Expect = e-135
Identities = 290/895 (32%), Positives = 465/895 (51%), Gaps = 60/895 (6%)
Query: 731 SVNEEQWV*HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDAL-CEACQKGKFTKVPFKA 789
SV E + H+RLGH S KI LS + + LP K + D+ C C K +PFK+
Sbjct: 550 SVKNESEMWHKRLGHPSFAKIDTLSDVLM---LPKQKINKDSSHCHVCHLSKQKHLPFKS 606
Query: 790 KNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFST 849
N + + EL+HID +GP ++ RY + IVDD+SR TW+ L +K + +VF +
Sbjct: 607 VNHIR-EKAFELVHIDTWGPFSVPTVDSYRYFLTIVDDFSRATWIYLLKQKSDVLTVFPS 665
Query: 850 FIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRT 909
F+ V+ + ++ VRSD+ E KF LF GI D CP TP+QN VVERK++
Sbjct: 666 FLKMVETQYHTKVCSVRSDNAHEL---KFNELFAKEGIKADHPCPETPEQNFVVERKHQH 722
Query: 910 LQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSF 969
L +AR ++ + G+ +W + V TA ++ NR+ I N+TPYE KP+ S +F
Sbjct: 723 LLNVARALMFQSGIPLEYWGDCVLTAVFLINRLLSPVINNETPYERLTKGKPDYSSLKAF 782
Query: 970 GCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRF-DDKLDS 1028
GC+CY + KFD ++ C+ LGY KG++ + + ++ S HV F +D
Sbjct: 783 GCLCYCSTSPKSRTKFDPRAKACIFLGYPMGYKGYKLLDIETYSVSISRHVIFYEDIFPF 842
Query: 1029 DQSKLVEK----FADLSISVSDKGKALEEAEPEEDDPEEAGPSDS------QPRK-KSRI 1077
S + + F + + + + L + D P S S +P+ + R
Sbjct: 843 ASSNITDAAKDFFPHIYLPAPNNDEHLPLVQSSSDAPHNHDESSSMIFVPSEPKSTRQRK 902
Query: 1078 IASHPKELILGNKDEPVRTRSAFRP----------SEETLLSMKGLVSLIEPKSIDEALQ 1127
+ SH ++ N + P T+++ P SE + + + P+ EA
Sbjct: 903 LPSHLQDFHCYN-NTPTTTKTSPYPLTNYISYSYLSEPFGAFINIITATKLPQKYSEARL 961
Query: 1128 DKDWILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAKG 1187
DK W AM +E++ F + WS+ P +G KW+ K G + R+KARLVAKG
Sbjct: 962 DKVWNDAMGKEISAFVRTGTWSICDLPAGKVAVGCKWIITIKFLADGSIERHKARLVAKG 1021
Query: 1188 YSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPP 1247
Y+QQEGID+ TF PVA++ +++L+S + LHQ+D+ +A LNG + EE+Y+ PP
Sbjct: 1022 YTQQEGIDFFNTFSPVAKMVTVKVLLSLAPKMKWYLHQLDISNALLNGDLEEEIYMKLPP 1081
Query: 1248 GFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDIL 1307
G+ + + G + +P A + D TLF K L
Sbjct: 1082 GYSE-------------IQGQEVSPNA---------------KCHGDHTLFVKAQDGFFL 1113
Query: 1308 IVQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGTYIHQSKY 1367
+V +YVD I+ S ++ E + + + F++ +GE K+FLGI++ + +G + Q KY
Sbjct: 1114 VVLVYVDDILIASTTEAASAELTSQLSSFFQLRDLGEPKFFLGIEIARNADGISLCQRKY 1173
Query: 1368 TKELLKKFNMTESTIAKTPMHPTCILEKEDASGKVCQKLYRCMIGSLLYLTASRPDNLFS 1427
+LL + ++ + PM P L K+ + K YR ++G L YL +RPD F+
Sbjct: 1174 VLDLLASSDFSDCKPSSIPMEPNQKLSKDTGTLLEDGKQYRRILGKLQYLCLTRPDINFA 1233
Query: 1428 VHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAGDRT*RK 1487
V A++ S P + HL A+ +ILRYLKGT GL Y + + L G+ D D+ R+
Sbjct: 1234 VSKLAQYSSAPTDIHLQALHKILRYLKGTIGQGLFYGADTNFDLRGFSDSDWQTCPDTRR 1293
Query: 1488 STFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQI-LESN 1546
G F+G++LVSW SK+Q + +S+A+AEY + ++ + +++W+ + L ++I
Sbjct: 1294 CVTGFAIFVGNSLVSWRSKKQDVVSMSSAEAEYRAMSVATKELIWLGYILTAFKIPFTHP 1353
Query: 1547 IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWAD 1601
+YCDN AA+ ++ N + H R KHIE H +R+ ++ G+L FV TD++ AD
Sbjct: 1354 AYLYCDNEAALHIANNSVFHERTKHIENDCHKVRECIEAGILKTIFVRTDNQLAD 1408
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 477 bits (1227), Expect = e-134
Identities = 302/902 (33%), Positives = 475/902 (52%), Gaps = 47/902 (5%)
Query: 740 HRRLGHASMRKISQLSKLNLVRGLPTLKFSSDALCEACQKGKFTKVPFKAKNVVSTSRPL 799
HRRLGH S + + L K L L K S CE C GK ++ F T L
Sbjct: 420 HRRLGHMSQKNMDLLLKKGL---LDKKKVSKLETCEDCIYGKAKRIGFNLAQH-DTREKL 475
Query: 800 ELLHIDLFG-PVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVFSTFIAQVQNEK 858
E +H DL+G P S+G +Y + +DDY+R + FL KDE+ F + V+N+
Sbjct: 476 EYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDEAFDKFVEWANLVENQT 535
Query: 859 ACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKNRTLQEMARTML 918
RI +R+D+G EF N F+ GI +C TPQQNGV ER NRTL E R+ML
Sbjct: 536 DKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNGVAERMNRTLMEKVRSML 595
Query: 919 QEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFHSFGCVCYVLNT 978
+ G+ K FWAE +T + N+ + + P + W P SY FGC+ +V +T
Sbjct: 596 SDSGLPKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSPIYSYLRRFGCIAFV-HT 654
Query: 979 KDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL---DSDQSKLVE 1035
D K + ++ K +L+GY KG++ + + K S +V F + D QSK E
Sbjct: 655 DDG--KLNPRAKKGILVGYPIGVKGYKIWLLEEKKCVVSRNVIFQENASYKDMMQSKDAE 712
Query: 1036 K---------FADLS------ISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIAS 1080
K + DL I+ +E P P ++ II S
Sbjct: 713 KDENEAPPSSYLDLDLDHEEVITSGGDDPIVEAQSPFNPSPATTQTYSEGVNSETDIIQS 772
Query: 1081 HPKELILGNKDEPVRTRSAFRPSEETLLSMKGLVSL-----IEPKSIDEALQDKDWI--- 1132
++ ++D R+ R +E L+ + L + IEP EA + +W
Sbjct: 773 PLSYQLVRDRDRRT-IRAPVRFDDEDYLA-EALYTTEDSGEIEPADYSEAKRSMNWNKWK 830
Query: 1133 LAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKG-DVVRNKARLVAKGYSQQ 1191
LAM EE+ KN W++VK+PQ VIG++W+++ KL G + R KARLVAKGY+Q+
Sbjct: 831 LAMNEEMESQIKNHTWTVVKRPQHQKVIGSRWIYKFKLGIPGVEEGRFKARLVAKGYAQR 890
Query: 1192 EGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYISEEVYVHQPPGFED 1251
+GIDY E F PV + +IR+L+S ++ L Q+DVK+AFL+G + E++Y+ P G+E+
Sbjct: 891 KGIDYHEIFAPVVKHVSIRILMSIVAQEDLELEQLDVKTAFLHGELKEKIYMVPPEGYEE 950
Query: 1252 EKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILI-VQ 1310
D V L KSLYGLKQAP+ W E+ ++++ E F+R D+ + K D + +
Sbjct: 951 MFKEDEVCLLNKSLYGLKQAPKQWNEKFNAYMSEIGFIRSLYDSCAYIKELSDGSRVYLL 1010
Query: 1311 IYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTPEGT--YIHQSKYT 1368
+YVD ++ + N+ + + + F+M +G K LG+++ + E ++ Q+ Y
Sbjct: 1011 LYVDDMLVAAKNKEDISQLKEELSQRFDMKDLGAAKRILGMEIIRNREENTLWLSQNGYL 1070
Query: 1369 KELLKKFNMTESTIAKTPMHP-----TCILEKEDASGKVCQKL-YRCMIGSLLY-LTASR 1421
++L+ +NM ES TP+ +EK++ + + Y +GS++Y + +R
Sbjct: 1071 NKILETYNMAESKHVVTPLGAHLKMRAATVEKQEQDEDYMKSIPYSSAVGSIMYAMIGTR 1130
Query: 1422 PDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSEYKLSGYCDVDYAG 1481
PD + V + +R+ S P H VK +LRY+KG+ L YK++S++K+ GYCD D+A
Sbjct: 1131 PDLAYPVGIISRYMSQPAREHWLGVKWVLRYIKGSLGTKLQYKRSSDFKVVGYCDADHAA 1190
Query: 1482 DRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICSTQMLWMKHQLEDYQ 1541
+ R+S G LG + +SW S +Q + LST +AEY+S + +WMK L+++
Sbjct: 1191 CKDRRRSITGLVFTLGGSTISWKSGQQRVVALSTTEAEYMSLTEAVKEAVWMKGLLKEFG 1250
Query: 1542 ILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHKWAD 1601
+ ++ I+CD+ +AI+LSKN + H R KHI+V+Y +IRD + G + +DT+ AD
Sbjct: 1251 YEQKSVEIFCDSQSAIALSKNNVHHERTKHIDVRYQYIRDIIANGDGDVVKIDTEKNPAD 1310
Query: 1602 IY 1603
I+
Sbjct: 1311 IF 1312
Score = 47.0 bits (110), Expect = 9e-05
Identities = 55/233 (23%), Positives = 105/233 (44%), Gaps = 35/233 (15%)
Query: 16 FDGQR-FEYWKDRLESFF--LGFDADLWDIIVDGYERPVDAEGKKIPRSEMTAEQKK--- 69
FDG+ + WK++L + LG L + D +K+ ++T E++K
Sbjct: 11 FDGRGDYTMWKEKLMAHLDILGLSVAL---------KEEDDLVEKVAEMQLTEEEEKEEV 61
Query: 70 -----LYAQHHKARAILLSAISYEEYQKITDLEFAN---GIFESLKMSHEGNKKVKESKT 121
L + KAR+ ++ +++ +KI + A G+ + L MS ++ +
Sbjct: 62 LRRELLEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQK-- 119
Query: 122 LSLIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLV 181
QK SF M N SIE F ++A + N + +D I ++ SLP+ + L
Sbjct: 120 ----QKLYSFKMSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLR 175
Query: 182 TSIELTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSK--YEKAKSE 232
+++ +SL+E+++ + EL+ KKSI +++ + K K+E
Sbjct: 176 DTLKYGLGRVTLSLDEVVAAIYSKELELGS----NKKSIKGQAEGLFVKEKTE 224
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 465 bits (1197), Expect = e-131
Identities = 281/855 (32%), Positives = 462/855 (53%), Gaps = 49/855 (5%)
Query: 789 AKNVVSTSRPLELLHIDLFGPVKTE-SIGGKRYGMVIVDDYSRWTWVKFLSRKDESHSVF 847
AK+V T L+ +H DL+G SIG +Y + +DD++R TW+ F+ KDE+ S F
Sbjct: 476 AKHV--TKDKLDYVHSDLWGSTNVPFSIGKCQYFITFIDDFTRRTWIYFIRTKDEAFSKF 533
Query: 848 STFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSCPRTPQQNGVVERKN 907
+ Q++N++ ++ + +D+G EF N +F+S G+ +C TPQQNGV ER N
Sbjct: 534 VEWKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCAYTPQQNGVAERMN 593
Query: 908 RTLQEMARTMLQEIGMAKHFWAEVVNTACYIQNRISVRRILNKTPYELWKNIKPNISYFH 967
RT+ R ML E G+ K FWAE +TA ++ N+ I P E W P+
Sbjct: 594 RTIMNKVRCMLSESGLGKQFWAEAASTAVFLINKSPSSSIEFDIPEEKWTGHPPDYKILK 653
Query: 968 SFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL- 1026
FG V Y+ + + +L+ ++ K + LGY D K F+ + + + S + F +
Sbjct: 654 KFGSVAYIHSDQGKLNP---RAKKGIFLGYPDGVKRFKVWLLEDRKCVVSRDIVFQENQM 710
Query: 1027 -----DSDQSKLVEKFADLSISVSDKGKALEEAEPEEDDPEEAGPSDSQPRKKSRIIASH 1081
+D S+ ++ ++ ++ + L+ ++++ E G + +Q + + AS
Sbjct: 711 YKELQKNDMSEEDKQLTEVERTLIE----LKNLSADDENQSEGGDNSNQEQASTTRSASK 766
Query: 1082 PKELILGNKDEP---------------VRTRSAFRPSEETL----LSMKGLVSLIEPKSI 1122
K++ + D+ +R F +++L L+M + EP++
Sbjct: 767 DKQVEETDSDDDCLENYLLARDRIRRQIRAPQRFVEEDDSLVGFALTMTEDGEVYEPETY 826
Query: 1123 DEALQDKD---WILAMEEEVNEFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKG-DVVR 1178
+EA++ + W A EE++ KND W ++ KP+ VIG KW+F+ K G + R
Sbjct: 827 EEAMRSPECEKWKQATIEEMDSMKKNDTWDVIDKPEGKRVIGCKWIFKRKAGIPGVEPPR 886
Query: 1179 NKARLVAKGYSQQEGIDYTETFVPVARLEAIRLLISFSVNHNIVLHQMDVKSAFLNGYIS 1238
KARLVAKG+SQ+EGIDY E F PV + +IR L+S V ++ L Q+DVK+AFL+G +
Sbjct: 887 YKARLVAKGFSQREGIDYQEIFSPVVKHVSIRYLLSIVVQFDMELEQLDVKTAFLHGNLD 946
Query: 1239 EEVYVHQPPGFEDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLF 1298
E + + QP G+EDE + + V LKKSLYGLKQ+PR W +R SF++ + + R K + ++
Sbjct: 947 EYILMSQPEGYEDEDSTEKVCLLKKSLYGLKQSPRQWNQRFDSFMINSGYQRSKYNPCVY 1006
Query: 1299 CKTYKDDILI-VQIYVDKIIFGSANQSLCKEFSKMMQAEFEMSMMGELKYFLGIQVDQTP 1357
+ D I + +YVD ++ S N+ ++ + + EFEM +G + LG+++ +
Sbjct: 1007 TQQLNDGSYIYLLLYVDDMLIASQNKDQIQKLKESLNREFEMKDLGPARKILGMEITRNR 1066
Query: 1358 EGTYIH--QSKYTKELLKKFNMTESTIAKTPMHPTCIL----EKEDASGKVCQKL--YRC 1409
E + QS+Y +L+ F M +S +++TP+ L EK A KL Y
Sbjct: 1067 EQGILDLSQSEYVAGVLRAFGMDQSKVSQTPLGAHFKLRAANEKTLARDAEYMKLVPYPN 1126
Query: 1410 MIGSLLY-LTASRPDNLFSVHLCARFQSDPRETHLTAVKRILRYLKGTTNLGLMYKKTSE 1468
IGS++Y + SRPD + V + +RF S P + H AVK ++RY+KGT + L +KK +
Sbjct: 1127 AIGSIMYSMIGSRPDLAYPVGVVSRFMSKPSKEHWQAVKWVMRYMKGTQDTCLRFKKDDK 1186
Query: 1469 YKLSGYCDVDYAGDRT*RKSTFGNCQFLGSNLVSWASKRQSTIELSTAKAEYISAAICST 1528
+++ GYCD DYA D R+S G G N +SW S Q + LST +AEY++ A
Sbjct: 1187 FEIRGYCDSDYATDLDRRRSITGFVFTAGGNTISWKSGLQRVVALSTTEAEYMALAEAVK 1246
Query: 1529 QMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVL 1588
+ +W++ + + + + CD+ +AI+LSKN + H R KHI+V+YHFIR+ + G +
Sbjct: 1247 EAIWLRGLAAEMGFEQDAVEVMCDSQSAIALSKNSVHHERTKHIDVRYHFIREKIADGEI 1306
Query: 1589 LLKFVDTDHKWADIY 1603
+ + T ADI+
Sbjct: 1307 QVVKISTTWNPADIF 1321
Score = 42.0 bits (97), Expect = 0.003
Identities = 61/309 (19%), Positives = 120/309 (38%), Gaps = 57/309 (18%)
Query: 11 AKPPMFDGQR-FEYWKDRLESFF--LGFDADLWD---IIVDGYERPVDAEGKKIPRSEMT 64
A+ FDG + WK++L + LG L + +V+ + G + P + +
Sbjct: 6 AEVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATS 65
Query: 65 A-EQKKLYAQHHKARAILLSAISYEEYQKITDLEFANGIFESLKMSHEGNKKVKESKTLS 123
E K L + KAR+ ++ ++ +K+ + A G+ + L +
Sbjct: 66 KLEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNR---IY 122
Query: 124 LIQKYESFIMEPNESIEEMFSRFQLLVAGIRPLNKSYTTKDHVIRVIRSLPESWMPLVTS 183
L Q+ + M N ++EE + F L++ + + +D I ++ SLP + L +
Sbjct: 123 LKQRLYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKET 182
Query: 184 IELTRDVERMSLEELISILKCHELKRSEMQDLRKKSIALKSKYEKAKSEKSKALQAEAEE 243
++ + + LEE+ S + RS++ +L LK
Sbjct: 183 LKYCKTT--LHLEEITSAI------RSKILELGASGKLLK-------------------- 214
Query: 244 SEEASEDSDEDELTLISKRLNHIWKHKQSKYRGSGKAKGKSESSGQKKSSIKEVTCFECK 303
++ D L + Q + R + KG +++ + KS TC+ C
Sbjct: 215 -------NNSDGLFV------------QDRGRSETRGKGPNKNKSRSKSKGAGKTCWICG 255
Query: 304 ESGHYKSDC 312
+ GH+K C
Sbjct: 256 KEGHFKKQC 264
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,464,156
Number of Sequences: 26719
Number of extensions: 1622381
Number of successful extensions: 8740
Number of sequences better than 10.0: 438
Number of HSP's better than 10.0 without gapping: 159
Number of HSP's successfully gapped in prelim test: 286
Number of HSP's that attempted gapping in prelim test: 7135
Number of HSP's gapped (non-prelim): 1292
length of query: 1622
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1509
effective length of database: 8,299,349
effective search space: 12523717641
effective search space used: 12523717641
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0346.14


