

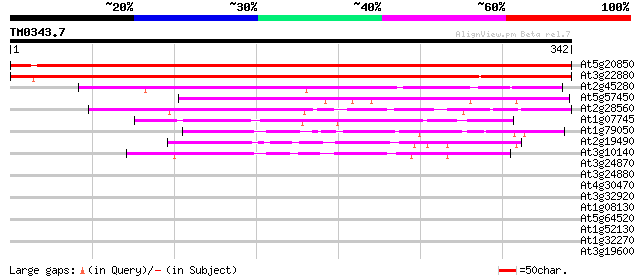
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0343.7
(342 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g20850 RAD51 homolog (AtRAD51) 600 e-172
At3g22880 meiotic recombination protein (AtDMC1) 338 2e-93
At2g45280 RAD51Calpha DNA repair-recombination factor (RAD51Calpha) 125 3e-29
At5g57450 putative protein 96 4e-20
At2g28560 putative RAD51B-like DNA repair protein 74 2e-13
At1g07745 unknown protein 72 4e-13
At1g79050 nuclear-encoded chloroplast DNA repair protein like 54 2e-07
At2g19490 putative recA protein 51 8e-07
At3g10140 RecA protein 49 3e-06
At3g24870 hypothetical protein 34 0.13
At3g24880 hypothetical protein 32 0.50
At4g30470 cinnamoyl-CoA reductase - like protein 31 0.86
At3g32920 recA, putative 31 1.1
At1g08130 DNA ligase like protein 30 1.5
At5g64520 putative protein 30 1.9
At1g52130 28 5.5
At1g32270 syntaxin, putative 28 5.5
At3g19600 hypothetical protein 28 9.5
>At5g20850 RAD51 homolog (AtRAD51)
Length = 342
Score = 600 bits (1546), Expect = e-172
Identities = 301/342 (88%), Positives = 324/342 (94%), Gaps = 3/342 (0%)
Query: 1 MEQQRHQKTAQQQPQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYTPR 60
MEQ+R+Q QQQ + EE QHGP P+EQLQA+GIAS+DVKKL+DAG+CTVE VAYTPR
Sbjct: 4 MEQRRNQNAVQQQ---DDEETQHGPFPVEQLQAAGIASVDVKKLRDAGLCTVEGVAYTPR 60
Query: 61 KDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGI 120
KDLLQIKGIS+AKVDKIVEAASKLVP+GFTSAS+LH QR+ IIQIT+GSRELDK+L+GGI
Sbjct: 61 KDLLQIKGISDAKVDKIVEAASKLVPLGFTSASQLHAQRQEIIQITSGSRELDKVLEGGI 120
Query: 121 ETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADR 180
ETGSITELYGEFRSGKTQLCHTLCVTCQLP+DQGGGEGKAMYIDAEGTFRPQRLLQIADR
Sbjct: 121 ETGSITELYGEFRSGKTQLCHTLCVTCQLPMDQGGGEGKAMYIDAEGTFRPQRLLQIADR 180
Query: 181 FGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGE 240
FGLNGADVLENVAYARAYNTDHQSRLLLEAASMM+ETRFA++IVDSATALYRTDFSGRGE
Sbjct: 181 FGLNGADVLENVAYARAYNTDHQSRLLLEAASMMIETRFALLIVDSATALYRTDFSGRGE 240
Query: 241 LSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATT 300
LSARQMHLAKFLRSLQKLADEFGVA+VITNQVV+QVDGSA+FAGPQ KPIGGNIMAHATT
Sbjct: 241 LSARQMHLAKFLRSLQKLADEFGVAVVITNQVVAQVDGSALFAGPQFKPIGGNIMAHATT 300
Query: 301 TRLALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
TRLALRKGR EERICKVISSPCL EAEARFQI EGV+D KD
Sbjct: 301 TRLALRKGRAEERICKVISSPCLPEAEARFQISTEGVTDCKD 342
>At3g22880 meiotic recombination protein (AtDMC1)
Length = 344
Score = 338 bits (868), Expect = 2e-93
Identities = 177/344 (51%), Positives = 240/344 (69%), Gaps = 3/344 (0%)
Query: 1 MEQQRHQKTAQQQ--PQEEGEEIQHGPLPIEQLQASGIASIDVKKLKDAGICTVESVAYT 58
M + ++T+Q Q +EE +E + I++L A GI + DVKKL++AGI T +
Sbjct: 2 MASLKAEETSQMQLVEREENDEDEDLFEMIDKLIAQGINAGDVKKLQEAGIHTCNGLMMH 61
Query: 59 PRKDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDG 118
+K+L IKG+SEAKVDKI EAA K+V G+ + S+ ++R+S+++ITTG + LD +L G
Sbjct: 62 TKKNLTGIKGLSEAKVDKICEAAEKIVNFGYMTGSDALIKRKSVVKITTGCQALDDLLGG 121
Query: 119 GIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIA 178
GIET +ITE +GEFRSGKTQL HTLCVT QLP + GG GK YID EGTFRP R++ IA
Sbjct: 122 GIETSAITEAFGEFRSGKTQLAHTLCVTTQLPTNMKGGNGKVAYIDTEGTFRPDRIVPIA 181
Query: 179 DRFGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGR 238
+RFG++ VL+N+ YARAY +HQ LLL A+ M E F ++IVDS AL+R DF+GR
Sbjct: 182 ERFGMDPGAVLDNIIYARAYTYEHQYNLLLGLAAKMSEEPFRILIVDSIIALFRVDFTGR 241
Query: 239 GELSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHA 298
GEL+ RQ LA+ L L K+A+EF VA+ +TNQV++ G + P+ KP GG+++AHA
Sbjct: 242 GELADRQQKLAQMLSRLIKIAEEFNVAVYMTNQVIADPGGGMFISDPK-KPAGGHVLAHA 300
Query: 299 TTTRLALRKGRGEERICKVISSPCLAEAEARFQILAEGVSDVKD 342
T RL RKG+G+ R+CKV +P LAEAEA FQI G++D KD
Sbjct: 301 ATIRLLFRKGKGDTRVCKVYDAPNLAEAEASFQITQGGIADAKD 344
>At2g45280 RAD51Calpha DNA repair-recombination factor (RAD51Calpha)
Length = 363
Score = 125 bits (314), Expect = 3e-29
Identities = 98/323 (30%), Positives = 157/323 (48%), Gaps = 36/323 (11%)
Query: 43 KLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAA-------SKLVPMGFTSASEL 95
KL AG + S+A DL + I+E + +I++ A S+ + G +A ++
Sbjct: 36 KLISAGYTCLSSIASVSSSDLARDANITEEEAFEILKLANQSCCNGSRSLINGAKNAWDM 95
Query: 96 HVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGG 155
+ ES+ +ITT +LD IL GGI +TE+ G GKTQ+ L V Q+P + GG
Sbjct: 96 LHEEESLPRITTSCSDLDNILGGGISCRDVTEIGGVPGIGKTQIGIQLSVNVQIPRECGG 155
Query: 156 GEGKAMYIDAEGTFRPQRLLQIAD--------------------RFGLNGADVLENVAYA 195
GKA+YID EG+F +R LQIA+ + + D+LEN+ Y
Sbjct: 156 LGGKAIYIDTEGSFMVERALQIAEACVEDMEEYTGYMHKHFQANQVQMKPEDILENIFYF 215
Query: 196 RAYNTDHQSRLLLEAASMMVETR-FAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRS 254
R + Q L+ + E + V+IVDS T +R D+ +L+ R L++
Sbjct: 216 RVCSYTEQIALVNHLEKFISENKDVKVVIVDSITFHFRQDYD---DLAQRTRVLSEMALK 272
Query: 255 LQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERI 314
KLA +F +A+V+ NQV ++ + Q+ G+ +H+ T R+ L G+ER
Sbjct: 273 FMKLAKKFSLAVVLLNQVTTKFSEGSF----QLALALGDSWSHSCTNRVIL-YWNGDERY 327
Query: 315 CKVISSPCLAEAEARFQILAEGV 337
+ SP L A A + + + G+
Sbjct: 328 AYIDKSPSLPSASASYTVTSRGL 350
>At5g57450 putative protein
Length = 304
Score = 95.5 bits (236), Expect = 4e-20
Identities = 79/284 (27%), Positives = 126/284 (43%), Gaps = 46/284 (16%)
Query: 104 QITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYI 163
++TTG LD L GGI S+TE+ E GKTQLC L + QLP+ GG G ++Y+
Sbjct: 20 KLTTGCEILDGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLCTQLPISHGGLNGSSLYL 79
Query: 164 DAEGTFRPQRLLQIADRFGLNGADVLEN--------VAYARAYNTDHQSRLL--LEAASM 213
+E F +RL Q++ F + + N V ++ DH ++ ++
Sbjct: 80 HSEFPFPFRRLHQLSHTFHQSNPSIYANYNDNPCDHVFVQNVHSVDHLFDIMPRIDGFVG 139
Query: 214 MVETRF--AVMIVDSATALYRTDFSGR-GELSARQMHLAKFLRSLQKLADEFGVAIVITN 270
+TRF ++++DS AL+R++F +L R K L++LA +F +AIVITN
Sbjct: 140 NSKTRFPLKLIVLDSVAALFRSEFDNTPSDLKKRSSLFFKISGKLKQLASKFDLAIVITN 199
Query: 271 QVVSQVDGS-------------AVFAGPQVKPIGGNIMAHATTTRLALRK---------- 307
QV V+ S +G +V P G A+ +R + +
Sbjct: 200 QVTDLVETSDGLSGLRIGNLRYLYSSGRRVVPSLGLAWANCVNSRFFISRSDGSIVKDRS 259
Query: 308 ----------GRGEERICKVISSPCLAEAEARFQILAEGVSDVK 341
R +R ++ SP L + F I EG+ V+
Sbjct: 260 EKDENCSSSVSRSAKRRLDIVFSPYLPGSSCEFMITREGICAVQ 303
>At2g28560 putative RAD51B-like DNA repair protein
Length = 353
Score = 73.6 bits (179), Expect = 2e-13
Identities = 81/322 (25%), Positives = 134/322 (41%), Gaps = 50/322 (15%)
Query: 49 ICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELH----VQRESII- 103
I T + +L+++ + ++ + S+ S S L V+ E +
Sbjct: 24 IITAKDALSMTEFELMELLDVGMKEIRSAISFISEATSPPCQSVSSLFFFKKVENEHLSG 83
Query: 104 QITTGSRELDKILDGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYI 163
+ T + LD L GGI G +TEL G GK+Q C L ++ P+ GG +G+ +YI
Sbjct: 84 HLPTHLKGLDDTLCGGIPFGVLTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYI 143
Query: 164 DAEGTFRPQRLLQIA-----DRFGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETR 218
D E F +R++++ + F L G + + V A + E + +++ +
Sbjct: 144 DVESKFSSRRVIEMGLESFPEVFHLKG--MAQEVVCGTAI-------CIQELKNSILQNQ 194
Query: 219 FAVMIVDSATALYRTDFSGRGELSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQ--- 275
++++DS TAL SG + A++ + S K ++TNQV SQ
Sbjct: 195 VKLLVIDSMTAL----LSGENKPGAQRQPQLGWHISFLKYE-------LVTNQVRSQNRD 243
Query: 276 ---------------VDGSAVFAGPQVKPIGGNIMAHATTTRLALRKGRGEERICKVISS 320
D + + V +G N AHA T RL L G +RI KV S
Sbjct: 244 ETSQYSFQAKVKDEFKDNTKTYDSHLVAALGIN-WAHAVTIRLVLEAKSG-KRIIKVAKS 301
Query: 321 PCLAEAEARFQILAEGVSDVKD 342
P F I + G+S + D
Sbjct: 302 PMSPPLAFPFHITSAGISLLSD 323
>At1g07745 unknown protein
Length = 296
Score = 72.0 bits (175), Expect = 4e-13
Identities = 61/239 (25%), Positives = 100/239 (41%), Gaps = 23/239 (9%)
Query: 77 IVEAASKLVPMGFTSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGK 136
++E + + G +LH + ++ +TG +E D +L GG G +TEL G SGK
Sbjct: 38 LIERQCRPLVNGLKLLEDLHRNKHTL---STGDKETDSLLQGGFREGQLTELVGPSSSGK 94
Query: 137 TQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQI--ADRFGLNGADVLENVAY 194
TQ C + G+ +Y+D +F +R+ Q + G V+ +
Sbjct: 95 TQFCMQAAASV-----AENHLGRVLYLDTGNSFSARRIAQFICSSSDATLGQKVMSRILC 149
Query: 195 ARAY------NTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSARQMHL 248
Y +T + L + E+R +++VDS ++L G G M
Sbjct: 150 HTVYDIYTLFDTLQDLEITLRLQMNVNESRLRLLVVDSISSLITPILGGSGSQGRALMVA 209
Query: 249 AKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRK 307
+L L+KLA E +AI++TN V A G + KP G RL+L +
Sbjct: 210 IGYL--LKKLAHEHSIAILVTNHTV-----GAGGEGGKTKPALGETWKSIPHVRLSLSR 261
>At1g79050 nuclear-encoded chloroplast DNA repair protein like
Length = 439
Score = 53.5 bits (127), Expect = 2e-07
Identities = 71/254 (27%), Positives = 109/254 (41%), Gaps = 45/254 (17%)
Query: 106 TTGSRELDKILDGGIETGSITELYGEFRSGKTQLC-HTLCVTCQLPLDQGGGEGKAMYID 164
++G LD L GG+ G + E+YG SGKT L H + +L G AM +D
Sbjct: 118 SSGILTLDLALGGGLPKGRVVEIYGPESSGKTTLALHAIAEVQKL-------GGNAMLVD 170
Query: 165 AEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMI- 223
AE F P + G DV EN+ + N + LE A M + +I
Sbjct: 171 AEHAFDPAYSKAL-------GVDV-ENLIVCQPDN----GEMALETADRMCRSGAVDLIC 218
Query: 224 VDSATALY-RTDFSGRGELSARQMHL-----AKFLRSLQKLADEFGVAIVITNQVVSQVD 277
VDS +AL R + GE+ +QM L ++ LR + A + G ++ NQ+ ++
Sbjct: 219 VDSVSALTPRAEI--EGEIGMQQMGLQARLMSQALRKMSGNASKAGCTLIFLNQIRYKI- 275
Query: 278 GSAVFAGPQVKPIGGNIMAHATTTRLALR------KGRGEE------RICKVISSPCLAE 325
+ P+V GG + + RL +R +G+E R+ S
Sbjct: 276 -GVYYGNPEVTS-GGIALKFFASVRLEIRSAGKIKSSKGDEDIGLRARVRVQKSKVSRPY 333
Query: 326 AEARFQIL-AEGVS 338
+A F+I+ EGVS
Sbjct: 334 KQAEFEIMFGEGVS 347
>At2g19490 putative recA protein
Length = 376
Score = 51.2 bits (121), Expect = 8e-07
Identities = 61/230 (26%), Positives = 99/230 (42%), Gaps = 36/230 (15%)
Query: 97 VQRESIIQITTGSRELDKILD-GGIETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQGG 155
V ++ +TGS LD L GG+ G + E+YG SGKT L + Q QGG
Sbjct: 31 VSPRNVPVFSTGSFALDVALGVGGLPKGRVVEIYGPEASGKTTLALHVIAEAQ---KQGG 87
Query: 156 GEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLLLEAASMMV 215
+++DAE +A G+N ++L + D + L +++
Sbjct: 88 ---TCVFVDAEHALDSS----LAKAIGVNTENLLLS-------QPDCGEQALSLVDTLIR 133
Query: 216 ETRFAVMIVDSATALYRTDFSGRGELSARQ--MHLAKFLR----SLQKLADEFGVA---I 266
V++VDS AL +GEL H+A R +L+KL+ ++ +
Sbjct: 134 SGSVDVIVVDSVAALVP-----KGELEGEMGDAHMAMQARLMSQALRKLSHSLSLSQTLL 188
Query: 267 VITNQVVSQVDGSAVFAGPQVKPIGGNIMAHATTTRLALRK----GRGEE 312
+ NQV S++ F GP GGN + + RL +++ +GEE
Sbjct: 189 IFINQVRSKLSTFGGFGGPTEVTCGGNALKFYASMRLNIKRIGLIKKGEE 238
>At3g10140 RecA protein
Length = 389
Score = 49.3 bits (116), Expect = 3e-06
Identities = 65/250 (26%), Positives = 104/250 (41%), Gaps = 40/250 (16%)
Query: 72 AKVDKIVEAASKLVPMGFTSASELHVQR----ESIIQITTGSRELDKILD-GGIETGSIT 126
A+ D + A + F S+L +QR + I+TGS LD L GG+ G +
Sbjct: 60 AEKDTALHLALSQLSGDFDKDSKLSLQRFYRKRRVSVISTGSLNLDLALGVGGLPKGRMV 119
Query: 127 ELYGEFRSGKTQLC-HTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNG 185
E+YG+ SGKT L H + +L G Y+DAE P +A+ G+N
Sbjct: 120 EVYGKEASGKTTLALHIIKEAQKL-------GGYCAYLDAENAMDP----SLAESIGVNT 168
Query: 186 ADVLENVAYARAYNTDHQSRLLLEAASMMVET-RFAVMIVDSATALYRTDFSGRGELSA- 243
++L + + +L ++ ++ V++VDS AL + + EL A
Sbjct: 169 EELL--------ISRPSSAEKMLNIVDVLTKSGSVDVIVVDSVAAL-----APQCELDAP 215
Query: 244 -----RQMHLAKFLRSLQKLADEFGVA---IVITNQVVSQVDGSAVFAGPQVKPIGGNIM 295
R ++L+K+ G + IV NQV S V + F + GGN +
Sbjct: 216 VGERYRDTQSRIMTQALRKIHYSVGYSQTLIVFLNQVRSHVKSNMHFPHAEEVTCGGNAL 275
Query: 296 AHATTTRLAL 305
RL +
Sbjct: 276 PFHAAIRLKM 285
>At3g24870 hypothetical protein
Length = 1841
Score = 33.9 bits (76), Expect = 0.13
Identities = 41/188 (21%), Positives = 75/188 (39%), Gaps = 51/188 (27%)
Query: 1 MEQQRHQKTAQQQPQEEGEE----IQHGPLPIEQ--------------LQASGIASIDVK 42
+E Q H+ TA+ Q +E+ E +Q G + +E SGI V
Sbjct: 442 IENQSHRSTAEMQTKEKSSETEKRLQDGLVVLENDSKVGSILSENPSSTLCSGIPQASV- 500
Query: 43 KLKDAGICTV----------ESVAYTPRKDLLQIKGISEAKV---DKIVEAASKLVPMGF 89
D CTV E++ + P D + + + E + +I++A K +
Sbjct: 501 ---DTSSCTVGNSLLSGTDIEALKHQPSSDAVMLDTVKEDAILEEARIIQAKKKRIAELS 557
Query: 90 TSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGK----TQLCHTLCV 145
+ + V+ +S + D +L+ E + + + R K TQ+CH + +
Sbjct: 558 CGTAPVEVREKS---------QWDFVLE---EMAWLANDFAQERLWKMTAATQICHRVAL 605
Query: 146 TCQLPLDQ 153
TCQL ++
Sbjct: 606 TCQLRFEE 613
>At3g24880 hypothetical protein
Length = 1843
Score = 32.0 bits (71), Expect = 0.50
Identities = 40/188 (21%), Positives = 74/188 (39%), Gaps = 51/188 (27%)
Query: 1 MEQQRHQKTAQQQPQEEGEE----IQHGPLPIEQ--------------LQASGIASIDVK 42
+E Q H+ TA+ Q +E+ E +Q G + +E SGI V
Sbjct: 442 IENQSHRSTAEMQTKEKSSETEKRLQDGLVVLENDSKVGSILSENPSSTLCSGIPQASV- 500
Query: 43 KLKDAGICTV----------ESVAYTPRKDLLQIKGISEAKV---DKIVEAASKLVPMGF 89
D CTV E++ + P D + + + E + +I++A K +
Sbjct: 501 ---DTSSCTVGNSLLSGTDIEALKHQPSSDAVMLDTVKEDAILEEARIIQAKKKRIAELS 557
Query: 90 TSASELHVQRESIIQITTGSRELDKILDGGIETGSITELYGEFRSGK----TQLCHTLCV 145
+ + V+ +S + D +L+ E + + + R K Q+CH + +
Sbjct: 558 CGTAPVEVREKS---------QWDFVLE---EMAWLANDFAQERLWKMTAAAQICHRVAL 605
Query: 146 TCQLPLDQ 153
TCQL ++
Sbjct: 606 TCQLRFEE 613
>At4g30470 cinnamoyl-CoA reductase - like protein
Length = 303
Score = 31.2 bits (69), Expect = 0.86
Identities = 25/91 (27%), Positives = 44/91 (47%), Gaps = 3/91 (3%)
Query: 30 QLQASGI-ASIDVKKLKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPMG 88
Q+ +G+ A +DVK L D I E V+ R +E + K+VE+ S L+PM
Sbjct: 212 QMYENGVLAYVDVKFLADVHIRAFEDVSACGRYFCFNQIVNTEEEALKLVESLSPLIPMP 271
Query: 89 FTSASELHVQRESIIQITTGSRELDKILDGG 119
+E+H + + + +L K+++ G
Sbjct: 272 PRYENEMH--GSEVYEERLRNNKLSKLVEAG 300
>At3g32920 recA, putative
Length = 229
Score = 30.8 bits (68), Expect = 1.1
Identities = 18/44 (40%), Positives = 24/44 (53%), Gaps = 1/44 (2%)
Query: 97 VQRESIIQITTGSRELDKILD-GGIETGSITELYGEFRSGKTQL 139
V ++ +TGS LD L GG+ G + E+YG SGKT L
Sbjct: 7 VSPRNVPVFSTGSFALDVALGVGGLPKGRLVEIYGPEASGKTAL 50
>At1g08130 DNA ligase like protein
Length = 790
Score = 30.4 bits (67), Expect = 1.5
Identities = 20/57 (35%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query: 173 RLLQIADRFGLNGADVLENVAYARAYNTDH-----QSRLLLEAASMMVETRFAVMIV 224
RLLQ R G +G VL + A YN +H ++ LE A+ +V+ F V+ V
Sbjct: 332 RLLQAKLRLGFSGQTVLAALGQAAVYNEEHSKPPPNTKSPLEEAAKIVKQVFTVLPV 388
>At5g64520 putative protein
Length = 403
Score = 30.0 bits (66), Expect = 1.9
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 5/63 (7%)
Query: 120 IETGSITELYGEFRSGKTQLCHTLCVTCQLPLDQG----GGEGK-AMYIDAEGTFRPQRL 174
+ G++ E+ G S KTQ+ ++C LP GG GK +++D + F RL
Sbjct: 40 LRAGNVVEITGASTSAKTQILIQAAISCILPKTWNGIHYGGLGKLVLFLDLDCRFDVLRL 99
Query: 175 LQI 177
Q+
Sbjct: 100 SQM 102
>At1g52130
Length = 483
Score = 28.5 bits (62), Expect = 5.5
Identities = 21/78 (26%), Positives = 39/78 (49%), Gaps = 7/78 (8%)
Query: 232 RTDFSGRGELSARQMHLAKFLRSLQKLADEFGVAIVITNQVVSQVDGSAVFAGPQVKPIG 291
R D+ G++ +R+ H K + ++Q E G + N+V++ V+G A +
Sbjct: 345 RFDYDDDGKVESRE-HGPKIVAAVQ----EGGFVLDYPNEVITSVEGIATVVNTGLSFST 399
Query: 292 GNIMAHATTTRLALRKGR 309
GN+M + T + + KGR
Sbjct: 400 GNVMIKSLTFKTS--KGR 415
>At1g32270 syntaxin, putative
Length = 416
Score = 28.5 bits (62), Expect = 5.5
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query: 44 LKDAGICTVESVAYTPRKDLLQIKGISEAKVDKIVEAASKLVPMGFTSASELHVQRES 101
+KD E+ R+D+ Q K I++AK+ K EAA K F A + V+RE+
Sbjct: 164 VKDTSANLREASETDHRRDVAQSKKIADAKLAKDFEAALK----EFQKAQHITVERET 217
>At3g19600 hypothetical protein
Length = 601
Score = 27.7 bits (60), Expect = 9.5
Identities = 40/194 (20%), Positives = 69/194 (34%), Gaps = 12/194 (6%)
Query: 124 SITELYGEFRSGKTQLCHTLCVTCQLPLDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGL 183
S + L G +CH +C+ C+ + + +G+A +G + + F
Sbjct: 319 SSSSLSSSSSCGHWYICHGICIGCKSTVKK--SQGRAFDYIFDGLQLSHEAVALTKCFTT 376
Query: 184 NGADVLENVAYARAYNTDHQSRLLLEAASMMVETRFAVMIVDSATALYRTDFSGRGELSA 243
+ + E + + DH + S+ ++ + SAT G+
Sbjct: 377 KLSCLNEKKLHL-VLDLDHTLLHTVMVPSLSQAEKYLIEEAGSATRDDLWKIKAVGDPME 435
Query: 244 RQMHLAKFLRSLQKLADEFGVAIVIT-------NQVVSQVDGSAVFAGPQVKPIGGNIMA 296
L FLR K A+EF V T QV+ +D ++ G +V I
Sbjct: 436 FLTKLRPFLRDFLKEANEFFTMYVYTKGSRVYAKQVLELIDPKKLYFGDRV--ITKTESP 493
Query: 297 HATTTRLALRKGRG 310
H T L + RG
Sbjct: 494 HMKTLDFVLAEERG 507
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,724,642
Number of Sequences: 26719
Number of extensions: 268278
Number of successful extensions: 787
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 764
Number of HSP's gapped (non-prelim): 18
length of query: 342
length of database: 11,318,596
effective HSP length: 100
effective length of query: 242
effective length of database: 8,646,696
effective search space: 2092500432
effective search space used: 2092500432
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0343.7


