

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0343.3
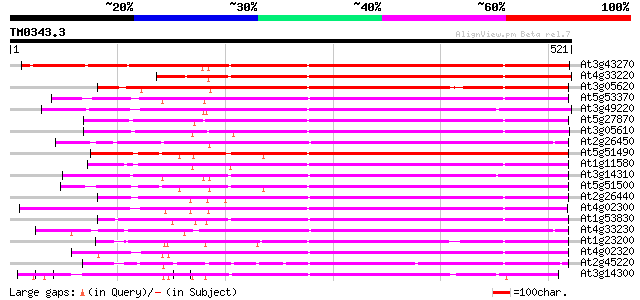
(521 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g43270 pectinesterase like protein 536 e-152
At4g33220 pectinesterase like protein 446 e-125
At3g05620 putative pectinesterase 417 e-117
At5g53370 pectinesterase 402 e-112
At3g49220 pectinesterase - like protein 398 e-111
At5g27870 pectin methyl-esterase - like protein 371 e-103
At3g05610 putative pectinesterase 367 e-102
At2g26450 putative pectinesterase 365 e-101
At5g51490 pectinesterase 365 e-101
At1g11580 unknown protein 360 1e-99
At3g14310 pectin methylesterase like protein 359 2e-99
At5g51500 pectinesterase 356 2e-98
At2g26440 putative pectinesterase 356 2e-98
At4g02300 putative pectinesterase 355 3e-98
At1g53830 unknown protein 354 6e-98
At4g33230 pectinesterase - like protein 354 8e-98
At1g23200 putative pectinesterase 354 8e-98
At4g02320 pectinesterase - like protein 349 2e-96
At2g45220 pectinesterase like protein 348 4e-96
At3g14300 putative pectin methylesterase 347 7e-96
>At3g43270 pectinesterase like protein
Length = 527
Score = 536 bits (1380), Expect = e-152
Identities = 283/521 (54%), Positives = 368/521 (70%), Gaps = 20/521 (3%)
Query: 12 ILLLIMSLQPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKV 71
+ L+I+SL C+ A + T+ + CLRV P F ++A V++ + +S F K
Sbjct: 14 LFLIIISL--CSAHKEAFSSTDLVQMECLRVPPLEFAEAAKTVVDAITKAVAIVSKFDK- 70
Query: 72 NQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLS 131
+ +R+SNAI DC++L D A E L W ISA+Q+ G K+N TG++ SDLRTWISA LS
Sbjct: 71 -KAGKSRVSNAIVDCVDLLDSAAEELSWIISASQSPNG-KDNSTGDVGSDLRTWISAALS 128
Query: 132 YQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPS---------NISNS 182
Q+TC+DGFEGTN +K V+GGL +V + V+N L + P S P ++S
Sbjct: 129 NQDTCLDGFEGTNGIIKKIVAGGLSKVGTTVRNLLTMVHSPPSKPKPKPIKAQTMTKAHS 188
Query: 183 G--EFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHV 240
G +FPSWVK D KLLQ D AD VVAADG+GN+TT+ DAV AP+Y+ RYVIHV
Sbjct: 189 GFSKFPSWVKPGDRKLLQTDNITV-ADAVVAADGTGNFTTISDAVLAAPDYSTKRYVIHV 247
Query: 241 KKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARD 300
K+G+Y E VEIKKKKWNIM+VGDG+D T I+GNRS+ D TTFR+ATFAVS GFIARD
Sbjct: 248 KRGVYVENVEIKKKKWNIMMVGDGIDATVITGNRSF-IDGWTTFRSATFAVSGRGFIARD 306
Query: 301 ITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFIC 360
ITF+NTAGPE QAVA+RSD+DL VFYRC ++GYQD+LYAHS RQF++EC I GTVDFI
Sbjct: 307 ITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLYAHSMRQFFRECIITGTVDFIF 366
Query: 361 GNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQ 420
G+A AVFQ+C I+ K+ L +QKN+I AQ R PN+ +GF+ I D D + +
Sbjct: 367 GDATAVFQSCQIKAKQGLPNQKNSITAQGRKDPNEPTGFTIQFSNIAADTDLLLNLNTTA 426
Query: 421 TFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRV 480
T+LGRPWK +SRTVFM+++MS+ I+P GWLEW N DTL++GEY N GPG+ + RV
Sbjct: 427 TYLGRPWKLYSRTVFMQNYMSDAINPVGWLEWNGNFAL-DTLYYGEYMNSGPGASLDRRV 485
Query: 481 KWRGYHVLNNS-QAMDFTVNQFIKGDLWLPSTGVSYNAGLL 520
KW GYHVLN S +A +FTV+Q I+G+LWLPSTG+++ AGL+
Sbjct: 486 KWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTGITFIAGLV 526
>At4g33220 pectinesterase like protein
Length = 404
Score = 446 bits (1146), Expect = e-125
Identities = 227/409 (55%), Positives = 292/409 (70%), Gaps = 29/409 (7%)
Query: 137 IDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNISNSG------------- 183
++GF+GT+ VK V+G LDQ+ S+++ ELL V P+ P +S G
Sbjct: 1 MEGFDGTSGLVKSLVAGSLDQLYSMLR-ELLPLVQPEQKPKAVSKPGPIAKGPKAPPGRK 59
Query: 184 ----------EFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNM 233
+FP WV+ +D KLL+ + DV VA DG+GN+T +MDA+K AP+Y+
Sbjct: 60 LRDTDEDESLQFPDWVRPDDRKLLESNGRT--YDVSVALDGTGNFTKIMDAIKKAPDYSS 117
Query: 234 TRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSA 293
TR+VI++KKG+Y E VEIKKKKWNI+++GDG+DVT ISGNRS+ D TTFR+ATFAVS
Sbjct: 118 TRFVIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNRSF-IDGWTTFRSATFAVSG 176
Query: 294 IGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIR 353
GF+ARDITF+NTAGPE QAVALRSDSDLSVF+RC ++GYQD+LY H+ RQFY+EC I
Sbjct: 177 RGFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECTIT 236
Query: 354 GTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQ 413
GTVDFI G+ VFQNC I K+ L +QKNTI AQ R NQ SGFS I D D
Sbjct: 237 GTVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADADLV 296
Query: 414 PSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPG 473
P + +T+LGRPWK +SRTVF+ + MS+++ PEGWLEW + DTLF+GE+ N GPG
Sbjct: 297 PYLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNADFAL-DTLFYGEFMNYGPG 355
Query: 474 SGVANRVKWRGYHVLNNS-QAMDFTVNQFIKGDLWLPSTGVSYNAGLLV 521
SG+++RVKW GYHV NNS QA +FTV+QFIKG+LWLPSTGV+++ GL +
Sbjct: 356 SGLSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGLYI 404
>At3g05620 putative pectinesterase
Length = 543
Score = 417 bits (1071), Expect = e-117
Identities = 219/454 (48%), Positives = 290/454 (63%), Gaps = 32/454 (7%)
Query: 82 AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSD------------LRTWISAV 129
AI DC EL + L WS+ + NK +G G + D L+TW+SA
Sbjct: 104 AIEDCKELVGFSVTELAWSML-----EMNKLHGGGGIDLDDGSHDAAAAGGNLKTWLSAA 158
Query: 130 LSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNISNSGEF---P 186
+S Q+TC++GFEGT ++ + G L QV LV N L + ++ P S + P
Sbjct: 159 MSNQDTCLEGFEGTERKYEELIKGSLRQVTQLVSNVLDMYTQLNALPFKASRNESVIASP 218
Query: 187 SWVKAEDEKLLQQDAPPPP-ADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIY 245
W+ DE L+ + P + VVA DG G Y T+ +A+ +AP ++ RYVI+VKKG+Y
Sbjct: 219 EWLTETDESLMMRHDPSVMHPNTVVAIDGKGKYRTINEAINEAPNHSTKRYVIYVKKGVY 278
Query: 246 KEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFEN 305
KE +++KKKK NIM+VGDG+ T I+G+R++ +TTFRTAT AVS GFIA+DITF N
Sbjct: 279 KENIDLKKKKTNIMLVGDGIGQTIITGDRNF-MQGLTTFRTATVAVSGRGFIAKDITFRN 337
Query: 306 TAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAA 365
TAGP+N+QAVALR DSD S FYRC ++GYQD+LYAHS RQFY++C I GT+DFI GN AA
Sbjct: 338 TAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAHSLRQFYRDCEIYGTIDFIFGNGAA 397
Query: 366 VFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGR 425
V QNC I + PL QK TI AQ R PNQ +GF N ++ QP T+LGR
Sbjct: 398 VLQNCKIYTRVPLPLQKVTITAQGRKSPNQNTGFVIQNSYVLAT---QP------TYLGR 448
Query: 426 PWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGY 485
PWK +SRTV+M ++MS+++ P GWLEW N DTL++GEY N GPG + RVKW GY
Sbjct: 449 PWKLYSRTVYMNTYMSQLVQPRGWLEWFGNFAL-DTLWYGEYNNIGPGWRSSGRVKWPGY 507
Query: 486 HVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
H+++ A+ FTV FI G WLP+TGV++ AGL
Sbjct: 508 HIMDKRTALSFTVGSFIDGRRWLPATGVTFTAGL 541
>At5g53370 pectinesterase
Length = 587
Score = 402 bits (1032), Expect = e-112
Identities = 215/496 (43%), Positives = 298/496 (59%), Gaps = 33/496 (6%)
Query: 40 LRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKW 99
+ +S N + +K + ++Y P R+ +A CLEL D + + L
Sbjct: 107 IHISFNATLQKFSKALYTSSTITYTQMP---------PRVRSAYDSCLELLDDSVDALTR 157
Query: 100 SISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGF---EGTNSNVKDHVSGGLD 156
++S+ G++++ SD+ TW+S+ ++ +TC DGF EG VKD V G +
Sbjct: 158 ALSSVVVVSGDESH------SDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVK 211
Query: 157 QVISLVKNELLLQVVPDSDPSNI-----------SNSGEFPSWVKAEDEKLLQQDAPPPP 205
+ +V N L + D S + + E P+W+K ED +LL
Sbjct: 212 DLSEMVSNCLAIFAGKVKDLSGVPVVNNRKLLGTEETEELPNWLKREDRELLGTPTSAIQ 271
Query: 206 ADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEY-VEIKKKKWNIMIVGDG 264
AD+ V+ DGSG + T+ +A+K APE++ R+VI+VK G Y+E +++ +KK N+M +GDG
Sbjct: 272 ADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVGRKKTNLMFIGDG 331
Query: 265 MDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLS 324
T I+G +S D++TTF TATFA + GFI RD+TFEN AGP QAVALR D +
Sbjct: 332 KGKTVITGGKSIA-DDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQAVALRVGGDHA 390
Query: 325 VFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNT 384
V YRC I GYQD+LY HS RQF++EC I GTVDFI GNAA + Q+C I +KP+ QK T
Sbjct: 391 VVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIYARKPMAQQKIT 450
Query: 385 IAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMI 444
I AQ+R PNQ +G S H C ++ PD + S GS T+LGRPWK +SR V+M S M + I
Sbjct: 451 ITAQNRKDPNQNTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVVYMMSDMGDHI 510
Query: 445 HPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNS-QAMDFTVNQFIK 503
P GWLEW D+L++GEY NKG GSG+ RVKW GYHV+ ++ +A FTV QFI
Sbjct: 511 DPRGWLEWNGPFAL-DSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEASKFTVAQFIS 569
Query: 504 GDLWLPSTGVSYNAGL 519
G WLPSTGVS+ +GL
Sbjct: 570 GSSWLPSTGVSFFSGL 585
>At3g49220 pectinesterase - like protein
Length = 598
Score = 398 bits (1022), Expect = e-111
Identities = 221/509 (43%), Positives = 307/509 (59%), Gaps = 30/509 (5%)
Query: 30 TITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISDCLEL 89
++ +F G S + + N ++ + Y+ + + V D R +A C+EL
Sbjct: 103 SLMDFPGSLAASSSKDLIHVTVNMTLHHFSHALYSSASLSFV--DMPPRARSAYDSCVEL 160
Query: 90 FDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNSN-VK 148
D + + L ++S+ + D+ TW+SA L+ +TC +GF+G + VK
Sbjct: 161 LDDSVDALSRALSSVVSSSAKPQ--------DVTTWLSAALTNHDTCTEGFDGVDDGGVK 212
Query: 149 DHVSGGLDQVISLVKNEL-LLQVVPDSDPSN---ISN---------SGEFPSWVKAEDEK 195
DH++ L + LV N L + D D I N +FP W++ ++ +
Sbjct: 213 DHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEEREEKFPRWMRPKERE 272
Query: 196 LLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEY-VEIKKK 254
+L+ AD++V+ DG+G T+ +A+K AP+ + R +I+VK G Y+E +++ +K
Sbjct: 273 ILEMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKAGRYEENNLKVGRK 332
Query: 255 KWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQA 314
K N+M VGDG T ISG +S DNITTF TA+FA + GFIARDITFEN AGP QA
Sbjct: 333 KINLMFVGDGKGKTVISGGKSI-FDNITTFHTASFAATGAGFIARDITFENWAGPAKHQA 391
Query: 315 VALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQP 374
VALR +D +V YRC I GYQD+LY HS RQF++EC I GTVDFI GNAA V QNC I
Sbjct: 392 VALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNAAVVLQNCSIYA 451
Query: 375 KKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTV 434
+KP+ QKNTI AQ+R PNQ +G S H ++ D Q +NGS QT+LGRPWK FSRTV
Sbjct: 452 RKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYLGRPWKLFSRTV 511
Query: 435 FMESFMSEMIHPEGWLEWETNTTYD-DTLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQ 492
+M S++ +H GWLEW NTT+ DTL++GEY N GPGSG+ RV W GY V+N+ ++
Sbjct: 512 YMMSYIGGHVHTRGWLEW--NTTFALDTLYYGEYLNSGPGSGLGQRVSWPGYRVINSTAE 569
Query: 493 AMDFTVNQFIKGDLWLPSTGVSYNAGLLV 521
A FTV +FI G WLPSTGVS+ AGL +
Sbjct: 570 ANRFTVAEFIYGSSWLPSTGVSFLAGLSI 598
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 371 bits (953), Expect = e-103
Identities = 188/461 (40%), Positives = 269/461 (57%), Gaps = 16/461 (3%)
Query: 69 TKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISA 128
T + D R A+ C EL D A L S + +K LR W+SA
Sbjct: 108 TMIELQKDPRAKMALDQCKELMDYAIGELSKSFEELGKFEFHK---VDEALVKLRIWLSA 164
Query: 129 VLSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQV----------VPDSDPSN 178
+S+++TC+DGF+GT N + + L + L N L + +P+ +
Sbjct: 165 TISHEQTCLDGFQGTQGNAGETIKKALKTAVQLTHNGLAMVTEMSNYLGQMQIPEMNSRR 224
Query: 179 ISNSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVI 238
+ S EFPSW+ A +LL D+VVA DGSG Y T+ +A+ P+ T +V+
Sbjct: 225 LL-SQEFPSWMDARARRLLNAPMSEVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVV 283
Query: 239 HVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIA 298
H+K+GIYKEYV++ + +++ +GDG D T ISG++SY D ITT++TAT A+ FIA
Sbjct: 284 HIKEGIYKEYVQVNRSMTHLVFIGDGPDKTVISGSKSY-KDGITTYKTATVAIVGDHFIA 342
Query: 299 RDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDF 358
++I FENTAG QAVA+R +D S+FY C GYQD+LYAHS RQFY++C I GT+DF
Sbjct: 343 KNIAFENTAGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDF 402
Query: 359 ICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGS 418
+ G+AAAVFQNC + +KPL +Q I A R P +++GF CTI+ +PD+
Sbjct: 403 LFGDAAAVFQNCTLLVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQ 462
Query: 419 IQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVAN 478
+T+LGRPWK +SRT+ M +F+ + + PEGW W +TLF+ E QN GPG+ +
Sbjct: 463 SKTYLGRPWKEYSRTIIMNTFIPDFVPPEGWQPWLGEFGL-NTLFYSEVQNTGPGAAITK 521
Query: 479 RVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
RV W G L++ + + FT Q+I+GD W+P GV Y GL
Sbjct: 522 RVTWPGIKKLSDEEILKFTPAQYIQGDAWIPGKGVPYILGL 562
>At3g05610 putative pectinesterase
Length = 669
Score = 367 bits (943), Expect = e-102
Identities = 192/464 (41%), Positives = 271/464 (58%), Gaps = 18/464 (3%)
Query: 69 TKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISA 128
T + D+R A+ C EL D A + L S + + + +LR W+SA
Sbjct: 109 TIMELQKDSRTRMALDQCKELMDYALDELSNSFEELGKFEFHLLD---EALINLRIWLSA 165
Query: 129 VLSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLL----------QVVPDSDPSN 178
+S++ETC++GF+GT N + + L I L N L + +P +
Sbjct: 166 AISHEETCLEGFQGTQGNAGETMKKALKTAIELTHNGLAIISEMSNFVGQMQIPGLNSRR 225
Query: 179 ISNSGEFPSWVKAEDEKLLQQDAPPPPA--DVVVAADGSGNYTTVMDAVKDAPEYNMTRY 236
+ G FPSWV KLLQ A D+VVA DGSG Y T+ +A++ P+ T +
Sbjct: 226 LLAEG-FPSWVDQRGRKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTF 284
Query: 237 VIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGF 296
V+H+K G+YKEYV++ K +++ +GDG D T ISGN++Y D ITT+RTAT A+ F
Sbjct: 285 VVHIKAGLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNY-KDGITTYRTATVAIVGNYF 343
Query: 297 IARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTV 356
IA++I FENTAG QAVA+R SD S+F+ C GYQD+LY HS RQF+++C I GT+
Sbjct: 344 IAKNIGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTI 403
Query: 357 DFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSN 416
DF+ G+AAAVFQNC + +KPL +Q I A R P +++GF F CTI +PD+
Sbjct: 404 DFLFGDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVK 463
Query: 417 GSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGV 476
+ + +LGRPWK +SRT+ M +F+ + + P+GW W + TLF+ E QN GPGS +
Sbjct: 464 ETSKAYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPWLGDFGL-KTLFYSEVQNTGPGSAL 522
Query: 477 ANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGLL 520
ANRV W G L+ + FT Q+I+GD W+P GV Y GLL
Sbjct: 523 ANRVTWAGIKTLSEEDILKFTPAQYIQGDDWIPGKGVPYTTGLL 566
>At2g26450 putative pectinesterase
Length = 496
Score = 365 bits (938), Expect = e-101
Identities = 200/485 (41%), Positives = 284/485 (58%), Gaps = 19/485 (3%)
Query: 43 SPNRFIDSANKVINL-LQNVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSI 101
+P F+ SA + +N L V + NQD+ +AI C L + A E ++
Sbjct: 20 NPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDD----KDAIEQCKLLVEDAKEE---TV 72
Query: 102 SATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNSNVKDHVSGGLDQVISL 161
++ + N + DL +W+SAV+SYQETC+DGFE N+K V ++ L
Sbjct: 73 ASLNKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFE--EGNLKSEVKTSVNSSQVL 130
Query: 162 VKNELLLQVVPDSDPSNISNSGE------FPSWVKAEDEKLLQQ-DAPPPPADVVVAADG 214
N L L + S + E PSWV +D ++L+ D + VA DG
Sbjct: 131 TSNSLALIKTFTENLSPVMKVVERHLLDDIPSWVSNDDRRMLRAVDVKALKPNATVAKDG 190
Query: 215 SGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNR 274
SG++TT+ DA++ PE RY+I+VK+GIY EYV + KKK N+ +VGDG T ++GN+
Sbjct: 191 SGDFTTINDALRAMPEKYEGRYIIYVKQGIYDEYVTVDKKKANLTMVGDGSQKTIVTGNK 250
Query: 275 SYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGY 334
S+ I TF TATF GF+A+ + F NTAGPE QAVA+R SD S+F C +GY
Sbjct: 251 SHAK-KIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSIFLNCRFEGY 309
Query: 335 QDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPN 394
QD+LYA++ RQ+Y+ C I GT+DFI G+AAA+FQNC I +K L QKNT+ AQ R
Sbjct: 310 QDTLYAYTHRQYYRSCVIVGTIDFIFGDAAAIFQNCNIFIRKGLPGQKNTVTAQGRVDKF 369
Query: 395 QTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWET 454
QT+GF HNC I + D +P +++LGRPWKN+SRT+ MES + +I P GWL W+
Sbjct: 370 QTTGFVVHNCKIAANEDLKPVKEEYKSYLGRPWKNYSRTIIMESKIENVIDPVGWLRWQE 429
Query: 455 NTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPSTGVS 514
DTL++ EY NKG +RVKW G+ V+N +A+++TV F++GD W+ ++G
Sbjct: 430 TDFAIDTLYYAEYNNKGSSGDTTSRVKWPGFKVINKEEALNYTVGPFLQGD-WISASGSP 488
Query: 515 YNAGL 519
GL
Sbjct: 489 VKLGL 493
>At5g51490 pectinesterase
Length = 536
Score = 365 bits (936), Expect = e-101
Identities = 194/453 (42%), Positives = 279/453 (60%), Gaps = 19/453 (4%)
Query: 76 DNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQET 135
D++ ++DC++L+ L ++ G + T D +TW+S L+ ET
Sbjct: 94 DSKKQAVLADCIDLYGDTIMQLNRTLHGVSPKAGAAKSCT---DFDAQTWLSTALTNTET 150
Query: 136 CIDGFEGTNSNVKDHVSGGLD--QVISLVKNELLLQ---VVPDSDPSNISNSGEFPSWVK 190
C G ++ NV D ++ + ++ L+ N L + + + + +N FP+W+
Sbjct: 151 CRRG--SSDLNVTDFITPIVSNTKISHLISNCLAVNGALLTAGNKGNTTANQKGFPTWLS 208
Query: 191 AEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMT--RYVIHVKKGIYKEY 248
+D++LL+ A++VVA DGSG++ TV A+ A +T R+VI+VK+GIY+E
Sbjct: 209 RKDKRLLRAVR----ANLVVAKDGSGHFNTVQAAIDVAGRRKVTSGRFVIYVKRGIYQEN 264
Query: 249 VEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAG 308
+ ++ +IM+VGDGM T I+G RS TT+ +AT + + FIA+ ITF NTAG
Sbjct: 265 INVRLNNDDIMLVGDGMRSTIITGGRSV-QGGYTTYNSATAGIEGLHFIAKGITFRNTAG 323
Query: 309 PENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQ 368
P QAVALRS SDLS+FY+C I+GYQD+L HSQRQFY+EC I GTVDFI GNAAAVFQ
Sbjct: 324 PAKGQAVALRSSSDLSIFYKCSIEGYQDTLMVHSQRQFYRECYIYGTVDFIFGNAAAVFQ 383
Query: 369 NCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWK 428
NCLI P++PLK Q N I AQ R P Q +G S HN I+P PD +P G+++T++GRPW
Sbjct: 384 NCLILPRRPLKGQANVITAQGRADPFQNTGISIHNSRILPAPDLKPVVGTVKTYMGRPWM 443
Query: 429 NFSRTVFMESFMSEMIHPEGWLEWETNTTYD-DTLFFGEYQNKGPGSGVANRVKWRGYHV 487
FSRTV +++++ ++ P GW W + + DTLF+ EY+N GP S RV W+G+HV
Sbjct: 444 KFSRTVVLQTYLDNVVSPVGWSPWIEGSVFGLDTLFYAEYKNTGPASSTRWRVSWKGFHV 503
Query: 488 LNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
L S A FTV +FI G WLP TG+ + +GL
Sbjct: 504 LGRASDASAFTVGKFIAGTAWLPRTGIPFTSGL 536
>At1g11580 unknown protein
Length = 557
Score = 360 bits (923), Expect = 1e-99
Identities = 198/452 (43%), Positives = 273/452 (59%), Gaps = 12/452 (2%)
Query: 73 QDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSY 132
+ N R +DC E+ D++ + + S+ + GN N + S++ TW+S+VL+
Sbjct: 113 RSNGVRDKAGFADCEEMMDVSKDRMMSSMEELRG--GNYNLES---YSNVHTWLSSVLTN 167
Query: 133 QETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLL--QVVPDSDPSNISNSGEFPSWVK 190
TC++ + N K V L+ ++S + L + V+P D + S FPSW+
Sbjct: 168 YMTCLESISDVSVNSKQIVKPQLEDLVSRARVALAIFVSVLPARDDLKMIISNRFPSWLT 227
Query: 191 AEDEKLLQQDAPP--PPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEY 248
A D KLL+ A+VVVA DG+G + TV +AV APE + TRYVI+VKKG+YKE
Sbjct: 228 ALDRKLLESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKET 287
Query: 249 VEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAG 308
++I KKK N+M+VGDG D T I+G+ + D TTFR+AT A + GF+A+DI F+NTAG
Sbjct: 288 IDIGKKKKNLMLVGDGKDATIITGSLNV-IDGSTTFRSATVAANGDGFMAQDIWFQNTAG 346
Query: 309 PENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQ 368
P QAVALR +D +V RC I YQD+LY H+ RQFY++ I GTVDFI GN+A VFQ
Sbjct: 347 PAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQ 406
Query: 369 NCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWK 428
NC I + P QKN + AQ R NQ + S C I D P GS++TFLGRPWK
Sbjct: 407 NCDIVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWK 466
Query: 429 NFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVL 488
+SRTV M+SF+ I P GW W+ TL++GEY N GPG+ + RV W+G+ V+
Sbjct: 467 LYSRTVIMQSFIDNHIDPAGWFPWDGEFAL-STLYYGEYANTGPGADTSKRVNWKGFKVI 525
Query: 489 NNS-QAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
+S +A FTV + I+G LWL TGV++ L
Sbjct: 526 KDSKEAEQFTVAKLIQGGLWLKPTGVTFQEWL 557
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 359 bits (921), Expect = 2e-99
Identities = 210/505 (41%), Positives = 283/505 (55%), Gaps = 41/505 (8%)
Query: 50 SANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQG 109
S N I +++ + + K + R A+ DCLE D + L ++ +
Sbjct: 94 SVNLTITAVEHNYFTVKKLIKKRKGLTPREKTALHDCLETIDETLDELHETVEDLHLYPT 153
Query: 110 NKNNGTGNLSSDLRTWISAVLSYQETCIDGF--EGTNSNVKDHVSGGLDQVISLVKNEL- 166
K + DL+T IS+ ++ QETC+DGF + + V+ + G V + N L
Sbjct: 154 KKT--LREHAGDLKTLISSAITNQETCLDGFSHDDADKQVRKALLKGQIHVEHMCSNALA 211
Query: 167 LLQVVPDSDPSN--------------------------ISNSGE-----FPSWVKAEDEK 195
+++ + D+D +N I+ +GE +P+W+ A D +
Sbjct: 212 MIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAGELDSEGWPTWLSAGDRR 271
Query: 196 LLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKK 255
LLQ AD VAADGSG + TV AV APE + RYVIH+K G+Y+E VE+ KKK
Sbjct: 272 LLQGSGVK--ADATVAADGSGTFKTVAAAVAAAPENSNKRYVIHIKAGVYRENVEVAKKK 329
Query: 256 WNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAV 315
NIM +GDG T I+G+R+ D TTF +AT A F+ARDITF+NTAGP QAV
Sbjct: 330 KNIMFMGDGRTRTIITGSRNVV-DGSTTFHSATVAAVGERFLARDITFQNTAGPSKHQAV 388
Query: 316 ALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPK 375
ALR SD S FY C + YQD+LY HS RQF+ +C I GTVDFI GNAA V Q+C I +
Sbjct: 389 ALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFIFGNAAVVLQDCDIHAR 448
Query: 376 KPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVF 435
+P QKN + AQ RT PNQ +G C I D Q GS T+LGRPWK +S+TV
Sbjct: 449 RPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSFPTYLGRPWKEYSQTVI 508
Query: 436 MESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLN-NSQAM 494
M+S +S++I PEGW EW T T +TL + EY N G G+G ANRVKWRG+ V+ ++A
Sbjct: 509 MQSAISDVIRPEGWSEW-TGTFALNTLTYREYSNTGAGAGTANRVKWRGFKVITAAAEAQ 567
Query: 495 DFTVNQFIKGDLWLPSTGVSYNAGL 519
+T QFI G WL STG ++ GL
Sbjct: 568 KYTAGQFIGGGGWLSSTGFPFSLGL 592
>At5g51500 pectinesterase
Length = 540
Score = 356 bits (913), Expect = 2e-98
Identities = 201/481 (41%), Positives = 285/481 (58%), Gaps = 28/481 (5%)
Query: 48 IDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNH 107
+D A + L N S N + F K ++DC+ L+ L ++ +
Sbjct: 79 MDRAVSAWDKLTNSSKNCTDFKK---------QAVLADCINLYGDTVMQLNRTLQGVSSK 129
Query: 108 QGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTNSNVKDHVSGGLD--QVISLVKNE 165
G + D +TW+S L+ ETC G ++ NV D + + ++ L+ N
Sbjct: 130 TGRRCT-----DFDAQTWLSTALTNTETCRRG--SSDLNVSDFTTPIVSNTKISHLISNC 182
Query: 166 LLLQVVPDSDPSNISNSGE---FPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVM 222
L + + N S +G+ FP+WV ++ +LLQ + A++VVA DGSG++ TV
Sbjct: 183 LAVNGALLTAGKNDSTTGDSKGFPTWVSRKERRLLQLQSVR--ANLVVAKDGSGHFKTVQ 240
Query: 223 DAVKDAPEYNMT--RYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDN 280
A+ A +T R+VI+VK+GIY+E + ++ NIM+VGDGM T I+G RS
Sbjct: 241 AAIDVAGRRKVTSGRFVIYVKRGIYQENLNVRLNNDNIMLVGDGMRYTIITGGRSV-KGG 299
Query: 281 ITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYA 340
TT+ +AT + + FIA+ I F+NTAGP QAVALRS SDLS+FYRC I+GYQD+L
Sbjct: 300 YTTYSSATAGIEGLHFIAKGIAFQNTAGPAKGQAVALRSSSDLSIFYRCSIEGYQDTLMV 359
Query: 341 HSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFS 400
HSQRQFY+EC I GTVDFI GNAA VFQNC+I P+ PLK Q N I AQ RT Q +G S
Sbjct: 360 HSQRQFYRECYIYGTVDFIFGNAAVVFQNCIILPRLPLKGQANVITAQGRTDLFQNTGIS 419
Query: 401 FHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYD- 459
HN IIP PD +P S++T++GRPW +SRTV +++++ ++ P GW W +TY
Sbjct: 420 IHNSIIIPAPDLKPVVRSVKTYMGRPWMMYSRTVVLKTYIDSVVSPVGWSPWTKGSTYGL 479
Query: 460 DTLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAG 518
DTLF+ EY+N GP S RV+W+G+HVL+ S A F+V +FI G WLP +G+ + +
Sbjct: 480 DTLFYAEYKNIGPASSTRWRVRWKGFHVLSKASDASAFSVGKFIAGTAWLPGSGIPFTSE 539
Query: 519 L 519
L
Sbjct: 540 L 540
>At2g26440 putative pectinesterase
Length = 547
Score = 356 bits (913), Expect = 2e-98
Identities = 191/451 (42%), Positives = 265/451 (58%), Gaps = 20/451 (4%)
Query: 82 AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFE 141
++ DC +L + LK SIS Q+ + +D R ++SA L+ + TC++G E
Sbjct: 104 SLQDCKDLHHITSSFLKRSISKIQDGVNDSRK-----LADARAYLSAALTNKITCLEGLE 158
Query: 142 GTNSNVKDHVSGGLDQVISLVKNEL--LLQVVPDSDPSNISNS------GEFPSWVKAED 193
+ +K + + N L L + ++P N+ G FP WV +D
Sbjct: 159 SASGPLKPKLVTSFTTTYKHISNSLSALPKQRRTTNPKTGGNTKNRRLLGLFPDWVYKKD 218
Query: 194 EKLLQQ-----DAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEY 248
+ L+ D P +VVAADG+GN++T+ +A+ AP + R +I+VK+G+Y E
Sbjct: 219 HRFLEDSSDGYDEYDPSESLVVAADGTGNFSTINEAISFAPNMSNDRVLIYVKEGVYDEN 278
Query: 249 VEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAG 308
++I K NI+++GDG DVT I+GNRS G D TTFR+AT AVS GF+ARDI NTAG
Sbjct: 279 IDIPIYKTNIVLIGDGSDVTFITGNRSVG-DGWTTFRSATLAVSGEGFLARDIMITNTAG 337
Query: 309 PENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQ 368
PE QAVALR ++D YRC I GYQD+LY HS RQFY+EC I GT+D+I GNAA VFQ
Sbjct: 338 PEKHQAVALRVNADFVALYRCVIDGYQDTLYTHSFRQFYRECDIYGTIDYIFGNAAVVFQ 397
Query: 369 NCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWK 428
C I K P+ Q I AQSR ++ +G S NC+I+ D S+ ++++LGRPW+
Sbjct: 398 GCNIVSKLPMPGQFTVITAQSRDTQDEDTGISMQNCSILASEDLFNSSNKVKSYLGRPWR 457
Query: 429 NFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVL 488
FSRTV MES++ E I GW +W DTL++GEY N GPGS RV W G+H++
Sbjct: 458 EFSRTVVMESYIDEFIDGSGWSKWNGGEAL-DTLYYGEYNNNGPGSETVKRVNWPGFHIM 516
Query: 489 NNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
A +FT +FI GD WL ST Y+ G+
Sbjct: 517 GYEDAFNFTATEFITGDGWLGSTSFPYDNGI 547
>At4g02300 putative pectinesterase
Length = 532
Score = 355 bits (912), Expect = 3e-98
Identities = 204/522 (39%), Positives = 298/522 (57%), Gaps = 23/522 (4%)
Query: 10 TLILLLIMSLQPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPF- 68
+LI LI ++ S + T + L+ + P I N I + S N S
Sbjct: 20 SLIFFLIFLSTVVSSQSPSYTTHKTQRLTETKTIPELIIADLNLTILKVNLASSNFSDLQ 79
Query: 69 TKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISA 128
T++ + + A DCL L D +L+ ++S ++ N D+ ++
Sbjct: 80 TRLFPNLTHYERCAFEDCLGLLDDTISDLETAVSDLRSSSLEFN--------DISMLLTN 131
Query: 129 VLSYQETCIDGFEGT----NSNVKDHVSGGLDQVISLVKNEL-----LLQVVPDSDPSNI 179
V++YQ+TC+DGF + N+++ + L ++I + N L +LQV+ PS
Sbjct: 132 VMTYQDTCLDGFSTSDNENNNDMTYELPENLKEIILDISNNLSNSLHMLQVISRKKPSPK 191
Query: 180 SNSG--EFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYV 237
S+ E+PSW+ D++LL+ ++ VA DG+GN+TT+ DAV AP + TR++
Sbjct: 192 SSEVDVEYPSWLSENDQRLLEAPVQETNYNLSVAIDGTGNFTTINDAVFAAPNMSETRFI 251
Query: 238 IHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFI 297
I++K G Y E VE+ KKK IM +GDG+ T I NRS D +TF+T T V G+I
Sbjct: 252 IYIKGGEYFENVELPKKKTMIMFIGDGIGKTVIKANRSR-IDGWSTFQTPTVGVKGKGYI 310
Query: 298 ARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVD 357
A+DI+F N+AGP QAVA RS SD S FYRC GYQD+LY HS +QFY+EC I GT+D
Sbjct: 311 AKDISFVNSAGPAKAQAVAFRSGSDHSAFYRCEFDGYQDTLYVHSAKQFYRECDIYGTID 370
Query: 358 FICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNG 417
FI GNAA VFQN + +KP K AQSR +Q +G S NC I+ PD P
Sbjct: 371 FIFGNAAVVFQNSSLYARKPNPGHKIAFTAQSRNQSDQPTGISILNCRILAAPDLIPVKE 430
Query: 418 SIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVA 477
+ + +LGRPW+ +SRTV ++SF+ ++IHP GWLE + + +TL++GEY N+GPG+ +A
Sbjct: 431 NFKAYLGRPWRKYSRTVIIKSFIDDLIHPAGWLEGKKDFAL-ETLYYGEYMNEGPGANMA 489
Query: 478 NRVKWRGY-HVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAG 518
RV W G+ + N ++A FTV FI G WL STG+ ++ G
Sbjct: 490 KRVTWPGFRRIENQTEATQFTVGPFIDGSTWLNSTGIPFSLG 531
>At1g53830 unknown protein
Length = 587
Score = 354 bits (909), Expect = 6e-98
Identities = 201/465 (43%), Positives = 269/465 (57%), Gaps = 33/465 (7%)
Query: 82 AISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFE 141
A+ DCLE D + L ++ HQ K + DL+T IS+ ++ Q TC+DGF
Sbjct: 129 ALHDCLETIDETLDELH--VAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFS 186
Query: 142 GTNSNVKD---------HVSGGLDQVISLVKNELLLQVV------PDSDPSNISN----- 181
+++ K HV ++++KN + S +N +N
Sbjct: 187 YDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKLKE 246
Query: 182 ------SGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTR 235
S +P W+ D +LLQ AD VA DGSG++TTV AV APE + R
Sbjct: 247 VTGDLDSDGWPKWLSVGDRRLLQGSTIK--ADATVADDGSGDFTTVAAAVAAAPEKSNKR 304
Query: 236 YVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIG 295
+VIH+K G+Y+E VE+ KKK NIM +GDG T I+G+R+ D TTF +AT A
Sbjct: 305 FVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVV-DGSTTFHSATVAAVGER 363
Query: 296 FIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGT 355
F+ARDITF+NTAGP QAVALR SD S FY+C + YQD+LY HS RQF+ +C I GT
Sbjct: 364 FLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGT 423
Query: 356 VDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPS 415
VDFI GNAAAV Q+C I ++P QKN + AQ R+ PNQ +G NC I D
Sbjct: 424 VDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAV 483
Query: 416 NGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSG 475
G+ T+LGRPWK +SRTV M+S +S++I PEGW EW + DTL + EY N+G G+G
Sbjct: 484 KGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFAL-DTLTYREYLNRGGGAG 542
Query: 476 VANRVKWRGYHVL-NNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
ANRVKW+GY V+ ++++A FT QFI G WL STG ++ L
Sbjct: 543 TANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>At4g33230 pectinesterase - like protein
Length = 609
Score = 354 bits (908), Expect = 8e-98
Identities = 207/516 (40%), Positives = 284/516 (54%), Gaps = 36/516 (6%)
Query: 25 PSSASTITEFSGLSCLRVSPNRFIDSANKVIN--LLQNVSYNLSPFTKVNQDNDNRLSNA 82
P+ +T+ + + P + SA +N L Q LS T+ D D A
Sbjct: 106 PTCQNTLKNETKKDTPQTDPRSLLKSAIVAVNDDLDQVFKRVLSLKTENKDDKD-----A 160
Query: 83 ISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEG 142
I+ C L D A E L S+ + ++ N + DL +W+SAV+SYQETC+DGFE
Sbjct: 161 IAQCKLLVDEAKEELGTSMKRIND---SEVNNFAKIVPDLDSWLSAVMSYQETCVDGFEE 217
Query: 143 TNSNVKDHVSGGLDQVISL------------------VKNELLLQVVPDSDPSNISNSGE 184
+ + QV++ VK LLL+ S+ +
Sbjct: 218 GKLKTEIRKNFNSSQVLTSNSLAMIKSLDGYLSSVPKVKTRLLLEA-----RSSAKETDH 272
Query: 185 FPSWVKAEDEKLLQQ-DAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKG 243
SW+ ++ ++L+ D + VA DGSGN+TT+ A+K P RY I++K G
Sbjct: 273 ITSWLSNKERRMLKAVDVKALKPNATVAKDGSGNFTTINAALKAMPAKYQGRYTIYIKHG 332
Query: 244 IYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITF 303
IY E V I KKK N+ +VGDG T ++GN+S+ I TF TATF GF+A+ + F
Sbjct: 333 IYDESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAK-KIRTFLTATFVAQGEGFMAQSMGF 391
Query: 304 ENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNA 363
NTAGPE QAVA+R SD SVF C +GYQD+LYA++ RQ+Y+ C I GTVDFI G+A
Sbjct: 392 RNTAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLYAYTHRQYYRSCVIIGTVDFIFGDA 451
Query: 364 AAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFL 423
AA+FQNC I +K L QKNT+ AQ R QT+GF HNCT+ P+ D +P +++L
Sbjct: 452 AAIFQNCDIFIRKGLPGQKNTVTAQGRVDKFQTTGFVIHNCTVAPNEDLKPVKAQFKSYL 511
Query: 424 GRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWR 483
GRPWK SRTV MES + ++I P GWL W+ DTL + EY+N GP A RVKW
Sbjct: 512 GRPWKPHSRTVVMESTIEDVIDPVGWLRWQETDFAIDTLSYAEYKNDGPSGATAARVKWP 571
Query: 484 GYHVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
G+ VLN +AM FTV F++G+ W+ + G GL
Sbjct: 572 GFRVLNKEEAMKFTVGPFLQGE-WIQAIGSPVKLGL 606
>At1g23200 putative pectinesterase
Length = 554
Score = 354 bits (908), Expect = 8e-98
Identities = 202/475 (42%), Positives = 277/475 (57%), Gaps = 56/475 (11%)
Query: 80 SNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDG 139
++A+ DCLEL++ + L NH ++ G + D +T +SA ++ Q+TC +G
Sbjct: 101 TSALFDCLELYEDTIDQL--------NHS-RRSYGQYSSPHDRQTSLSAAIANQDTCRNG 151
Query: 140 FEG---TNS-----------NVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNIS----- 180
F T+S N+ +S L + + E + + P + + S
Sbjct: 152 FRDFKLTSSYSKYFPVQFHRNLTKSISNSLAVTKAAAEAEAVAEKYPSTGFTKFSKQRSS 211
Query: 181 ------------NSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDA 228
+ +FPSW D KLL+ AD+VVA DGSG+YT++ AV A
Sbjct: 212 AGGGSHRRLLLFSDEKFPSWFPLSDRKLLEDSKTTAKADLVVAKDGSGHYTSIQQAVNAA 271
Query: 229 ---PEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFR 285
P N R VI+VK G+Y+E V IKK N+M++GDG+D T ++GNR+ D TTFR
Sbjct: 272 AKLPRRNQ-RLVIYVKAGVYRENVVIKKSIKNVMVIGDGIDSTIVTGNRNV-QDGTTTFR 329
Query: 286 TATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQ 345
+ATFAVS GFIA+ ITFENTAGPE QAVALRS SD SVFY C KGYQD+LY HS RQ
Sbjct: 330 SATFAVSGNGFIAQGITFENTAGPEKHQAVALRSSSDFSVFYACSFKGYQDTLYLHSSRQ 389
Query: 346 FYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCT 405
F + C I GTVDFI G+A A+ QNC I +KP+ QKNTI AQSR P++T+GF + T
Sbjct: 390 FLRNCNIYGTVDFIFGDATAILQNCNIYARKPMSGQKNTITAQSRKEPDETTGFVIQSST 449
Query: 406 IIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFG 465
+ + +T+LGRPW++ SRTVFM+ + ++ P GWL W + TL++G
Sbjct: 450 VAT---------ASETYLGRPWRSHSRTVFMKCNLGALVSPAGWLPWSGSFAL-STLYYG 499
Query: 466 EYQNKGPGSGVANRVKWRGYHVLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
EY N G G+ V+ RVKW GYHV+ ++A FTV F+ G+ W+ +TGV N GL
Sbjct: 500 EYGNTGAGASVSGRVKWPGYHVIKTVTEAEKFTVENFLDGNYWITATGVPVNDGL 554
>At4g02320 pectinesterase - like protein
Length = 518
Score = 349 bits (896), Expect = 2e-96
Identities = 190/480 (39%), Positives = 280/480 (57%), Gaps = 28/480 (5%)
Query: 58 LQNVSYNLSPFTKVNQDNDNRLSN----AISDCLELFDMAYENLKWSISATQNHQGNKNN 113
+ NV+ + S F+ + Q + LS+ A DCLEL D +L +IS ++H +N
Sbjct: 49 ISNVNLSSSNFSDLLQRLGSNLSHRDLCAFDDCLELLDDTVFDLTTAISKLRSHSPELHN 108
Query: 114 GTGNLSSDLRTWISAVLSYQETCIDGF------EGTNSN------VKDHVSGGLDQVISL 161
++ +SA ++ TC+DGF E N+N V + + L + S
Sbjct: 109 --------VKMLLSAAMTNTRTCLDGFASSDNDENLNNNDNKTYGVAESLKESLFNISSH 160
Query: 162 VKNEL-LLQVVPDSDPSNISNSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTT 220
V + L +L+ +P P + FP WV D LLQ ++VVA +G+GNYTT
Sbjct: 161 VSDSLAMLENIPGHIPGKVKEDVGFPMWVSGSDRNLLQDPVDETKVNLVVAQNGTGNYTT 220
Query: 221 VMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDN 280
+ +A+ AP + TR+VI++K G Y E +EI ++K IM +GDG+ T I NRSY D
Sbjct: 221 IGEAISAAPNSSETRFVIYIKCGEYFENIEIPREKTMIMFIGDGIGRTVIKANRSYA-DG 279
Query: 281 ITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYA 340
T F +AT V GFIA+D++F N AGPE QAVALRS SDLS +YRC + YQD++Y
Sbjct: 280 WTAFHSATVGVRGSGFIAKDLSFVNYAGPEKHQAVALRSSSDLSAYYRCSFESYQDTIYV 339
Query: 341 HSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFS 400
HS +QFY+EC I GTVDFI G+A+ VFQNC + ++P +QK AQ R + + +G S
Sbjct: 340 HSHKQFYRECDIYGTVDFIFGDASVVFQNCSLYARRPNPNQKIIYTAQGRENSREPTGIS 399
Query: 401 FHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDD 460
+ I+ PD P + + +LGRPW+ +SRTV M+SF+ +++ P GWL+W+ + +
Sbjct: 400 IISSRILAAPDLIPVQANFKAYLGRPWQLYSRTVIMKSFIDDLVDPAGWLKWKDDFAL-E 458
Query: 461 TLFFGEYQNKGPGSGVANRVKWRGYHVLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
TL++GEY N+GPGS + NRV+W G+ + +A F+V FI G+ WL ST + + L
Sbjct: 459 TLYYGEYMNEGPGSNMTNRVQWPGFKRIETVEEASQFSVGPFIDGNKWLNSTRIPFTLDL 518
>At2g45220 pectinesterase like protein
Length = 511
Score = 348 bits (893), Expect = 4e-96
Identities = 203/455 (44%), Positives = 274/455 (59%), Gaps = 28/455 (6%)
Query: 68 FTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWIS 127
FT + D R A DC++L+D+ + N + N L D +TW+S
Sbjct: 82 FTLGPKCRDTREKAAWEDCIKLYDLTVSKI--------NETMDPNVKCSKL--DAQTWLS 131
Query: 128 AVLSYQETCIDGFE--GTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNISNSGEF 185
L+ +TC GF G V +S + V +L+ N L + VP + + F
Sbjct: 132 TALTNLDTCRAGFLELGVTDIVLPLMS---NNVSNLLCNTLAINKVPFNYTPPEKDG--F 186
Query: 186 PSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIY 245
PSWVK D KLLQ P A VVA DGSGN+ T+ +A+ A R+VI+VK+G+Y
Sbjct: 187 PSWVKPGDRKLLQSSTPKDNA--VVAKDGSGNFKTIKEAIDAAS--GSGRFVIYVKQGVY 242
Query: 246 KEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFEN 305
E +EI+KK N+M+ GDG+ T I+G++S G TTF +AT A GFIAR ITF N
Sbjct: 243 SENLEIRKK--NVMLRGDGIGKTIITGSKSVG-GGTTTFNSATVAAVGDGFIARGITFRN 299
Query: 306 TAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAA 365
TAG N+QAVALRS SDLSVFY+C + YQD+LY HS RQFY++C + GTVDFI GNAAA
Sbjct: 300 TAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQFYRDCDVYGTVDFIFGNAAA 359
Query: 366 VFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGR 425
V QNC I ++P + + NTI AQ R+ PNQ +G HN + D +P GS +T+LGR
Sbjct: 360 VLQNCNIFARRP-RSKTNTITAQGRSDPNQNTGIIIHNSRVTAASDLRPVLGSTKTYLGR 418
Query: 426 PWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGY 485
PW+ +SRTVFM++ + +I P GWLEW+ N TLF+ E+QN GPG+ + RV W G+
Sbjct: 419 PWRQYSRTVFMKTSLDSLIDPRGWLEWDGNFAL-KTLFYAEFQNTGPGASTSGRVTWPGF 477
Query: 486 HVLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
VL + S+A FTV F+ G W+PS+ V + +GL
Sbjct: 478 RVLGSASEASKFTVGTFLAGGSWIPSS-VPFTSGL 511
>At3g14300 putative pectin methylesterase
Length = 968
Score = 347 bits (891), Expect = 7e-96
Identities = 207/525 (39%), Positives = 289/525 (54%), Gaps = 42/525 (8%)
Query: 8 TPTLILLLIMSLQ--PCTTPSSASTI----TEFSGLSCLRVSPNRFIDSANKVINLLQNV 61
TP+ +L + ++ P + SS S + T R+S D N ++ L
Sbjct: 454 TPSSVLRTVCNVTNYPASCISSISKLPLSKTTTDPKVLFRLSLQVTFDELNSIVGL---- 509
Query: 62 SYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNH-QGNKNNGTGNLSS 120
P + ND L +A+S C ++FD+A +++ +IS+ G K N +
Sbjct: 510 -----PKKLAEETNDEGLKSALSVCADVFDLAVDSVNDTISSLDEVISGGKKNLNSSTIG 564
Query: 121 DLRTWISAVLSYQETCIDGFEGTNSN--VKDHVSGGLDQVISLVKNELLL--QVVPDSDP 176
DL TW+S+ ++ TC D + N N + + + N L + QV+
Sbjct: 565 DLITWLSSAVTDIGTCGDTLDEDNYNSPIPQKLKSAMVNSTEFTSNSLAIVAQVLKKPSK 624
Query: 177 SNIS-------NSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAP 229
S I NS FP+WV+ +LLQ P V VAADGSG+ TV +AV P
Sbjct: 625 SRIPVQGRRLLNSNSFPNWVRPGVRRLLQAKNLTP--HVTVAADGSGDVRTVNEAVWRVP 682
Query: 230 EYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATF 289
+ T +VI+VK G Y E V +KK KWN+ I GDG D T ISG+ + D + TF T+TF
Sbjct: 683 KKGKTMFVIYVKAGTYVENVLMKKDKWNVFIYGDGRDKTIISGSTNMV-DGVRTFNTSTF 741
Query: 290 AVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKE 349
A GF+ +D+ NTAGPE QAVA RSDSD SV+YRC GYQD+LY HS RQ+Y+
Sbjct: 742 ATEGKGFMMKDMGIINTAGPEKHQAVAFRSDSDRSVYYRCSFDGYQDTLYTHSNRQYYRN 801
Query: 350 CRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPD 409
C + GTVDFI G VFQ C I+P++PL +Q NTI A+ NQ +G S H CTI P+
Sbjct: 802 CDVTGTVDFIFGAGTVVFQGCSIRPRQPLPNQFNTITAEGTQEANQNTGISIHQCTISPN 861
Query: 410 PDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDD---TLFFGE 466
N + T+LGRPWK FS+TV M+S + ++P GW+ W N+TYD T+F+ E
Sbjct: 862 -----GNVTATTYLGRPWKLFSKTVIMQSVIGSFVNPAGWIAW--NSTYDPPPRTIFYRE 914
Query: 467 YQNKGPGSGVANRVKWRGYH-VLNNSQAMDFTVNQFIKG-DLWLP 509
Y+N GPGS ++ RVKW GY + ++ +A FTV F++G D W+P
Sbjct: 915 YKNSGPGSDLSKRVKWAGYKPISSDDEAARFTVKYFLRGDDNWIP 959
Score = 52.0 bits (123), Expect = 8e-07
Identities = 35/131 (26%), Positives = 58/131 (43%), Gaps = 13/131 (9%)
Query: 41 RVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLKWS 100
R+S ID N ++ L P + +D L +A+S C L D+A + + +
Sbjct: 110 RLSLQVVIDELNSIVEL---------PKKLAEETDDEGLKSALSVCEHLLDLAIDRVNET 160
Query: 101 ISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFE---GTNSNVKDHVSGGLDQ 157
+SA + G K + DL TW+SA ++Y TC+D + TNS + + G+
Sbjct: 161 VSAMEVVDGKKILNAATID-DLLTWLSAAVTYHGTCLDALDEISHTNSAIPLKLKSGMVN 219
Query: 158 VISLVKNELLL 168
N L +
Sbjct: 220 STEFTSNSLAI 230
Score = 42.7 bits (99), Expect = 5e-04
Identities = 29/128 (22%), Positives = 60/128 (46%), Gaps = 13/128 (10%)
Query: 25 PSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQNVSYNLSPFTKVNQDNDNRLSNAIS 84
PSS +T E + R+S I+ N + L P + +D RL +++S
Sbjct: 292 PSSNTTDPE----ALFRLSLQVVINELNSIAGL---------PKKLAEETDDERLKSSLS 338
Query: 85 DCLELFDMAYENLKWSISATQNHQGNKNNGTGNLSSDLRTWISAVLSYQETCIDGFEGTN 144
C ++F+ A + + +IS + K + +++TW+SA ++ +TC+D + +
Sbjct: 339 VCGDVFNDAIDIVNDTISTMEEVGDGKKILKSSTIDEIQTWLSAAVTDHDTCLDALDELS 398
Query: 145 SNVKDHVS 152
N ++ +
Sbjct: 399 QNKTEYAN 406
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,599,396
Number of Sequences: 26719
Number of extensions: 568339
Number of successful extensions: 1888
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 85
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1486
Number of HSP's gapped (non-prelim): 133
length of query: 521
length of database: 11,318,596
effective HSP length: 104
effective length of query: 417
effective length of database: 8,539,820
effective search space: 3561104940
effective search space used: 3561104940
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0343.3


