

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0342b.11
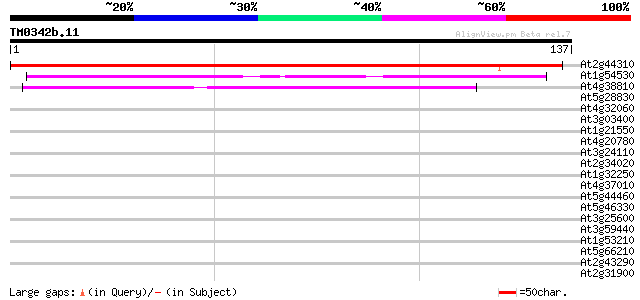
(137 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g44310 unknown protein 133 3e-32
At1g54530 hypothetical protein 57 2e-09
At4g38810 EF-Hand containing protein -like 50 4e-07
At5g28830 putative protein 39 0.001
At4g32060 unknown protein (At4g32060) 37 0.002
At3g03400 calmodulin-like protein 34 0.027
At1g21550 unknown protein 34 0.027
At4g20780 calcium-binding protein - like 33 0.046
At3g24110 hypothetical protein 33 0.046
At2g34020 unknown protein 32 0.079
At1g32250 calmodulin, putative 32 0.10
At4g37010 caltractin-like protein 32 0.13
At5g44460 calmodulin-like protein 31 0.18
At5g46330 receptor protein kinase 30 0.30
At3g25600 calmodulin, putative 30 0.30
At3g59440 calmodulin-like protein 30 0.39
At1g53210 unknown protein 30 0.39
At5g66210 calcium-dependent protein kinase 30 0.51
At2g43290 putative calcium binding protein 30 0.51
At2g31900 putative unconventional myosin 29 0.87
>At2g44310 unknown protein
Length = 142
Score = 133 bits (334), Expect = 3e-32
Identities = 65/137 (47%), Positives = 99/137 (71%), Gaps = 2/137 (1%)
Query: 1 MSVEILDGATIVNFLEDEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFG 60
M V ++DG+T+ +F++DEE F SV RFA LD NKDG+LS E+ K + +R+LE+HFG
Sbjct: 1 MGVVLIDGSTVRSFVDDEEQFKKSVDERFAALDLNKDGVLSRSELRKAFESMRLLESHFG 60
Query: 61 IDVKPDPDELGRVYKSLFIQFDHNLNGSVDLEEFKMETREMMLAMADGMGFLPVQMAL-- 118
+DV DEL +Y S+F +FD + +GSVDLEEF+ E ++++LA+ADG+G P+ M L
Sbjct: 61 VDVVTPQDELTNLYDSIFEKFDTDQSGSVDLEEFRSEMKKIVLAIADGLGSCPITMVLDD 120
Query: 119 EEDSILKRAVERECNKV 135
++D+ LK+A + E +K+
Sbjct: 121 DDDNFLKKAADLEASKL 137
>At1g54530 hypothetical protein
Length = 127
Score = 57.4 bits (137), Expect = 2e-09
Identities = 38/127 (29%), Positives = 68/127 (52%), Gaps = 9/127 (7%)
Query: 5 ILDGATIVNFLEDEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVK 64
++ T++ FL D ++F + F +LD +K+G+LS E+ + L + +E+ +V
Sbjct: 5 VITSKTLIGFLSDTKSFESITNDYFQILDLDKNGMLSPSELRQGLNNVVAVES----EVA 60
Query: 65 PDPDELGRVYKSLFIQFDHNLNGSVDLEEFKMETREMMLAMADGMGFLPVQMALEEDSIL 124
P DE VY ++F +F +L + F+ E++ AMA G+G PV M + D ++
Sbjct: 61 PG-DETDNVYNAIFERFGEDLVP----KNFRDLIAEILTAMARGIGNSPVIMVVHNDGLI 115
Query: 125 KRAVERE 131
+AV E
Sbjct: 116 MKAVLHE 122
>At4g38810 EF-Hand containing protein -like
Length = 375
Score = 50.1 bits (118), Expect = 4e-07
Identities = 30/111 (27%), Positives = 54/111 (48%), Gaps = 3/111 (2%)
Query: 4 EILDGATIVNFLEDEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDV 63
++LDG+ IV +E+E+ F+ V +F LD ++DG LS E+ + + G+
Sbjct: 13 QVLDGSDIVELVENEKVFDKFVEQKFQQLDQDEDGKLSVTEL---QPAVADIGAALGLPA 69
Query: 64 KPDPDELGRVYKSLFIQFDHNLNGSVDLEEFKMETREMMLAMADGMGFLPV 114
+ + +Y + +F H V EFK +++L MA G+ P+
Sbjct: 70 QGTSPDSDHIYSEVLNEFTHGSQEKVSKTEFKEVLSDILLGMAAGLKRDPI 120
>At5g28830 putative protein
Length = 324
Score = 38.5 bits (88), Expect = 0.001
Identities = 29/116 (25%), Positives = 51/116 (43%), Gaps = 7/116 (6%)
Query: 3 VEILDGATIVNFLEDEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGID 62
V ILDG + FLEDE+ F + N F LD G L E+ K L H G++
Sbjct: 103 VSILDGTMLKMFLEDEDDFAMLAENLFTDLDEEDKGKLCKSEIRKAL-------VHMGVE 155
Query: 63 VKPDPDELGRVYKSLFIQFDHNLNGSVDLEEFKMETREMMLAMADGMGFLPVQMAL 118
+ P + + + D + + + +F ++++ +AD + P+ + L
Sbjct: 156 MGVPPLSEFPILDDIIKKHDADSDEELGQAQFAELLQQVLQEIADVLHEKPITIVL 211
>At4g32060 unknown protein (At4g32060)
Length = 498
Score = 37.4 bits (85), Expect = 0.002
Identities = 24/82 (29%), Positives = 36/82 (43%), Gaps = 15/82 (18%)
Query: 29 FALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDHNLNGS 88
F L D + DGL+S++E + + L + E+ F + K FD + NG
Sbjct: 218 FMLFDVDNDGLISFKEYIFFVTLLSIPESSFAVAFK---------------MFDTDNNGE 262
Query: 89 VDLEEFKMETREMMLAMADGMG 110
+D EEFK M G+G
Sbjct: 263 IDKEEFKTVMSLMRSQHRQGVG 284
>At3g03400 calmodulin-like protein
Length = 137
Score = 33.9 bits (76), Expect = 0.027
Identities = 22/73 (30%), Positives = 32/73 (43%), Gaps = 13/73 (17%)
Query: 22 NISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQF 81
N+S+ + F DT+KDG +S+EE + L P +FIQ
Sbjct: 3 NMSLSDIFERFDTSKDGKISWEEFRDAIHAL-------------SPSIPSEKLVEMFIQL 49
Query: 82 DHNLNGSVDLEEF 94
D N +G VD +F
Sbjct: 50 DTNGDGQVDAAKF 62
>At1g21550 unknown protein
Length = 155
Score = 33.9 bits (76), Expect = 0.027
Identities = 23/72 (31%), Positives = 35/72 (47%), Gaps = 11/72 (15%)
Query: 24 SVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDH 83
++ F + D N DG +S EE+ L+ L G + + + GR+ + D
Sbjct: 89 AIARAFNVFDVNGDGYISAEELRDVLERL-------GFEEEAKAWDCGRMIRV----HDK 137
Query: 84 NLNGSVDLEEFK 95
NL+G VD EEFK
Sbjct: 138 NLDGFVDFEEFK 149
>At4g20780 calcium-binding protein - like
Length = 191
Score = 33.1 bits (74), Expect = 0.046
Identities = 23/75 (30%), Positives = 36/75 (47%), Gaps = 11/75 (14%)
Query: 29 FALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDHNLNGS 88
F + D N DG +S E+ L+ L + P+ E+ RV K + + D N +G
Sbjct: 125 FKVFDENGDGFISARELQTVLKKLGL----------PEGGEMERVEK-MIVSVDRNQDGR 173
Query: 89 VDLEEFKMETREMML 103
VD EFK R +++
Sbjct: 174 VDFFEFKNMMRTVVI 188
>At3g24110 hypothetical protein
Length = 229
Score = 33.1 bits (74), Expect = 0.046
Identities = 13/36 (36%), Positives = 25/36 (69%)
Query: 72 RVYKSLFIQFDHNLNGSVDLEEFKMETREMMLAMAD 107
R +S+F +D++ NG++D+EE K E+ L+++D
Sbjct: 56 RNIRSVFESYDNDTNGTIDIEELKKCLEELKLSLSD 91
>At2g34020 unknown protein
Length = 610
Score = 32.3 bits (72), Expect = 0.079
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 8/81 (9%)
Query: 15 LEDEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVY 74
+ D + S+ + F +D NKDG + E L+ L FG+ K D +
Sbjct: 319 IRDGQLSKESLKSLFDKIDRNKDGKIQISE-------LKDLTVEFGVFGKMKCD-INEFA 370
Query: 75 KSLFIQFDHNLNGSVDLEEFK 95
+L +FD + NG +D EF+
Sbjct: 371 STLLAEFDKDKNGELDENEFE 391
>At1g32250 calmodulin, putative
Length = 166
Score = 32.0 bits (71), Expect = 0.10
Identities = 26/78 (33%), Positives = 36/78 (45%), Gaps = 14/78 (17%)
Query: 17 DEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKS 76
DEE N + F D NKDG L+ E+ L+ L VKP PD+ +++
Sbjct: 10 DEEQIN-ELREIFRSFDRNKDGSLTQLELGSLLRAL---------GVKPSPDQ----FET 55
Query: 77 LFIQFDHNLNGSVDLEEF 94
L + D NG V+ EF
Sbjct: 56 LIDKADTKSNGLVEFPEF 73
>At4g37010 caltractin-like protein
Length = 167
Score = 31.6 bits (70), Expect = 0.13
Identities = 21/83 (25%), Positives = 39/83 (46%), Gaps = 12/83 (14%)
Query: 19 EAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLF 78
E N + A +D N+ G + ++E + ++ T FG + DEL + +K +
Sbjct: 58 EMNNQQINELMAEVDKNQSGAIDFDEFV------HMMTTKFG--ERDSIDELSKAFKII- 108
Query: 79 IQFDHNLNGSVDLEEFKMETREM 101
DH+ NG + + KM +E+
Sbjct: 109 ---DHDNNGKISPRDIKMIAKEL 128
>At5g44460 calmodulin-like protein
Length = 181
Score = 31.2 bits (69), Expect = 0.18
Identities = 25/78 (32%), Positives = 37/78 (47%), Gaps = 19/78 (24%)
Query: 18 EEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSL 77
EEAFN+ D + DG +S E+ K L+ L + P+ E+ +V K +
Sbjct: 113 EEAFNV--------FDEDGDGFISAVELQKVLKKLGL----------PEAGEIEQVEK-M 153
Query: 78 FIQFDHNLNGSVDLEEFK 95
+ D N +G VD EFK
Sbjct: 154 IVSVDSNHDGRVDFFEFK 171
>At5g46330 receptor protein kinase
Length = 1173
Score = 30.4 bits (67), Expect = 0.30
Identities = 20/67 (29%), Positives = 35/67 (51%), Gaps = 3/67 (4%)
Query: 18 EEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSL 77
++ FN S ++ D N G L + ++ +LQ LR+L+ + P P E+G + K L
Sbjct: 449 DDIFNCSNLETLSVADNNLTGTL--KPLIGKLQKLRILQVSYNSLTGPIPREIGNL-KDL 505
Query: 78 FIQFDHN 84
I + H+
Sbjct: 506 NILYLHS 512
>At3g25600 calmodulin, putative
Length = 161
Score = 30.4 bits (67), Expect = 0.30
Identities = 22/65 (33%), Positives = 31/65 (46%), Gaps = 13/65 (20%)
Query: 29 FALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDHNLNGS 88
FA D +KDG L+ E+ L+ L +KP D++ L Q D N NGS
Sbjct: 17 FARFDMDKDGSLTQLELAALLRSL---------GIKPRSDQISL----LLNQIDRNGNGS 63
Query: 89 VDLEE 93
V+ +E
Sbjct: 64 VEFDE 68
>At3g59440 calmodulin-like protein
Length = 195
Score = 30.0 bits (66), Expect = 0.39
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 13/80 (16%)
Query: 16 EDEEAFNISVCNRFALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYK 75
E E + + F + D N DG ++ EE+ L+ L + PD D + + K
Sbjct: 43 ESETESPVDLKRVFQMFDKNGDGRITKEELNDSLENLGIF--------MPDKDLIQMIQK 94
Query: 76 SLFIQFDHNLNGSVDLEEFK 95
D N +G VD+ EF+
Sbjct: 95 -----MDANGDGCVDINEFE 109
>At1g53210 unknown protein
Length = 585
Score = 30.0 bits (66), Expect = 0.39
Identities = 25/80 (31%), Positives = 37/80 (46%), Gaps = 9/80 (11%)
Query: 29 FALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDHNLNGS 88
F +D N DG LS EL+ L + + ID D D +G+V + FD L+
Sbjct: 308 FLTIDANNDGHLS----AAELKALIIGISFEDIDFDKD-DAVGKVLQD----FDKTLDEQ 358
Query: 89 VDLEEFKMETREMMLAMADG 108
VD EEF ++ ++ G
Sbjct: 359 VDQEEFVRGIKQWLIQAMGG 378
>At5g66210 calcium-dependent protein kinase
Length = 523
Score = 29.6 bits (65), Expect = 0.51
Identities = 18/67 (26%), Positives = 33/67 (48%), Gaps = 7/67 (10%)
Query: 32 LDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDHNLNGSVDL 91
+D+N DGL+ + E + + LE H D ++ ++ F +FD + +G +
Sbjct: 414 IDSNTDGLVDFTEFVAAALHVHQLEEH-------DSEKWQLRSRAAFEKFDLDKDGYITP 466
Query: 92 EEFKMET 98
EE +M T
Sbjct: 467 EELRMHT 473
>At2g43290 putative calcium binding protein
Length = 215
Score = 29.6 bits (65), Expect = 0.51
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 13/67 (19%)
Query: 29 FALLDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKPDPDELGRVYKSLFIQFDHNLNGS 88
F + D N DG ++ EE+ L+ + GI + PD D ++K D N +G
Sbjct: 70 FQMFDKNGDGRITKEELNDSLE-------NLGIYI-PDKDLTQMIHK-----IDANGDGC 116
Query: 89 VDLEEFK 95
VD++EF+
Sbjct: 117 VDIDEFE 123
>At2g31900 putative unconventional myosin
Length = 1490
Score = 28.9 bits (63), Expect = 0.87
Identities = 26/89 (29%), Positives = 42/89 (46%), Gaps = 11/89 (12%)
Query: 12 VNFLEDEEAFNISVCNRFAL--LDTNKDGLLSYEEMLKELQCLRVLETHFGIDVKP-DPD 68
VNF++ EEA + + + + L T + L+ E+M+++ C RV+ T G KP DP+
Sbjct: 266 VNFIKGEEADSSKLRDDKSRYHLQTAAELLMCNEKMMEDSLCKRVIVTPDGNITKPLDPE 325
Query: 69 ELG--------RVYKSLFIQFDHNLNGSV 89
VY LF +N S+
Sbjct: 326 SAASNRDALAKTVYSRLFDWIVDKINSSI 354
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,098,981
Number of Sequences: 26719
Number of extensions: 125956
Number of successful extensions: 478
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 422
Number of HSP's gapped (non-prelim): 109
length of query: 137
length of database: 11,318,596
effective HSP length: 89
effective length of query: 48
effective length of database: 8,940,605
effective search space: 429149040
effective search space used: 429149040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0342b.11


