

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.21
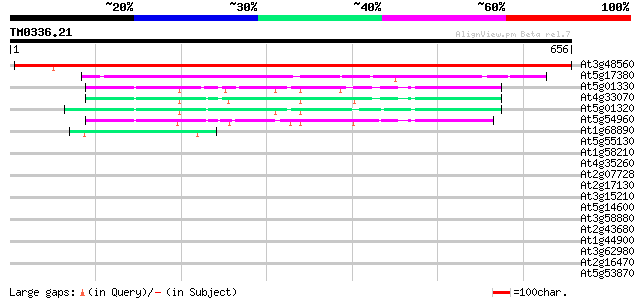
(656 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g48560 acetolactate synthase 1023 0.0
At5g17380 2-hydroxyphytanoyl-CoA lyase-like protein 166 4e-41
At5g01330 pyruvate decarboxylase-like protein 96 5e-20
At4g33070 pyruvate decarboxylase-1 (Pdc1) 91 2e-18
At5g01320 pyruvate decarboxylase-like protein 88 2e-17
At5g54960 pyruvate decarboxylase 86 7e-17
At1g68890 putative menaquinone biosynthesis protein 52 1e-06
At5g55130 molybdopterin synthase sulphurylase (gb|AAD18050.1) 36 0.059
At1g58210 hypothetical protein 34 0.23
At4g35260 NAD+ dependent isocitrate dehydrogenase subunit 1 34 0.29
At2g07728 putative protein 34 0.29
At2g17130 putative NAD+ dependent isocitrate dehydrogenase subun... 32 0.86
At3g15210 ethylene responsive element binding factor 4 32 1.5
At5g14600 unknown protein 31 1.9
At3g58880 putative protein 30 3.3
At2g43680 SF16 protein {Helianthus annuus} like protein 30 3.3
At1g44900 transcription factor, putative 30 3.3
At3g62980 transport inhibitor response 1 (TIR1) 30 4.3
At2g16470 unknown protein 30 4.3
At5g53870 predicted GPI-anchored protein 30 5.6
>At3g48560 acetolactate synthase
Length = 670
Score = 1023 bits (2645), Expect = 0.0
Identities = 510/660 (77%), Positives = 574/660 (86%), Gaps = 9/660 (1%)
Query: 6 AAKLAFTTVPSPSS-RPQNHVLKFTIPFPSYPNNSSPHSRCRSLQ------ISASLSNPT 58
++ ++F+T PSPSS + + +F++PF PN SS SR R ++ ISA L+ T
Sbjct: 11 SSSISFSTKPSPSSSKSPLPISRFSLPFSLNPNKSSSSSRRRGIKSSSPSSISAVLNTTT 70
Query: 59 PTAASASTAVAT--ENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQAL 116
+ S T E F SRF+ D+PRKGADILVEALERQGV VFAYPGGASMEIHQAL
Sbjct: 71 NVTTTPSPTKPTKPETFISRFAPDQPRKGADILVEALERQGVETVFAYPGGASMEIHQAL 130
Query: 117 TRSATIRNVLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLI 176
TRS++IRNVLPRHEQGG+FAAEGYARSSG PG+CIATSGPGATNLVSGLADALLDSVPL+
Sbjct: 131 TRSSSIRNVLPRHEQGGVFAAEGYARSSGKPGICIATSGPGATNLVSGLADALLDSVPLV 190
Query: 177 AITGQVPRRMIGTDAFQETPIVEVTRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPV 236
AITGQVPRRMIGTDAFQETPIVEVTRSITKHNYLV+D++DIPRI+ EAFFLATSGRPGPV
Sbjct: 191 AITGQVPRRMIGTDAFQETPIVEVTRSITKHNYLVMDVEDIPRIIEEAFFLATSGRPGPV 250
Query: 237 LIDIPKDIQQQLAVPNWDQPIKLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGS 296
L+D+PKDIQQQLA+PNW+Q ++LPGYM+R+P+ P D+ LEQ+VRL+ E KKPVLY GGG
Sbjct: 251 LVDVPKDIQQQLAIPNWEQAMRLPGYMSRMPKPPEDSHLEQIVRLISESKKPVLYVGGGC 310
Query: 297 LNSSEELRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAF 356
LNSS+EL RFVELTG+PVASTLMGLG+YP DE SL MLGMHGTVYANYAV+ SDLLLAF
Sbjct: 311 LNSSDELGRFVELTGIPVASTLMGLGSYPCDDELSLHMLGMHGTVYANYAVEHSDLLLAF 370
Query: 357 GVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGINRILESRG 416
GVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNK PHVSVC D+KLAL+G+N++LE+R
Sbjct: 371 GVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKTPHVSVCGDVKLALQGMNKVLENRA 430
Query: 417 VKGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQM 476
+ KLDF WR ELN QK +FPL FKTF EAIPPQYAI+VLDELT+G AIISTGVGQHQM
Sbjct: 431 EELKLDFGVWRNELNVQKQKFPLSFKTFGEAIPPQYAIKVLDELTDGKAIISTGVGQHQM 490
Query: 477 WAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELAT 536
WAAQFY YK+PRQWL+S GLGAMGFGLPAA+GA+VANP +VVDIDGDGSF+MNVQELAT
Sbjct: 491 WAAQFYNYKKPRQWLSSGGLGAMGFGLPAAIGASVANPDAIVVDIDGDGSFIMNVQELAT 550
Query: 537 IKVENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLGDPSNEKEVFPNMLKFADACGI 596
I+VENLPVK+LLLNNQHLGMV+QWEDRFYK+NRAHT+LGDP+ E E+FPNML FA ACGI
Sbjct: 551 IRVENLPVKVLLLNNQHLGMVMQWEDRFYKANRAHTFLGDPAQEDEIFPNMLLFAAACGI 610
Query: 597 PAARVTKKQDLRAAIQKMLDTPGPYLLDVIVPHQEHVLPMIPSNGTFQDVITEGDGRRSY 656
PAARVTKK DLR AIQ MLDTPGPYLLDVI PHQEHVLPMIPS GTF DVITEGDGR Y
Sbjct: 611 PAARVTKKADLREAIQTMLDTPGPYLLDVICPHQEHVLPMIPSGGTFNDVITEGDGRIKY 670
>At5g17380 2-hydroxyphytanoyl-CoA lyase-like protein
Length = 572
Score = 166 bits (420), Expect = 4e-41
Identities = 148/562 (26%), Positives = 266/562 (46%), Gaps = 37/562 (6%)
Query: 85 GADILVEALERQGVTDVFAYPGGASMEIHQALTRSAT--IRNVLPRHEQGGIFAAEGYAR 142
G ++ ++L GVT +F G + + +R+ IR + +EQ +AA Y
Sbjct: 14 GNVLVAKSLSHLGVTHMFGVVG---IPVTSLASRAMALGIRFIAFHNEQSAGYAASAYGY 70
Query: 143 SSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPIVEVTR 202
+G PG+ + SGPG + ++GL++A +++ P++ I+G +R +G FQE +E +
Sbjct: 71 LTGKPGILLTVSGPGCVHGLAGLSNAWVNTWPMVMISGSCDQRDVGRGDFQELDQIEAVK 130
Query: 203 SITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDI-QQQLAVPNWDQPI-KLP 260
+ +K + D+ +IP V+ A SGRPG +DIP D+ +Q+++ D+ + ++
Sbjct: 131 AFSKLSEKAKDVREIPDCVSRVLDRAVSGRPGGCYLDIPTDVLRQKISESEADKLVDEVE 190
Query: 261 GYMARLP-RSPSDALLEQVVRLLLECKKPVLYAGGGSLNS--SEELRRFVELTGVPVAST 317
P R + +E V LL + ++P++ G G+ S +EL++ VE+TG+P T
Sbjct: 191 RSRKEEPIRGSLRSEIESAVSLLRKAERPLIVFGKGAAYSRAEDELKKLVEITGIPFLPT 250
Query: 318 LMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAFGVRFDDRV-TGKLEAFASRAK 376
MG G P E S T + A+ K D+ L G R + + G+ + K
Sbjct: 251 PMGKGLLPDTHEFS-------ATAARSLAIGKCDVALVVGARLNWLLHFGESPKWDKDVK 303
Query: 377 IVHIDIDSAEIGKNKQPHVSVCADLKLALKGINR-ILESRGVKGKLDFRAWREELNEQKL 435
+ +D+ EI + ++PH+ + D K + +NR I + GK + +W E ++++
Sbjct: 304 FILVDVSEEEI-ELRKPHLGIVGDAKTVIGLLNREIKDDPFCLGKSN--SWVESISKKAK 360
Query: 436 R--FPLQFKTFEEAIP-----PQYAIQVLDELTNGNAIISTGVGQHQMWAAQ-FYKYKRP 487
++ + ++ +P P I+ G + + G + M + K P
Sbjct: 361 ENGEKMEIQLAKDVVPFNFLTPMRIIRDAILAVEGPSPVVVSEGANTMDVGRSVLVQKEP 420
Query: 488 RQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVKIL 547
R L + G MG GL + AAVA+P +VV ++GD F + E+ T+ NL V I+
Sbjct: 421 RTRLDAGTWGTMGVGLGYCIAAAVASPDRLVVAVEGDSGFGFSAMEVETLVRYNLAVVII 480
Query: 548 LLNNQHLGMVVQWEDRFYKSNRAHTYLGDPSNEKEVFPN--MLKFADACGIPAARVTKKQ 605
+ NN V DR + + DP+ V PN K +A G V
Sbjct: 481 VFNNGG----VYGGDRRGPEEISGPHKEDPAPTSFV-PNAGYHKLIEAFGGKGYIVETPD 535
Query: 606 DLRAAIQKMLDTPGPYLLDVIV 627
+L++A+ + P +++VI+
Sbjct: 536 ELKSALAESFAARKPAVVNVII 557
>At5g01330 pyruvate decarboxylase-like protein
Length = 592
Score = 96.3 bits (238), Expect = 5e-20
Identities = 122/526 (23%), Positives = 217/526 (41%), Gaps = 80/526 (15%)
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
L L + GVTD+F PG ++ + L + + N+ +E +AA+GYARS G+ G
Sbjct: 36 LSRRLVQAGVTDIFTVPGDFNLSLLDQLIANPELNNIGCCNELNAGYAADGYARSRGV-G 94
Query: 149 VCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPI--------VEV 200
C+ T G ++++ +A A +++P+I I G GT+ I +
Sbjct: 95 ACVVTFTVGGLSVLNAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRC 154
Query: 201 TRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPG-PVLIDIPKDIQQQLAVP-----NWD 254
+++T + +V ++D +++A +AT+ R PV I I ++ A+P ++
Sbjct: 155 FQTVTCYQAVVNHLEDAHEQIDKA--IATALRESKPVYISISCNL---AAIPHPTFASYP 209
Query: 255 QPIKLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLNSSEELRRFVEL---TG 311
P L ++ + +A +E + L + KPV+ GG L ++ FVEL +G
Sbjct: 210 VPFDLTPRLSN--KDCLEAAVEATLEFLNKAVKPVM-VGGPKLRVAKARDAFVELADASG 266
Query: 312 VPVASTLMGLGAYPTADENSLQMLGMH----GTVYANYAVDKSDLLLAFGVRFDDRVTGK 367
PVA G P EN +G + T++ + V+ +D + G F+D +
Sbjct: 267 YPVAVMPSAKGFVP---ENHPHFIGTYWGAVSTLFCSEIVESADAYIFAGPIFNDYSSVG 323
Query: 368 LEAFASRAKIVHIDIDSA------------------EIGKNKQPHVSVCADLKLALKGIN 409
+ K + + DS E+ K +P+ K A + +
Sbjct: 324 YSLLLKKEKAIIVHPDSVVVANGPTFGCVRMSEFFRELAKRVKPN-------KTAYENYH 376
Query: 410 RILESRGVKGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIIST 469
RI G K R + LR F+ ++ + + A+ I T
Sbjct: 377 RIFVPEGKPLKCKPR--------EPLRINAMFQHIQKMLSNETAV-----------IAET 417
Query: 470 GVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMM 529
G + Q K + + G++G+ + A +G A A P V+ GDGSF +
Sbjct: 418 G---DSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQATPEKRVLSFIGDGSFQV 474
Query: 530 NVQELATIKVENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLG 575
Q+++T+ I L+NN + V+ D Y + Y G
Sbjct: 475 TAQDVSTMIRNGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 520
>At4g33070 pyruvate decarboxylase-1 (Pdc1)
Length = 607
Score = 91.3 bits (225), Expect = 2e-18
Identities = 112/513 (21%), Positives = 205/513 (39%), Gaps = 54/513 (10%)
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
L L + GVTDVF+ PG ++ + L + + +E +AA+GYARS G+ G
Sbjct: 51 LARRLVQAGVTDVFSVPGDFNLTLLDHLMAEPDLNLIGCCNELNAGYAADGYARSRGV-G 109
Query: 149 VCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPI--------VEV 200
C+ T G ++++ +A A +++PLI I G GT+ I +
Sbjct: 110 ACVVTFTVGGLSVLNAIAGAYSENLPLICIVGGPNSNDYGTNRILHHTIGLPDFSQELRC 169
Query: 201 TRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDIQQQLAVPNWD---QPI 257
+++T + +V ++DD +++A A PV I + ++ A+P+ P+
Sbjct: 170 FQTVTCYQAVVNNLDDAHEQIDKAISTALK-ESKPVYISVSCNL---AAIPHHTFSRDPV 225
Query: 258 KLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLNSSEELRRFVELTGVPVAST 317
+ +A +E + L + KPV+ GG L ++ FVEL +
Sbjct: 226 PFSLAPRLSNKMGLEAAVEATLEFLNKAVKPVM-VGGPKLRVAKACDAFVELADASGYAL 284
Query: 318 LMGLGAYPTADENSLQMLGMH----GTVYANYAVDKSDLLLAFGVRFDDRVTGKLEAFAS 373
M A E+ +G + T + + V+ +D + G F+D +
Sbjct: 285 AMMPSAKGFVPEHHPHFIGTYWGAVSTPFCSEIVESADAYIFAGPIFNDYSSVGYSLLLK 344
Query: 374 RAKIVHIDIDSAEIGKNKQPHVSVCADL-----------KLALKGINRILESRGVKGKLD 422
+ K + + D + + +D + A + +RI G K +
Sbjct: 345 KEKAIVVQPDRITVANGPTFGCILMSDFFRELSKRVKRNETAYENYHRIFVPEGKPLKCE 404
Query: 423 FRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQFY 482
R + LR F+ ++ + + A+ I TG + Q
Sbjct: 405 SR--------EPLRVNTMFQHIQKMLSSETAV-----------IAETG---DSWFNCQKL 442
Query: 483 KYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENL 542
K + + G++G+ + A +G A A+P V+ GDGSF + VQ+++T+
Sbjct: 443 KLPKGCGYEFQMQYGSIGWSVGATLGYAQASPEKRVLAFIGDGSFQVTVQDISTMLRNGQ 502
Query: 543 PVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLG 575
I L+NN + V+ D Y + Y G
Sbjct: 503 KTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 535
>At5g01320 pyruvate decarboxylase-like protein
Length = 603
Score = 87.8 bits (216), Expect = 2e-17
Identities = 119/528 (22%), Positives = 213/528 (39%), Gaps = 36/528 (6%)
Query: 65 STAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRN 124
S AVAT ++ + L L + GVTDVF+ PG ++ + L + N
Sbjct: 23 SNAVATIQDSAPITTTSESTLGRHLSRRLVQAGVTDVFSVPGDFNLTLLDHLIAEPELNN 82
Query: 125 VLPRHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPR 184
+ +E +AA+GYARS G+ G C+ T G ++++ +A A +++P+I I G
Sbjct: 83 IGCCNELNAGYAADGYARSRGV-GACVVTFTVGGLSVLNAIAGAYSENLPVICIVGGPNS 141
Query: 185 RMIGTDAFQETPI--------VEVTRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPV 236
GT+ I + +++T + +V +++D +++A A PV
Sbjct: 142 NDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEQIDKAIATALK-ESKPV 200
Query: 237 LIDIPKDIQQQLAVPNWDQPIKLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGS 296
I I ++ P+ +A +E + L + KPV+ GG
Sbjct: 201 YISISCNLAATPHPTFARDPVPFDLTPRMSNTMGLEAAVEATLEFLNKAVKPVM-VGGPK 259
Query: 297 LNSSEELRRFVEL---TGVPVASTLMGLGAYPTADENSLQMLGMH----GTVYANYAVDK 349
L ++ F+EL +G P+A G P EN +G + T + + V+
Sbjct: 260 LRVAKASEAFLELADASGYPLAVMPSTKGLVP---ENHPHFIGTYWGAVSTPFCSEIVES 316
Query: 350 SDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGIN 409
+D + G F+D + + K + + D + + +D
Sbjct: 317 ADAYIFAGPIFNDYSSVGYSLLLKKEKAIIVHPDRVVVANGPTFGCVLMSD-------FF 369
Query: 410 RILESRGVKGKLDFRAW-REELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIIS 468
R L R + + + + R + E K PL+ K E + L++ A+I+
Sbjct: 370 RELAKRVKRNETAYENYERIFVPEGK---PLKCKPGEPLRVNAMFQHIQKMLSSETAVIA 426
Query: 469 -TGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSF 527
TG + Q K + + G++G+ + A +G A A P V+ GDGSF
Sbjct: 427 ETG---DSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQATPEKRVLSFIGDGSF 483
Query: 528 MMNVQELATIKVENLPVKILLLNNQHLGMVVQWEDRFYKSNRAHTYLG 575
+ Q+++T+ I L+NN + V+ D Y + Y G
Sbjct: 484 QVTAQDISTMIRNGQKAIIFLINNGGYTIEVEIHDGPYNVIKNWNYTG 531
>At5g54960 pyruvate decarboxylase
Length = 607
Score = 85.9 bits (211), Expect = 7e-17
Identities = 115/500 (23%), Positives = 204/500 (40%), Gaps = 48/500 (9%)
Query: 89 LVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLPRHEQGGIFAAEGYARSSGLPG 148
L L GVTDVF+ PG ++ + L ++ + +E +AA+GYARS G+ G
Sbjct: 51 LARRLVEIGVTDVFSVPGDFNLTLLDHLIAEPNLKLIGCCNELNAGYAADGYARSRGV-G 109
Query: 149 VCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQE--------TPIVEV 200
C+ T G ++++ +A A +++PLI I G GT+ T +
Sbjct: 110 ACVVTFTVGGLSVLNAIAGAYSENLPLICIVGGPNSNDYGTNRILHHTIGLPDFTQELRC 169
Query: 201 TRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDIQQQLAVPNWDQ---PI 257
+++T ++ ++++ +++ A A PV I I ++ + +P + + P
Sbjct: 170 FQAVTCFQAVINNLEEAHELIDTAISTALK-ESKPVYISISCNL-PAIPLPTFSRHPVPF 227
Query: 258 KLPGYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLNSSEELRRFVELTGVPVAST 317
LP M + DA +E L + KPVL GG + ++ FVEL ++
Sbjct: 228 MLP--MKVSNQIGLDAAVEAAAEFLNKAVKPVL-VGGPKMRVAKAADAFVELAD----AS 280
Query: 318 LMGLGAYPTA----DENSLQMLGMH----GTVYANYAVDKSDLLLAFGVRFDDRVTGKLE 369
GL P+A E+ +G + T + V+ +D L G F+D +
Sbjct: 281 GYGLAVMPSAKGQVPEHHKHFIGTYWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYS 340
Query: 370 AFASRAKIVHIDIDSAEIGKNKQPHVSVCAD----LKLALKGINRILESRGVKGKLDFRA 425
+ K + + D IG + D L +K N E+ + +
Sbjct: 341 LLLKKEKAIIVQPDRVTIGNGPAFGCVLMKDFLSELAKRIKHNNTSYENYHRIYVPEGKP 400
Query: 426 WREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQFYKYK 485
R+ NE LR + F+ + + + A+ + TG + Q K
Sbjct: 401 LRDNPNE-SLRVNVLFQHIQNMLSSESAV-----------LAETG---DSWFNCQKLKLP 445
Query: 486 RPRQWLTSSGLGAMGFGLPAAMGAAVANPGDVVVDIDGDGSFMMNVQELATIKVENLPVK 545
+ G++G+ + A +G A A P V+ GDGSF + Q+++T+
Sbjct: 446 EGCGYEFQMQYGSIGWSVGATLGYAQAMPNRRVIACIGDGSFQVTAQDVSTMIRCGQKTI 505
Query: 546 ILLLNNQHLGMVVQWEDRFY 565
I L+NN + V+ D Y
Sbjct: 506 IFLINNGGYTIEVEIHDGPY 525
>At1g68890 putative menaquinone biosynthesis protein
Length = 894
Score = 51.6 bits (122), Expect = 1e-06
Identities = 50/180 (27%), Positives = 72/180 (39%), Gaps = 9/180 (5%)
Query: 71 ENFTSRFSLDEPRKGA---DILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNVLP 127
E S F DE A ++E R G+T PG S + A +
Sbjct: 326 ETEVSNFLRDEANINAVWASAIIEECTRLGLTYFCVAPGSRSSHLAIAAANHPLTTCLAC 385
Query: 128 RHEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMI 187
E+ F A GYA+ S P V I +SG +NL+ + +A D +PL+ +T P +
Sbjct: 386 FDERSLAFHAIGYAKGSLKPAVIITSSGTAVSNLLPAVVEASEDFLPLLLLTADRPPELQ 445
Query: 188 GTDAFQETPIVEVTRSITKHNY-LVLDIDDIP-----RIVNEAFFLATSGRPGPVLIDIP 241
G A Q + S + + L D IP V+ A AT GPV ++ P
Sbjct: 446 GVGANQAINQINHFGSFVRFFFNLPPPTDLIPVRMVLTTVDSALHWATGSACGPVHLNCP 505
>At5g55130 molybdopterin synthase sulphurylase (gb|AAD18050.1)
Length = 464
Score = 36.2 bits (82), Expect = 0.059
Identities = 29/109 (26%), Positives = 48/109 (43%), Gaps = 14/109 (12%)
Query: 428 EELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTGVGQHQMWAAQFYKYKR- 486
EEL +K + T E + A+++ D ++NG++ ++ +H + Q Y+Y R
Sbjct: 16 EELKLKKAEIEHRISTLEAKLQDTAAVELYDAVSNGDSYLTAPELEHGLSPDQIYRYSRQ 75
Query: 487 ----------PRQWLTSSGL--GAMGFGLPAAMGAAVANPGDV-VVDID 522
L SS L GA G G PA + A G + ++D D
Sbjct: 76 LLLPSFAVEGQSNLLKSSVLVIGAGGLGSPALLYLAACGVGQLGIIDHD 124
>At1g58210 hypothetical protein
Length = 1195
Score = 34.3 bits (77), Expect = 0.23
Identities = 27/110 (24%), Positives = 45/110 (40%), Gaps = 15/110 (13%)
Query: 11 FTTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASASTAVAT 70
FT +P P++ P + T+P P NSSP + PTP + T + T
Sbjct: 18 FTPLPEPNTSPSTY--SKTLPKP----NSSP---------GTDGTFPTPFPLAVITPIKT 62
Query: 71 ENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRSA 120
+ +KG D+ ++ E G + V + G + H++ T A
Sbjct: 63 LKSVTLSDWWLTKKGKDLCIKGFESNGASGVRLFSSGTISKRHESTTLEA 112
>At4g35260 NAD+ dependent isocitrate dehydrogenase subunit 1
Length = 367
Score = 33.9 bits (76), Expect = 0.29
Identities = 34/130 (26%), Positives = 58/130 (44%), Gaps = 12/130 (9%)
Query: 201 TRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDIQQQLAVPNWDQPIKLP 260
TRS+T Y+ D PR V L GP++ + + + + + P + + +
Sbjct: 23 TRSVT---YMPRPGDGAPRAVT----LIPGDGIGPLVTNAVEQVMEAMHAPIFFEKYDVH 75
Query: 261 GYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLNSSEELRRFVELTGVPVASTLMG 320
G M+R+P +++ + V L K PV GGG + + +LR+ ++L V
Sbjct: 76 GEMSRVPPEVMESIRKNKVCLKGGLKTPV---GGGVSSLNVQLRKELDLFASLV--NCFN 130
Query: 321 LGAYPTADEN 330
L PT EN
Sbjct: 131 LPGLPTRHEN 140
>At2g07728 putative protein
Length = 286
Score = 33.9 bits (76), Expect = 0.29
Identities = 44/160 (27%), Positives = 64/160 (39%), Gaps = 11/160 (6%)
Query: 256 PIKLPGYMARLPRSPSDALLEQV--VRLLLECKKPVLYAGGGSLNSSEELRRFVELTGVP 313
P GY + S L++ + RLL ++ + G G L LR E TG+
Sbjct: 102 PTPSRGYFVTSVKPASKPLVKDLRQSRLLRHLRQTSVKGGCGRLRGCGRLR---EATGLR 158
Query: 314 VASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKSDLLLAFGVRFDDRVTGKLEAFAS 373
A+ LG A+E Q+ + A SDLLL RFD L S
Sbjct: 159 EAAGATSLGVCAYAEERK-QVQTSAPDLVAEEGSPSSDLLLPPRRRFDGETLDPLGKVTS 217
Query: 374 RA-----KIVHIDIDSAEIGKNKQPHVSVCADLKLALKGI 408
RA K+V + I S I + + VS ++ L +G+
Sbjct: 218 RAEYAQVKLVVMPISSFSISRTVKRKVSPESNCTLFEEGL 257
>At2g17130 putative NAD+ dependent isocitrate dehydrogenase subunit
2, IDH2
Length = 367
Score = 32.3 bits (72), Expect = 0.86
Identities = 29/109 (26%), Positives = 51/109 (46%), Gaps = 10/109 (9%)
Query: 201 TRSITKHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDIQQQLAVPNWDQPIKLP 260
TRS+T Y+ D PR V L GP++ + + + + + P + +P ++
Sbjct: 23 TRSVT---YMPRPGDGKPRPVT----LIPGDGVGPLVTNAVQQVMEAMHAPVYFEPFEVH 75
Query: 261 GYMARLPRSPSDALLEQVVRLLLECKKPVLYAGGGSLNSSEELRRFVEL 309
G M LP +++ + V L K PV GGG + + LR+ ++L
Sbjct: 76 GDMKSLPEGLLESIKKNKVCLKGGLKTPV---GGGVSSLNVNLRKELDL 121
>At3g15210 ethylene responsive element binding factor 4
Length = 222
Score = 31.6 bits (70), Expect = 1.5
Identities = 31/107 (28%), Positives = 47/107 (42%), Gaps = 11/107 (10%)
Query: 415 RGVKGKLDFRAWREELNEQKL-----RFPLQFKTFEEAIPPQYAIQVLDELTNGNAI--I 467
RG K K +F + E L++QK+ R P Q T + A PP V+ T GN +
Sbjct: 72 RGAKAKTNFPTFLE-LSDQKVPTGFARSPSQSSTLDCASPPTL---VVPSATAGNVPPQL 127
Query: 468 STGVGQHQMWAAQFYKYKRPRQWLTSSGLGAMGFGLPAAMGAAVANP 514
+G + RP +L G+G +G G P + +A +P
Sbjct: 128 ELSLGGGGGGSCYQIPMSRPVYFLDLMGIGNVGRGQPPPVTSAFRSP 174
>At5g14600 unknown protein
Length = 318
Score = 31.2 bits (69), Expect = 1.9
Identities = 35/135 (25%), Positives = 60/135 (43%), Gaps = 20/135 (14%)
Query: 146 LPGVCIATSGPGATNLVSGLADALLDSVPLIAITGQVPRRMIGTDAFQETPIVEVTRSIT 205
+PG + SG G+ +L + LA A+ + + + R + + F++T I
Sbjct: 107 VPGCVVLESGTGSGSLSTSLARAVAPTGHVYSFDFHEQRAVSAREDFEKTGI-------- 158
Query: 206 KHNYLVLDIDDIPRIVNEAFFLATSGRPGPVLIDIPKDIQQQLAVPNWDQPIKLPGYMAR 265
+ + +++ DI E F SG V +D+P Q LAVP+ + +K G +
Sbjct: 159 -SSLVTVEVRDIQ---GEGFPEKLSGLADSVFLDLP---QPWLAVPSAAKMLKQDGVLCS 211
Query: 266 LPRSPSDALLEQVVR 280
SP +EQV R
Sbjct: 212 F--SP---CIEQVQR 221
>At3g58880 putative protein
Length = 454
Score = 30.4 bits (67), Expect = 3.3
Identities = 25/84 (29%), Positives = 38/84 (44%), Gaps = 1/84 (1%)
Query: 18 SSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQI-SASLSNPTPTAASASTAVATENFTSR 76
S+ P L SYPN + + ++L I S SLS P + + + N+T+
Sbjct: 182 SACPALEALNLANVHGSYPNATVSIASLKTLTIKSVSLSGPAHVFSFDTPNLLCLNYTAL 241
Query: 77 FSLDEPRKGADILVEALERQGVTD 100
F D P + LVEA + +TD
Sbjct: 242 FEDDYPLVNLEYLVEAQIKFVLTD 265
>At2g43680 SF16 protein {Helianthus annuus} like protein
Length = 668
Score = 30.4 bits (67), Expect = 3.3
Identities = 29/85 (34%), Positives = 38/85 (44%), Gaps = 8/85 (9%)
Query: 4 APAAKLAFTTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRC-----RSLQISASLSNPT 58
+P A++ T P P S P+ V + P P P P S RS SAS + P
Sbjct: 117 SPRAEVPRTLSPKPPS-PRAEVPRSLSPKPPSPRADLPRSLSPKPFDRSKPSSASANAP- 174
Query: 59 PTAASASTAVATENFTSRFSLDEPR 83
PT AST V ++ T S+ PR
Sbjct: 175 PTLRPASTRVPSQRITPH-SVPSPR 198
>At1g44900 transcription factor, putative
Length = 928
Score = 30.4 bits (67), Expect = 3.3
Identities = 64/266 (24%), Positives = 105/266 (39%), Gaps = 54/266 (20%)
Query: 234 GPVLIDIPKDIQ---QQLAVPNWDQPIKLPGYMARLPRSPSDALLEQVVRLLLECKKPVL 290
GP +++ + I Q+L + + P +P RLPR LL L++C +P
Sbjct: 377 GPFTVNVEQTIYRNYQKLTIQ--ESPGTVPA--GRLPRHKEVILLND----LIDCARP-- 426
Query: 291 YAGGGSLNSSEELRRFVELTGVPVASTLMGLGAYPTADENSLQMLGMHGTVYANYAVDKS 350
EE +E+TG+ + + L +N + V ANY K
Sbjct: 427 ---------GEE----IEVTGIYTNNFDLSLNT-----KNGFPVFAT--VVEANYVTKKQ 466
Query: 351 DLLLAFGVRFDDRVTGKLEAFASRAKIVHIDIDSAEIGKNKQPHVSVCADLKLALKGINR 410
DL A+ + +D+ ++E + +IV I S I + H + L LA+ G
Sbjct: 467 DLFSAYKLTQEDKT--QIEELSKDPRIVERIIKS--IAPSIYGHEDIKTALALAMFG--- 519
Query: 411 ILESRGVKGKLDFRAWREELNEQKLRFPLQFKTFEEAIPPQYAIQVLDELTNGNAIISTG 470
+ + +KGK R ++N L P K+ Q+ V E T A+ +TG
Sbjct: 520 -GQEKNIKGK---HRLRGDINVLLLGDPGTAKS-------QFLKYV--EKTGQRAVYTTG 566
Query: 471 VGQHQM-WAAQFYKYKRPRQWLTSSG 495
G + A +K R+W G
Sbjct: 567 KGASAVGLTAAVHKDPVTREWTLEGG 592
>At3g62980 transport inhibitor response 1 (TIR1)
Length = 594
Score = 30.0 bits (66), Expect = 4.3
Identities = 28/108 (25%), Positives = 49/108 (44%), Gaps = 6/108 (5%)
Query: 71 ENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQA--LTRSATIRNVLPR 128
+ +TS SL+ +++ ALER V P S+++++A L + AT+ P+
Sbjct: 181 DTYTSLVSLNISCLASEVSFSALERL----VTRCPNLKSLKLNRAVPLEKLATLLQRAPQ 236
Query: 129 HEQGGIFAAEGYARSSGLPGVCIATSGPGATNLVSGLADALLDSVPLI 176
E+ G R G+ +A SG +SG DA+ +P +
Sbjct: 237 LEELGTGGYTAEVRPDVYSGLSVALSGCKELRCLSGFWDAVPAYLPAV 284
>At2g16470 unknown protein
Length = 670
Score = 30.0 bits (66), Expect = 4.3
Identities = 28/122 (22%), Positives = 53/122 (42%), Gaps = 6/122 (4%)
Query: 6 AAKLAFTTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTAASAS 65
AA++A TT+ P + ++P P+ ++P ++ R+ + S + P+P +A+ S
Sbjct: 283 AARIAPTTIEIPRNSQDTWSQGGSLPSPTPNQITTPTAKRRNFESRWSPTKPSPQSANQS 342
Query: 66 TAVATENFTSRFSLDEPRKGADILVEALERQGVTDVFAYPGGASMEIHQALTRSATIRNV 125
N++ S DI V + G YP I+ ++ SAT+ +
Sbjct: 343 M-----NYSVAQSGQSQTSRIDIPV-VVNSAGALQPQTYPIPTPDPINVSVNHSATLHSP 396
Query: 126 LP 127
P
Sbjct: 397 TP 398
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 29.6 bits (65), Expect = 5.6
Identities = 25/75 (33%), Positives = 31/75 (41%), Gaps = 4/75 (5%)
Query: 2 AAAPAAKLAFTTVPSPSSRPQNHVLKFTIPFPSYPNNSSPHSRCRSLQISASLSNPTPTA 61
A +P+ A PSPSS P P P P + SP S Q +A TP A
Sbjct: 266 AHSPSHSPATPKSPSPSSSPAQSP---ATPSPMTPQSPSPVSSPSPDQSAAPSDQSTPLA 322
Query: 62 ASAS-TAVATENFTS 75
S S T +N T+
Sbjct: 323 PSPSETTPTADNITA 337
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,625,111
Number of Sequences: 26719
Number of extensions: 636633
Number of successful extensions: 1649
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1628
Number of HSP's gapped (non-prelim): 28
length of query: 656
length of database: 11,318,596
effective HSP length: 106
effective length of query: 550
effective length of database: 8,486,382
effective search space: 4667510100
effective search space used: 4667510100
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0336.21


