

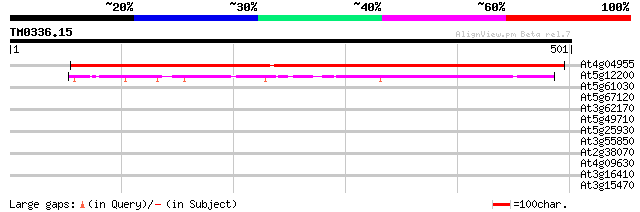
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.15
(501 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g04955 Dihydroorotase like protein 646 0.0
At5g12200 dihydropyrimidinase like protein 95 1e-19
At5g61030 RNA-binding protein - like 30 3.1
At5g67120 putative protein 30 4.1
At3g62170 PECTINESTERASE-like protein 30 4.1
At5g49710 putative protein 29 6.9
At5g25930 receptor-like protein kinase - like 29 6.9
At3g55850 unknown protein 29 6.9
At2g38070 unknown protein 29 6.9
At4g09630 putative protein 28 9.0
At3g16410 putative lectin 28 9.0
At3g15470 unknown protein 28 9.0
>At4g04955 Dihydroorotase like protein
Length = 506
Score = 646 bits (1666), Expect = 0.0
Identities = 309/442 (69%), Positives = 376/442 (84%), Gaps = 3/442 (0%)
Query: 55 SEDLEINEGKIVSITEGYGKQGNSKQEV-VIDYGEAVIMPGLIDVHVHLDEPGRTEWEGF 113
S +E+ G IVS+ + + + V VIDYGEAV+MPGLIDVHVHLD+PGR+EWEGF
Sbjct: 60 SGSVEVKGGIIVSVVKEVDWHKSQRSRVKVIDYGEAVLMPGLIDVHVHLDDPGRSEWEGF 119
Query: 114 VTGTRAAAAGGVTTVVDMPLNNHPTTVSQETLQLKLEAAENELHVDVGFWGGLVPENALN 173
+GT+AAAAGG+TT+VDMPLN+ P+TVS ETL+LK+EAA+N +HVDVGFWGGLVP+NALN
Sbjct: 120 PSGTKAAAAGGITTLVDMPLNSFPSTVSPETLKLKIEAAKNRIHVDVGFWGGLVPDNALN 179
Query: 174 TSILEGLLSAGVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPLLVHAEIQQDSK 233
+S LE LL AGVLGLKSFMCPSGINDFPMT I HIKEGLSVLAKY+RPLLVHAEI++D +
Sbjct: 180 SSALESLLDAGVLGLKSFMCPSGINDFPMTNITHIKEGLSVLAKYKRPLLVHAEIERDLE 239
Query: 234 NNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGPLEGAHVHIVHLSDSS 293
+E +DPR Y TYL TRP SWEE AI+ L+ VT++T GG EGAH+HIVHLSD+S
Sbjct: 240 --IEDGSENDPRSYLTYLKTRPTSWEEGAIRNLLSVTENTRIGGSAEGAHLHIVHLSDAS 297
Query: 294 VSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGDTRFKCSPPIRDAFNKEKLWEAVLE 353
SLDLIKEAK +GDS++VETC HYLAFS+EEIP+GDTRFKCSPPIRDA N+EKLWEA++E
Sbjct: 298 SSLDLIKEAKGKGDSVTVETCPHYLAFSAEEIPEGDTRFKCSPPIRDAANREKLWEALME 357
Query: 354 GHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPVTWSYGKKHGLTLEQLSLLW 413
G ID+L+SDHSPT PELKL+ +G+FLKAWGGISSLQF LP+TWSYGKK+G+TLEQ++ W
Sbjct: 358 GDIDMLSSDHSPTKPELKLMSDGNFLKAWGGISSLQFVLPITWSYGKKYGVTLEQVTSWW 417
Query: 414 SQKPATLAGIASKGAIVVGNHADIVVWHPELEFELNDDYPVFLKHPNLSAYMGRRLSGKV 473
S +P+ LAG+ SKGA+ VG HAD+VVW PE EF++++D+P+ KHP++SAY+GRRLSGKV
Sbjct: 418 SDRPSKLAGLHSKGAVTVGKHADLVVWEPEAEFDVDEDHPIHFKHPSISAYLGRRLSGKV 477
Query: 474 LDTFVRGNLVFKDGKHAPAACG 495
+ TFVRGNLVF +GKHA ACG
Sbjct: 478 VSTFVRGNLVFGEGKHASDACG 499
>At5g12200 dihydropyrimidinase like protein
Length = 531
Score = 94.7 bits (234), Expect = 1e-19
Identities = 119/454 (26%), Positives = 199/454 (43%), Gaps = 48/454 (10%)
Query: 53 NHSE--DLEINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHL--DEPGRT 108
+H E D+ + G IV++ K G+ + V+D +MPG ID H HL + G
Sbjct: 58 HHQELADVYVENGIIVAVQPNI-KVGD--EVTVLDATGKFVMPGGIDPHTHLAMEFMGTE 114
Query: 109 EWEGFVTGTRAAAAGGVTTVVD--MPLNNHPTTVSQETLQLKLEAAEN---ELHVDVGFW 163
+ F +G AA AGG T +D +P+N + L EA EN E +D GF
Sbjct: 115 TIDDFFSGQAAALAGGTTMHIDFVIPVNGN--------LVAGFEAYENKSRESCMDYGFH 166
Query: 164 GGLVPENALNTSILEGLLSA-GVLGLKSFMCPSGINDFPMTTIDHIKEGLSVLAKYRRPL 222
+ + + +E L+ G+ K F+ G M T D + EGL
Sbjct: 167 MAITKWDEGVSRDMEMLVKEKGINSFKFFLAYKGSL---MVTDDLLLEGLKRCKSLGALA 223
Query: 223 LVHAE----IQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGP 278
+VHAE + + K +EL G P ++ +RPP E A + + +
Sbjct: 224 MVHAENGDAVFEGQKRMIEL-GITGPEGHAL---SRPPVLEGEATARAIRLAR------- 272
Query: 279 LEGAHVHIVHLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDGD----TRFKC 334
+++VH+ S ++D I +A+ G + E L + D D +++
Sbjct: 273 FINTPLYVVHVM-SVDAMDEIAKARKSGQKVIGEPVVSGLILDDHWLWDPDFTIASKYVM 331
Query: 335 SPPIRDAFNKEKLWEAVLEGHIDLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDLPV 394
SPPIR + + L +A+ G + L+ +DH K L DF + G++ L+ + +
Sbjct: 332 SPPIRPVGHGKALQDALSTGILQLVGTDHCTFNSTQKALGLDDFRRIPNGVNGLEERMHL 391
Query: 395 TWSYGKKHG-LTLEQLSLLWSQKPATLAGI-ASKGAIVVGNHADIVVWHPELEFELNDDY 452
W + G L+ + S + A + I KGAI+ G+ ADI++ +P +E++
Sbjct: 392 IWDTMVESGQLSATDYVRITSTECARIFNIYPRKGAILAGSDADIIILNPNSSYEISS-- 449
Query: 453 PVFLKHPNLSAYMGRRLSGKVLDTFVRGNLVFKD 486
+ + Y GRR GKV T G +V+++
Sbjct: 450 KSHHSRSDTNVYEGRRGKGKVEVTIAGGRIVWEN 483
>At5g61030 RNA-binding protein - like
Length = 309
Score = 30.0 bits (66), Expect = 3.1
Identities = 22/60 (36%), Positives = 27/60 (44%), Gaps = 7/60 (11%)
Query: 72 YGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLD-EPGRTEWEGFVTGTRAAAAGGVTTVVD 130
Y +S +E YGE V D V LD E GR+ GFVT T + AA +D
Sbjct: 49 YSMDEDSLREAFTKYGEVV------DTRVILDRETGRSRGFGFVTFTSSEAASSAIQALD 102
>At5g67120 putative protein
Length = 272
Score = 29.6 bits (65), Expect = 4.1
Identities = 22/73 (30%), Positives = 32/73 (43%), Gaps = 6/73 (8%)
Query: 19 SPHKGSFLDQGETCEQLLRVGID------FEKRKADPYTLNHSEDLEINEGKIVSITEGY 72
S H G+F+ + ET EQ R + FE + YT S +++SIT+
Sbjct: 136 SSHGGTFIYEEETLEQYWRNWLQSSTNEQFETESLEEYTNPSSHGDIFTYEELLSITDET 195
Query: 73 GKQGNSKQEVVID 85
G + E VID
Sbjct: 196 GDERTGLSEEVID 208
>At3g62170 PECTINESTERASE-like protein
Length = 588
Score = 29.6 bits (65), Expect = 4.1
Identities = 20/83 (24%), Positives = 35/83 (42%)
Query: 24 SFLDQGETCEQLLRVGIDFEKRKADPYTLNHSEDLEINEGKIVSITEGYGKQGNSKQEVV 83
S DQG + L V D + + + + + + S G G N+ + V
Sbjct: 54 STTDQGSCAKTLDPVKSDDPSKLVKAFLMATKDAITKSSNFTASTEGGMGTNMNATSKAV 113
Query: 84 IDYGEAVIMPGLIDVHVHLDEPG 106
+DY + V+M L D+ ++E G
Sbjct: 114 LDYCKRVLMYALEDLETIVEEMG 136
>At5g49710 putative protein
Length = 208
Score = 28.9 bits (63), Expect = 6.9
Identities = 27/110 (24%), Positives = 46/110 (41%), Gaps = 10/110 (9%)
Query: 224 VHAEIQQDSKNNLELKGNDDPRVYSTYLNTRPPSWEEAAIKELVDVTKDTIHGGP----L 279
V +E +D K+ E G+ + R + + P +E L D+ K + +
Sbjct: 43 VSSEDDEDDKSTGEGSGSQESRGEVSSSDREDPEADE-----LYDLIKSRVECDDFLEKI 97
Query: 280 EGAHVHIV-HLSDSSVSLDLIKEAKLRGDSISVETCSHYLAFSSEEIPDG 328
E A V HL++ S S D++ E L D + Y+ E+I +G
Sbjct: 98 ESAQVSAPQHLAEDSSSWDVVSEDDLWDDETMAQREEDYVLVREEDIAEG 147
>At5g25930 receptor-like protein kinase - like
Length = 1005
Score = 28.9 bits (63), Expect = 6.9
Identities = 13/29 (44%), Positives = 17/29 (57%)
Query: 396 WSYGKKHGLTLEQLSLLWSQKPATLAGIA 424
W +GKK G T+E +L WSQ+ G A
Sbjct: 773 WLHGKKKGGTVEANNLTWSQRLNIAVGAA 801
>At3g55850 unknown protein
Length = 576
Score = 28.9 bits (63), Expect = 6.9
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 5/59 (8%)
Query: 51 TLNHSEDLEINEGKIV---SITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPG 106
+L ++ + I G+I+ S G G+ EV ++ +++PGLID HVHL G
Sbjct: 57 SLPFADSMAIRNGRILKVGSFATLKGFIGDGTMEVNLE--GKIVVPGLIDSHVHLISGG 113
>At2g38070 unknown protein
Length = 619
Score = 28.9 bits (63), Expect = 6.9
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 8/73 (10%)
Query: 71 GYGKQG-NSKQEVVIDYGEAVIMPGLIDV---HVHLDEPGRTEWEGFVTGTRAA---AAG 123
GYG++ ++ ID G + G + V +EP R W+G++ G AA
Sbjct: 301 GYGRRSCDTDPRFSIDAGRFSLDAGRVSVDDPRYSFEEP-RASWDGYLIGRAAAPMRMPS 359
Query: 124 GVTTVVDMPLNNH 136
++ V D P+ NH
Sbjct: 360 MLSVVEDSPVRNH 372
>At4g09630 putative protein
Length = 711
Score = 28.5 bits (62), Expect = 9.0
Identities = 28/96 (29%), Positives = 43/96 (44%), Gaps = 8/96 (8%)
Query: 303 KLRGDSISVETCSHYLAFSSEEIPDG-DTRFKCSPPIR-----DAFNKEKLWEAVLEGHI 356
+ R S +C L SS++I + T+ +P ++ D + E+ WE GH
Sbjct: 341 RARSHRFSGVSCEMKLLNSSQQIQEPLKTQNFAAPSLQYIQMEDKPDGEEQWEPKFAGHQ 400
Query: 357 DLLTSDHSPTVPELKLLKEGDFLKAWGGISSLQFDL 392
L + S V E K+ F+KA G+ S FDL
Sbjct: 401 SLQEREDSFLVQEQKI--HCGFVKAPEGLPSTGFDL 434
>At3g16410 putative lectin
Length = 619
Score = 28.5 bits (62), Expect = 9.0
Identities = 17/55 (30%), Positives = 29/55 (51%), Gaps = 7/55 (12%)
Query: 22 KGSFLDQGETCEQLLRVGIDFEKRKADPYTLNHSEDLEINEGKIVSITEGYGKQG 76
K +++ G+T E LR G+ +ADP+ +NH E+ +VS+ Y +G
Sbjct: 41 KFTYVKNGQTEEAPLR-GVKGRSFEADPFVINHPEE------HLVSVEGRYNPEG 88
>At3g15470 unknown protein
Length = 883
Score = 28.5 bits (62), Expect = 9.0
Identities = 23/90 (25%), Positives = 34/90 (37%), Gaps = 3/90 (3%)
Query: 50 YTLNHSEDLEINEGKIVSITEGYGKQGNSKQEVVIDYGEAVIMPGLIDVHVHLDEPGRTE 109
Y ++ SED + K S + N+K V + E DV + PG
Sbjct: 703 YVVSASEDSHVYIWKYESPASRPSRSNNNKNVTVTNSYEHFHSQ---DVSAAISWPGMAS 759
Query: 110 WEGFVTGTRAAAAGGVTTVVDMPLNNHPTT 139
E + T RA G + ++ NHP T
Sbjct: 760 TENWGTQNRAGFNGSTNNLDNISTANHPPT 789
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,940,461
Number of Sequences: 26719
Number of extensions: 537907
Number of successful extensions: 1297
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1288
Number of HSP's gapped (non-prelim): 12
length of query: 501
length of database: 11,318,596
effective HSP length: 103
effective length of query: 398
effective length of database: 8,566,539
effective search space: 3409482522
effective search space used: 3409482522
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0336.15


