

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0335b.2
(106 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
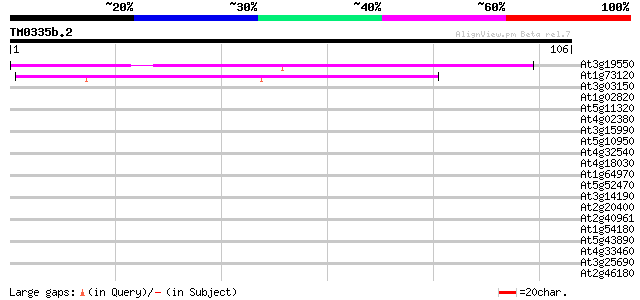
Score E
Sequences producing significant alignments: (bits) Value
At3g19550 unknown protein 80 2e-16
At1g73120 unknown protein 50 1e-07
At3g03150 unknown protein 37 0.002
At1g02820 unknown protein 33 0.031
At5g11320 putative protein 28 0.76
At4g02380 unknown protein 27 1.7
At3g15990 putative sulfate transporter 26 2.9
At5g10950 putative protein 26 3.8
At4g32540 dimethylaniline monooxygenase - like protein 26 3.8
At4g18030 unknown protein 26 3.8
At1g64970 unknown protein 26 3.8
At5g52470 fibrillarin homolog (Fbr1) 25 4.9
At3g14190 unknown protein 25 4.9
At2g20400 unknown protein 25 4.9
At2g40961 unknown protein 25 6.4
At1g54180 hypothetical protein 25 6.4
At5g43890 dimethylaniline monooxygenase-like 25 8.4
At4g33460 unknown protein 25 8.4
At3g25690 unknown protein 25 8.4
At2g46180 unknown protein 25 8.4
>At3g19550 unknown protein
Length = 110
Score = 79.7 bits (195), Expect = 2e-16
Identities = 45/102 (44%), Positives = 58/102 (56%), Gaps = 7/102 (6%)
Query: 1 MARGGITKSTLLILRGARRAENRVMKFSGTTAKAAAAESSEEGVPKIISG---KTEDSSA 57
M R GI K+ L+LR ++ + R +G ++KAA + + IS K
Sbjct: 1 MGRTGIAKAPKLLLRSWKQFQGR----AGISSKAAKSNPMIVDYFEDISDHNLKFSGEEE 56
Query: 58 IWVPHPRTGIYYPKGHECVMEDVPEGAARFTQTYWFRNDDGV 99
WVPHPRTGI++P G E VMEDVP GAA F T+W RN DGV
Sbjct: 57 SWVPHPRTGIFFPPGQESVMEDVPNGAASFDMTFWLRNVDGV 98
>At1g73120 unknown protein
Length = 109
Score = 50.4 bits (119), Expect = 1e-07
Identities = 31/86 (36%), Positives = 44/86 (51%), Gaps = 6/86 (6%)
Query: 2 ARGGITKSTLLI---LRGARRAENRVMKFSGTTAKAAAAESSEEGVPK---IISGKTEDS 55
+RG T +T+ + LR + ++ T K + S EE +G+T
Sbjct: 8 SRGAQTMNTMFVKPMLRKSIHKKSASHDIVRDTVKTEGSSSGEEVKTMRGFYGAGETSSP 67
Query: 56 SAIWVPHPRTGIYYPKGHECVMEDVP 81
++ WVPH TGIYYPKG E VM+DVP
Sbjct: 68 ASSWVPHEGTGIYYPKGQEKVMQDVP 93
>At3g03150 unknown protein
Length = 121
Score = 37.0 bits (84), Expect = 0.002
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 2/43 (4%)
Query: 54 DSSAIWVPHPRTGIYYPK--GHECVMEDVPEGAARFTQTYWFR 94
DS W PHP+TG++ P H E + A +T WFR
Sbjct: 66 DSDKYWSPHPKTGVFGPSTTEHSATAEGAHQDTAVLEETAWFR 108
>At1g02820 unknown protein
Length = 91
Score = 32.7 bits (73), Expect = 0.031
Identities = 22/72 (30%), Positives = 30/72 (41%), Gaps = 4/72 (5%)
Query: 16 GARRAENRVMKFS-GTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYYPKGHE 74
G+ + N V + AK A S K +G+ A WVP P+TG Y P E
Sbjct: 15 GSEKLSNAVFRRGFAAAAKTALDGSVSTAEMKKRAGEASSEKAPWVPDPKTGYYRP---E 71
Query: 75 CVMEDVPEGAAR 86
V E++ R
Sbjct: 72 TVSEEIDPAELR 83
>At5g11320 putative protein
Length = 411
Score = 28.1 bits (61), Expect = 0.76
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query: 28 SGTTAKAAAAESSEEGVPKIISGKTEDSSAIWV--PHPRTGIYYPKGHECVMEDVP 81
+G + A AA S GVP +I +T+ +++W + R ++ PK H C + +P
Sbjct: 22 AGPSGLAVAACLSNRGVPSVILERTDCLASLWQKRTYDRLKLHLPK-HFCELPLMP 76
>At4g02380 unknown protein
Length = 97
Score = 26.9 bits (58), Expect = 1.7
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Query: 9 STLLILRG-ARRAENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGI 67
S + RG A A + G + A+A ++GV + T+ S WVP P+TG
Sbjct: 20 SNAIFRRGYAATAAQGSVSSGGRSGAVASAVMKKKGVEE----STQKIS--WVPDPKTGY 73
Query: 68 YYPK 71
Y P+
Sbjct: 74 YRPE 77
>At3g15990 putative sulfate transporter
Length = 653
Score = 26.2 bits (56), Expect = 2.9
Identities = 12/21 (57%), Positives = 15/21 (71%)
Query: 13 ILRGARRAENRVMKFSGTTAK 33
ILR AR ENR+ + +GTT K
Sbjct: 551 ILRWAREEENRIKENNGTTLK 571
>At5g10950 putative protein
Length = 395
Score = 25.8 bits (55), Expect = 3.8
Identities = 14/41 (34%), Positives = 23/41 (55%), Gaps = 2/41 (4%)
Query: 17 ARRAENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSA 57
+ A+ +V+K + A A SS+E PK++ KT +S A
Sbjct: 233 SEEAKPKVLKSCNSNADEVAENSSDEDEPKVL--KTNNSKA 271
>At4g32540 dimethylaniline monooxygenase - like protein
Length = 414
Score = 25.8 bits (55), Expect = 3.8
Identities = 17/59 (28%), Positives = 31/59 (51%), Gaps = 5/59 (8%)
Query: 28 SGTTAKAAAAESSEEGVPKIISGKTEDSSAIW--VPHPRTGIYYPKGHECVME--DVPE 82
+G + A +A S GVP +I +++ +++W + R ++ PK H C + D PE
Sbjct: 26 AGPSGLATSACLSSRGVPSLILERSDSIASLWKSKTYDRLRLHLPK-HFCRLPLLDFPE 83
>At4g18030 unknown protein
Length = 621
Score = 25.8 bits (55), Expect = 3.8
Identities = 23/86 (26%), Positives = 38/86 (43%), Gaps = 9/86 (10%)
Query: 14 LRGARRAENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYYPKGH 73
L G+ R N + +G AAA ES + V +I +++ ++ GIY+
Sbjct: 454 LIGSTRYRNVMDMNAGLGGFAAALESPKSWVMNVIPTINKNTLSVVYERGLIGIYH---- 509
Query: 74 ECVMEDVPEGAARFTQTYWFRNDDGV 99
D EG + + +TY F + GV
Sbjct: 510 -----DWCEGFSTYPRTYDFIHASGV 530
>At1g64970 unknown protein
Length = 348
Score = 25.8 bits (55), Expect = 3.8
Identities = 19/67 (28%), Positives = 30/67 (44%), Gaps = 3/67 (4%)
Query: 7 TKSTLLILRGARRAENRVMKFSGTTAKAAAAESSE---EGVPKIISGKTEDSSAIWVPHP 63
+KS+LL + + + G A AAAA S+E +G+ + + + IW H
Sbjct: 24 SKSSLLFRSPSSSSSVSMTTTRGNVAVAAAATSTEALRKGIAEFYNETSGLWEEIWGDHM 83
Query: 64 RTGIYYP 70
G Y P
Sbjct: 84 HHGFYDP 90
>At5g52470 fibrillarin homolog (Fbr1)
Length = 308
Score = 25.4 bits (54), Expect = 4.9
Identities = 20/74 (27%), Positives = 28/74 (37%), Gaps = 5/74 (6%)
Query: 28 SGTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTGIYYPKGHECVMED---VPEGA 84
SG + A G P+ G S I PH G++ KG E + VP A
Sbjct: 42 SGPRGRGRGAPRGRGGPPR--GGMKGGSKVIVEPHRHAGVFIAKGKEDALVTKNLVPGEA 99
Query: 85 ARFTQTYWFRNDDG 98
+ +N+DG
Sbjct: 100 VYNEKRISVQNEDG 113
>At3g14190 unknown protein
Length = 193
Score = 25.4 bits (54), Expect = 4.9
Identities = 12/27 (44%), Positives = 18/27 (66%)
Query: 16 GARRAENRVMKFSGTTAKAAAAESSEE 42
GAR+A N + SG AKAAA+ +++
Sbjct: 44 GARKALNDITNKSGIHAKAAASSKNKQ 70
>At2g20400 unknown protein
Length = 397
Score = 25.4 bits (54), Expect = 4.9
Identities = 10/22 (45%), Positives = 13/22 (58%), Gaps = 1/22 (4%)
Query: 59 WVPHPRTGIYYPKGH-ECVMED 79
W+P P IY+P G +MED
Sbjct: 113 WIPSPLPHIYFPSGSPNLIMED 134
>At2g40961 unknown protein
Length = 351
Score = 25.0 bits (53), Expect = 6.4
Identities = 15/51 (29%), Positives = 24/51 (46%)
Query: 16 GARRAENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSSAIWVPHPRTG 66
GA E+R++ + ++ AA SE V + +GK E + P P G
Sbjct: 88 GADGNESRIVVTKSSDSRFPAARLSEIPVKQSETGKFEHMKVVIKPRPTKG 138
>At1g54180 hypothetical protein
Length = 170
Score = 25.0 bits (53), Expect = 6.4
Identities = 14/36 (38%), Positives = 17/36 (46%), Gaps = 7/36 (19%)
Query: 39 SSEEGVPKIISGKTE-------DSSAIWVPHPRTGI 67
SSEEG P +SG+TE D WV G+
Sbjct: 115 SSEEGTPTSMSGRTESIVFMEDDEVKEWVAQVEPGV 150
>At5g43890 dimethylaniline monooxygenase-like
Length = 424
Score = 24.6 bits (52), Expect = 8.4
Identities = 13/46 (28%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Query: 28 SGTTAKAAAAESSEEGVPKIISGKTEDSSAIWV--PHPRTGIYYPK 71
+G + A AA EEGVP ++ + + +++W + R ++ PK
Sbjct: 30 AGPSGLATAACLREEGVPFVVLERADCIASLWQKRTYDRIKLHLPK 75
>At4g33460 unknown protein
Length = 271
Score = 24.6 bits (52), Expect = 8.4
Identities = 14/39 (35%), Positives = 20/39 (50%), Gaps = 4/39 (10%)
Query: 38 ESSEEGVPK----IISGKTEDSSAIWVPHPRTGIYYPKG 72
ES + GV K +I+ K D +A+WV H + Y G
Sbjct: 198 ESDQMGVIKAVKDLINAKKGDVTALWVTHRLEELKYADG 236
>At3g25690 unknown protein
Length = 939
Score = 24.6 bits (52), Expect = 8.4
Identities = 11/20 (55%), Positives = 14/20 (70%)
Query: 38 ESSEEGVPKIISGKTEDSSA 57
ES +EG P +IS T +SSA
Sbjct: 670 ESKKEGAPSLISSGTGNSSA 689
>At2g46180 unknown protein
Length = 725
Score = 24.6 bits (52), Expect = 8.4
Identities = 15/52 (28%), Positives = 23/52 (43%)
Query: 5 GITKSTLLILRGARRAENRVMKFSGTTAKAAAAESSEEGVPKIISGKTEDSS 56
GI++ L + A EN +K + +AA S + K+ TED S
Sbjct: 278 GISRENLKEVNKALEKENNELKLKRSELEAALEASQKSTSRKLFPKSTEDLS 329
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.131 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,305,575
Number of Sequences: 26719
Number of extensions: 85497
Number of successful extensions: 227
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 215
Number of HSP's gapped (non-prelim): 21
length of query: 106
length of database: 11,318,596
effective HSP length: 82
effective length of query: 24
effective length of database: 9,127,638
effective search space: 219063312
effective search space used: 219063312
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0335b.2


