

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0335a.7
(632 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
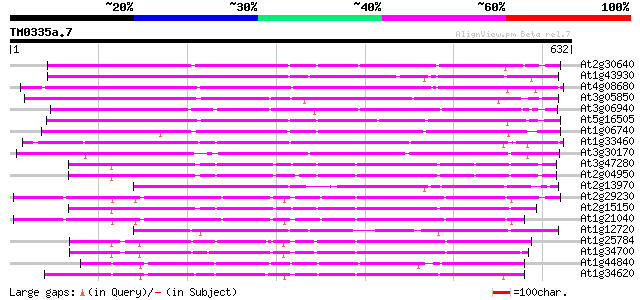
Score E
Sequences producing significant alignments: (bits) Value
At2g30640 Mutator-like transposase 266 3e-71
At1g43930 hypothetical protein 253 2e-67
At4g08680 putative MuDR-A-like transposon protein 245 6e-65
At3g05850 unknown protein 235 5e-62
At3g06940 putative mudrA protein 234 1e-61
At5g16505 mutator-like transposase-like protein (MQK4.25) 231 7e-61
At1g06740 mudrA-like protein 223 3e-58
At1g33460 mutator transposase MUDRA, putative 222 6e-58
At3g30170 unknown protein 207 2e-53
At3g47280 putative protein 202 5e-52
At2g04950 putative transposase of transposon FARE2.9 (cds1) 199 4e-51
At2g13970 Mutator-like transposase 195 6e-50
At2g29230 Mutator-like transposase 190 2e-48
At2g15150 putative transposase of FARE2.2 (CDS1) 188 9e-48
At1g21040 hypothetical protein 176 4e-44
At1g12720 hypothetical protein 174 1e-43
At1g25784 mutator-like transposase, putative 173 3e-43
At1g34700 hypothetical protein 169 4e-42
At1g44840 hypothetical protein 169 6e-42
At1g34620 Mutator-like protein 165 8e-41
>At2g30640 Mutator-like transposase
Length = 754
Score = 266 bits (679), Expect = 3e-71
Identities = 169/591 (28%), Positives = 286/591 (47%), Gaps = 31/591 (5%)
Query: 43 LGMDFSSLLQFKDALHDHFVTIGREFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETF 102
+GM+FS +L + AL D + + E + +K+D R KC S+GCP+ I C+K+ TF
Sbjct: 182 VGMEFSDVLACRRALRDAAIALRFEMQTVKSDKTRFTAKCNSEGCPWRIHCAKLPGLPTF 241
Query: 103 RIKTLVGEHNCARV--FDNKSANVRWVKLKLLNKVSTMNKISTNEIVDDFRSNHGIGITR 160
I+T+ G H C + + A+V+WV + ++ EI++ HGI +T
Sbjct: 242 TIRTIHGSHTCGGISHLGHHQASVQWVADAVTERLKVNPHCKPKEILEQIHQVHGITLTY 301
Query: 161 YRAWKGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFY 220
+AW+GK+ + V G+ E Y LL R+ +++R+ G+ ++ S + F +F+
Sbjct: 302 KQAWRGKERIMAAVRGSYEEDYRLLPRYCDQIRRTNPGSVAVVH----GSPVDGSFQQFF 357
Query: 221 MCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDS 280
+ GFL CRP IGLD LK+K+ G LL A D FPLAF ++ E S
Sbjct: 358 ISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGAVFPLAFAIISEENDSS 417
Query: 281 WMWFMELLMD--DIDPERSRRWVFISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKTK 338
W WF+ L +++ E + +S + + +++ + + H LCV L +++T+
Sbjct: 418 WQWFLSELRQLLEVNSENMPKLTILSSRDQSIVDGV-DTNFPTAFHGLCVHCLTESVRTQ 476
Query: 339 FGGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRC 398
F +L+ +L+ AAK + ++ KM I + A W+ S W + F +R
Sbjct: 477 FNNSILV-NLVWEAAKCLTDFEFEGKMGEIAQISPEAASWIRNIQHSQWATYCFE-GTRF 534
Query: 399 DVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRI 458
+ N+SE+ N+ + A PI+ M+E IR +M F + E + G ++P + +
Sbjct: 535 GHLTANVSESLNSWVQDASGLPIIQMLESIRRQLMTLFNERRETSMQWSGMLVPSAERHV 594
Query: 459 LHEVDHIKNWIPRVVGAQKYEVRHTITLE-RHVISLKDKSCTCRFWELNGFPCRHAASAL 517
L ++ + + ++EV +T E + ++ ++ ++C CR WEL G PC HA +AL
Sbjct: 595 LEAIEECRLYPVHKANEAQFEV---MTSEGKWIVDIRCRTCYCRGWELYGLPCSHAVAAL 651
Query: 518 TFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQWIVT--------PHDPVCILPPMY 569
+ +F + + Y TY + P+P +W T + + I PP
Sbjct: 652 LACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESGEDGEEIVIRPPRD 711
Query: 570 KRSAGRPKKLRRRDPYENDPNPTKLQRGGATWKCSRCKQYGHNKRGCKNPV 620
R RPKK RR E+ ++ R CSRC Q GH + C P+
Sbjct: 712 LRQPPRPKK--RRSQGEDRGRQKRVVR------CSRCNQAGHFRTTCTAPI 754
>At1g43930 hypothetical protein
Length = 946
Score = 253 bits (646), Expect = 2e-67
Identities = 168/592 (28%), Positives = 284/592 (47%), Gaps = 28/592 (4%)
Query: 43 LGMDFSSLLQFKDALHDHFVTIGREFEYMKNDSIRCRVKCRSK--GCPFVILCSKVGNKE 100
L F++ FK A+ + + + + ++ S+R KC C + CS K+
Sbjct: 239 LAKTFNNPEDFKIAVLRYSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQ 298
Query: 101 TFRIKTLVGEHNCARVFDNKSANVRWVKLKLLNKVSTMNKISTNEIVDDFRSNHGIGITR 160
++K H+C R +K R + ++ KI+ E+VD+ + + + +T
Sbjct: 299 KMQVKVYKDGHSCVRSGYSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYKLTVTE 358
Query: 161 YRAWKGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGNTCKIN-FESPRSTLFPRFSRF 219
+ K K + ++E + + + +W + ++ + G T +I P RF R
Sbjct: 359 DQCSKAKTMVMRERQATHQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQRFLRL 418
Query: 220 YMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKD 279
++C K + CRP IG+DG LK +G LLAA RD + + P+A+ VVE E D
Sbjct: 419 FICFKSQKESWKQTCRPIIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVEIENDD 478
Query: 280 SWMWFMELLMDDIDPERSRRWVFISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKTKF 339
+W WF+ +L +D + R ISD+Q GL++ ++ +EHR C RH+ N K +
Sbjct: 479 NWDWFVRMLSRTLDLQDGRNVAIISDKQSGLVKAI-HSVIPQAEHRQCARHIMENWK-RN 536
Query: 340 GGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCD 399
+ L+ L ++ E + M +K N SA++ L++ +W R F + S C+
Sbjct: 537 SHDMELQRLFWKIVRSYTEGEFGAHMRALKSYNASAFELLLKTLPVTWSRAFFKIGSCCN 596
Query: 400 VVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRIL 459
+NNLSE+FN I AR KP+L M+E IR M R EK + ++ R KR
Sbjct: 597 DNLNNLSESFNRTIREARRKPLLDMLEDIRRQCMVR----NEKRYVIADRLRTRFTKRAH 652
Query: 460 HEVDHI-------KNWIPRVVGAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRH 512
E++ + + W+ R K+E++ ++ + + + +C C W++ G PC H
Sbjct: 653 AEIEKMIAGSQVCQRWMAR---HNKHEIK-VGPVDSFTVDMNNNTCGCMKWQMTGIPCIH 708
Query: 513 AASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQWIVTPHDPVCILPPMYKR- 571
AAS + + +V D Y + TY + + P+ G W + V +LPP ++R
Sbjct: 709 AASVIIGKRQKVEDYVSDWYTTSMWKQTYNDGIGPVQGKLLWPTV--NKVGVLPPPWRRG 766
Query: 572 SAGRPKK-LRRRDPYE----NDPNPTKLQRGGATWKCSRCKQYGHNKRGCKN 618
+ GRPK RR+ +E + + T+L R CS C+ GHNK+GCKN
Sbjct: 767 NPGRPKNHARRKGVFESSTASSSSTTELSRLHRVMTCSNCQGEGHNKQGCKN 818
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 245 bits (625), Expect = 6e-65
Identities = 164/627 (26%), Positives = 305/627 (48%), Gaps = 24/627 (3%)
Query: 13 VPTDSEVEDVPRRRFPKFVKEKMGQDFKFSLGMDFSSLLQFKDALHDHFVTIGREFEYMK 72
+P S+ ++ P + ++ MG KF +G F + ++FK+A+ + + G +
Sbjct: 83 IPESSDEDEDPILVRDRRIRRAMGD--KFVIGSTFFTGIEFKEAILEVTLKHGHNVVQDR 140
Query: 73 NDSIRCRVKCRSKG-CPFVILCSKVGNKETFRIKTLVGEHNCARVFDNKSANVRWVKLKL 131
+ + KC G C + + CS +++ F +KT H+C+ + +
Sbjct: 141 WEKEKISFKCGMGGKCKWRVYCSYDPDRQLFVVKTSCTWHSCSPNGKCQILKSPVIARLF 200
Query: 132 LNKVSTMNKISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFSNE 191
L+K+ K +I + + + T + +G+ LA++ ++ E Q+ + + E
Sbjct: 201 LDKLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLALKMLKKEYEEQFAHIRGYVEE 260
Query: 192 LRKRAAGNTCKIN-FESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHR 250
+ + G+ I+ + + + F+RFY+C + + + CRP IGLDG LK +
Sbjct: 261 IHSQNPGSVAFIDTYRNEKGE--DVFNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVK 318
Query: 251 GILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQKGL 310
G+LL AV D N Q +P+A+ VV+SE ++W+WF++ + D++ E R+V +SD+ KGL
Sbjct: 319 GVLLTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGL 378
Query: 311 MEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKD 370
+ K++ L +EHR+CV+H+ N+K +L+ L+ A + E + + + ++
Sbjct: 379 LSAVKQE-LPNAEHRMCVKHIVENLKKNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRC 437
Query: 371 ANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRT 430
+E+ Y ++ + +W R + L S C+ V NN +E+FN+ I AR K ++ M+E IR
Sbjct: 438 YDEALYNDVLNEEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRR 497
Query: 431 YVMGRFPQMMEKLHKHHGQIMPRPLKRI-LHEVDHIKNWIPRVVGAQKYEVRHTITLERH 489
M R + +K +H G+ LK + L + D ++ + R +EV I H
Sbjct: 498 QGMTRIVKRNKKSLRHEGRFTKYALKMLALEKTDADRSKVYRCTHG-VFEV--YIDENGH 554
Query: 490 VISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLP 549
+ + C+C W+++G PC H+ A+ G D + ++ + + + + +YE PL
Sbjct: 555 RVDIPKTQCSCGKWQISGIPCEHSYGAMIEAGLDAENYISEFFSTDLWRDSYETATMPLR 614
Query: 550 GPNQWIVTPH-----DPVCILPPMYKRSAGRPKKLRRRDPYENDPNP-------TKLQRG 597
GP W+ + + P ILP K + + K R + E+ KL +
Sbjct: 615 GPKYWLNSSYGLVTAPPEPILPGRKKEKSKKEKFARIKGKNESPKKKKRKKNEVEKLGKK 674
Query: 598 GATWKCSRCKQYGHNKRGCKNPVPEEQ 624
G C C + GHN CK P+E+
Sbjct: 675 GKIIHCKSCGEAGHNALRCKK-FPKEK 700
>At3g05850 unknown protein
Length = 777
Score = 235 bits (600), Expect = 5e-62
Identities = 161/615 (26%), Positives = 286/615 (46%), Gaps = 33/615 (5%)
Query: 17 SEVEDVPRRRFPKFVKEK--MGQDFKFSLGMDFSSLLQFKDALHDHFVTIGREFEYMKND 74
S + +VP + K + Q+ +G F ++ +F++AL + + F Y KND
Sbjct: 178 SSIVEVPNPKGDILTKARTQQWQNTITGVGQRFKNVGEFREALRKYAIANQFGFRYKKND 237
Query: 75 SIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNC--ARVFDNKSANVRWVKLKLL 132
S R VKC+++GCP+ I S++ + IK + H C A + + WV +
Sbjct: 238 SHRVTVKCKAEGCPWRIHASRLSTTQLICIKKMNPTHTCEGAGGINGLQTSRSWVASIIK 297
Query: 133 NKVSTMNKISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFSNEL 192
K+ +IV D + +GI + ++AW+GK++A ++++G+ + Y L F ++
Sbjct: 298 EKLKVFPNYKPKDIVSDIKEEYGIQLNYFQAWRGKEIAREQLQGSYKDGYKQLPLFCEKI 357
Query: 193 RKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGI 252
+ G+ + S F R ++ GFL CRP + LD LK+K++G
Sbjct: 358 METNPGSLATFTTKEDSS-----FHRVFVSFHASVHGFLEACRPLVFLDSMQLKSKYQGT 412
Query: 253 LLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQKGLME 312
LLAA + D +++ FPLAF VV++ET D+W WF+ L + F++D+QK L E
Sbjct: 413 LLAATSVDGDDEVFPLAFAVVDAETDDNWEWFLLQLRSLL--STPCYITFVADRQKNLQE 470
Query: 313 VFKEDMLHGSEHRLCVRH----LYANMKTKFGGGV--LLRDLMMGAAKATYEAAWKEKML 366
+ + S H C+R+ L ++K F + L+ D AA A +++ +
Sbjct: 471 SIPK-VFEKSFHAYCLRYLTDELIKDLKGPFSHEIKRLIVDDFYSAAYAPRADSFERHVE 529
Query: 367 LIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMME 426
IK + AY W+++K +A+ +R + + ++ E F + A D PI M++
Sbjct: 530 NIKGLSPEAYDWIVQKSQPDHWANAYFRGARYNHMTSHSGEPFFSWASDANDLPITQMVD 589
Query: 427 WIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRVVGAQKYEVRHTITL 486
IR +MG ++ +G + P + E + ++ V + +
Sbjct: 590 VIRGKIMGLIHVRRISANEANGNLTPS--MEVKLEKESLRAQTVHVAPSADNNLFQVRGE 647
Query: 487 ERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVT 546
++++ + C+C+ W+L G PC HA + + + G +P + + Y +TY +
Sbjct: 648 TYELVNMAECDCSCKGWQLTGLPCHHAVAVINYYGRNPYDYCSKYFTVAYYRSTYAQSIN 707
Query: 547 PLP---GPNQWIVTPHDPVCILPPMYKRSAGRPKKLRRRDPYENDPNPTKLQRGGATWKC 603
P+P G + V + PP +R GRP P + P ++R +C
Sbjct: 708 PVPLLEGEMCRESSGGSAVTVTPPPTRRPPGRP-------PKKKTPAEEVMKR---QLQC 757
Query: 604 SRCKQYGHNKRGCKN 618
SRCK GHNK CK+
Sbjct: 758 SRCKGLGHNKSTCKD 772
>At3g06940 putative mudrA protein
Length = 749
Score = 234 bits (596), Expect = 1e-61
Identities = 154/583 (26%), Positives = 272/583 (46%), Gaps = 33/583 (5%)
Query: 47 FSSLLQFKDALHDHFVTIGREFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKT 106
F++ L+F+DALH + V G ++Y KNDS R VKC+++GCP+ I S++ + IK
Sbjct: 186 FNTFLEFRDALHKYSVAHGFAYKYKKNDSHRVSVKCKAQGCPWRITASRLSTTQLICIKK 245
Query: 107 LVGEHNCAR--VFDNKSANVRWVKLKLLNKVSTMNKISTNEIVDDFRSNHGIGITRYRAW 164
+ H C R + A WV+ L K+ +I +D + +GI + +AW
Sbjct: 246 MNPRHTCERAVIKPGYRATRGWVRTILKEKLKAFPDYKPKDIAEDIKKEYGIQLNYSQAW 305
Query: 165 KGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPR-FSRFYMCL 223
+ K++A ++++G+ + Y+ L ++++ G+ F + + F R F FY +
Sbjct: 306 RAKEIAREQLQGSYKEAYSQLPLICEKIKETNPGSIA--TFMTKEDSSFHRLFISFYASI 363
Query: 224 DGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMW 283
G K+G RP + LD L +K++G++L A A DA + FP+AF +V+SET+++W+W
Sbjct: 364 SGFKQG----SRPLLFLDNAILNSKYQGVMLVATASDAEDGIFPVAFAIVDSETEENWLW 419
Query: 284 FMELLMDDIDPERSRRWVFISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGG-- 341
F+E L + SR F++D Q GL + + H C+ L + G
Sbjct: 420 FLEQLKTAL--SESRIITFVADFQNGLKNAIAQVFEKDAHHAYCLGQLAEKLNVDLKGQF 477
Query: 342 -----GVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYS 396
+L D A AT + + IK + AY W++E W F
Sbjct: 478 SHEARRYMLND-FYSVAYATTPVGYYLALENIKSISPDAYNWVIESEPHHWANALFQ-GE 535
Query: 397 RCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLK 456
R + + +N + F + + A + PI M++ R +M K + + P +
Sbjct: 536 RYNKMNSNFGKDFYSWVSEAHEFPITQMIDEFRAKMMQSIYTRQVKSREWVTTLTPSNEE 595
Query: 457 RILHEVDHIKNWIPRVVGAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRHAASA 516
++ E++ + ++V + ++ + C C+ W L G PC HA +
Sbjct: 596 KLQKEIELAQLLQVSSPEGSLFQVNGGES-SVSIVDINQCDCDCKTWRLTGLPCSHAIAV 654
Query: 517 LTFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQWIV--TPHDPVCILPPMYKRSAG 574
+ P ++ E++ Y + P+P ++ ++ P VC+ PP +R+ G
Sbjct: 655 IGCIEKSPYEYCSTYLTVESHRLMYAESIQPVPNMDRMMLDDPPEGLVCVTPPPTRRTPG 714
Query: 575 RPKKLRRRDPYENDPNPTKLQRGGATWKCSRCKQYGHNKRGCK 617
RP K+++ +P D +LQ CS+CK GHNK+ CK
Sbjct: 715 RP-KIKKVEPL--DMMKRQLQ-------CSKCKGLGHNKKTCK 747
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 231 bits (590), Expect = 7e-61
Identities = 150/589 (25%), Positives = 272/589 (45%), Gaps = 27/589 (4%)
Query: 42 SLGMDFSSLLQFKDALHDHFVTIGREFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKET 101
++G +F + + L D + + + +K+D R KC +GCP+ I +K +T
Sbjct: 26 AVGKEFPDVETCRRTLKDLAIALHFDLRIVKSDRSRFIAKCSKEGCPWRIHAAKCPGVQT 85
Query: 102 FRIKTLVGEHNCARVFD--NKSANVRWVKLKLLNKVSTMNKISTNEIVDDFRSNHGIGIT 159
F ++TL EH C V D ++ A+V WV + ++ + EI+ D R HG+ ++
Sbjct: 86 FTVRTLNSEHTCEGVRDLHHQQASVGWVARSVEARIRDNPQYKPKEILQDIRDEHGVAVS 145
Query: 160 RYRAWKGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRF 219
+AW+GK+ ++ + G E Y L + +++ G+ ++ P + F R
Sbjct: 146 YMQAWRGKERSMAALHGTYEEGYRFLPAYCEQIKLVNPGSFASVSALGPENC----FQRL 201
Query: 220 YMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKD 279
++ C GF + CRP + LD HLK K+ G +L A A DA++ FPLA +V++E+ +
Sbjct: 202 FIAYRACISGFFSSCRPLLELDRAHLKGKYLGAILCAAAVDADDGLFPLAIAIVDNESDE 261
Query: 280 SWMWFMELL--MDDIDPERSRRWVFISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKT 337
+W WF+ L + ++ + + +S++Q ++E E + H C+R++ N +
Sbjct: 262 NWSWFLSELRKLLGMNTDSMPKLTILSERQSAVVEAV-ETHFPTAFHGFCLRYVSENFRD 320
Query: 338 KFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSR 397
F L+ ++ A A A ++ K + + ++ QW W F
Sbjct: 321 TFKNTKLV-NIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAVAYFQGVRY 379
Query: 398 CDVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKR 457
+ +N A+ + PI+ MME IR + F E + ++P +R
Sbjct: 380 GHFGLGITEVLYNWALE-CHELPIIQMMEHIRHQISSWFDNRRELSMGWNSILVPSAERR 438
Query: 458 ILHEVDHIKNWIPRVVGAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRHAASAL 517
I V + + +V+ A + E T +++ ++ + C+CR W++ G PC HAA+AL
Sbjct: 439 ITEAVADARCY--QVLRANEVEFEIVSTERTNIVDIRTRDCSCRRWQIYGLPCAHAAAAL 496
Query: 518 TFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQWIVTPH------DPVCILPPMYKR 571
G + F + C+ +Y TY + +P + W + + I PP +R
Sbjct: 497 ISCGRNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESLRIRPPKTRR 556
Query: 572 SAGRPKKLRRRDPYENDPNPTKLQRGGATWKCSRCKQYGHNKRGCKNPV 620
GRPKK R EN P ++ +C RC GH+++ C P+
Sbjct: 557 PPGRPKKKVVR--VENLKRPKRIV------QCGRCHLLGHSQKKCTQPI 597
>At1g06740 mudrA-like protein
Length = 726
Score = 223 bits (568), Expect = 3e-58
Identities = 154/595 (25%), Positives = 262/595 (43%), Gaps = 29/595 (4%)
Query: 36 GQDFKFSLGMDFSSLLQFKDALHDHFVTIGREFEYMKNDSIRCRVKCRSKGCPFVILCSK 95
G D + +GM+FS + A+ + +++ E +K+D R KC SKGCP+ I C+K
Sbjct: 151 GTDHEMVVGMEFSDAYACRRAIKNAAISLRFEMRTIKSDKTRFTAKCNSKGCPWRIHCAK 210
Query: 96 VGNKETFRIKTLVGEHNCARV--FDNKSANVRWVKLKLLNKVSTMNKISTNEIVDDFRSN 153
V N TF I+T+ G H C + ++ A+V+WV + K+ EI+++
Sbjct: 211 VSNAPTFTIRTIHGSHTCGGISHLGHQQASVQWVADVVAEKLKENPHFKPKEILEEIYRV 270
Query: 154 HGIGITRYRAWKGKQ-----LAIQEVEGAIEMQYTLLWRFSNELRKRAAGNTCKINFESP 208
HGI ++ +AW+GK+ L + G+ E +Y LL ++ +E+R+ G+ ++
Sbjct: 271 HGISLSYKQAWRGKERIMATLRGSTLRGSFEEEYRLLPQYCDEIRRSNPGSVAVVHV--- 327
Query: 209 RSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVARDANEQYFPL 268
+ + F ++ GFL CRP I LD LK+K+ G LL A D + FPL
Sbjct: 328 -NPIDGCFQHLFISFQASISGFLNACRPLIALDSTVLKSKYPGTLLLATGFDGDGAVFPL 386
Query: 269 AFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQKGLMEVFKEDMLHGSEHRLCV 328
AF +V E D+W F+ L +D + + S ++ + V E + H C+
Sbjct: 387 AFAIVNEENDDNWHRFLSELRKILDENMPKLTILSSGERPVVDGV--EANFPAAFHGFCL 444
Query: 329 RHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWC 388
+L + +F VL+ DL AA +K K+ I+ + A W+ K + W
Sbjct: 445 HYLTERFQREFQSSVLV-DLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWA 503
Query: 389 RHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHG 448
F + N ++E+ + + PI+ ME I +++ + E
Sbjct: 504 SSYFEGTRFGQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLINMLKERRETSLHWSN 563
Query: 449 QIMPRPLKRILHEVDHIKNWIPRVVGAQKYEVRHTITLERHVISLKDKSCTCRFWELNGF 508
++P K++L ++ + RV A + E V+++++ SC C W++ G
Sbjct: 564 VLVPSAEKQMLAAIEQSR--AHRVYRANEAEFEVMTCEGNVVVNIENCSCLCGRWQVYGL 621
Query: 509 PCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQWIVTPHD---PVCIL 565
PC HA AL D ++ + C+ E Y Y + P+ QW + I
Sbjct: 622 PCSHAVGALLSCEEDVYRYTESCFTVENYRRAYAETLEPISDKVQWKENDSERDSENVIK 681
Query: 566 PPMYKRSAGRPKKLRRRDPYENDPNPTKLQRGGATWKCSRCKQYGHNKRGCKNPV 620
P + A R +++R D R C RC Q GH + C P+
Sbjct: 682 TPKAMKGAPRKRRVRAED----------RDRVRRVVHCGRCNQTGHFRTTCTAPM 726
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 222 bits (565), Expect = 6e-58
Identities = 171/627 (27%), Positives = 288/627 (45%), Gaps = 33/627 (5%)
Query: 15 TDSEVED--VPRRRFPKFVKEKMGQDFKFSLGMDFSSLLQFKDALHDHFVT--IGREFEY 70
+D+E ED + R R K ++ D + ++G F + +FK+ + + + I +
Sbjct: 153 SDNEEEDHVITRDR-----KIRLASDDRLAIGRTFFTGFEFKEVVLHYAMKHRINAKQNR 207
Query: 71 MKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNCARVFDNKSANVRWVKLK 130
+ D I R R K C + + S ++ + +KT +H+C K R +
Sbjct: 208 WEKDKISFRCAQR-KECEWYVYASYSHERQLWVLKTKCLDHSCTSNGKCKLLKRRVIGRL 266
Query: 131 LLNKVSTMNKISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFSN 190
++K+ +I + + T + G+ LA++ ++ Q+ L +
Sbjct: 267 FMDKLRLQPNFMPLDIQRHIKEQWKLVSTIGQVQDGRLLALKWLKEEYAQQFAHLRGYVA 326
Query: 191 ELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHR 250
E+ G+T ++ + F+R Y+C K F CRP IG+DG LK +
Sbjct: 327 EILSTNKGSTAIVDTIRDANEN-DVFNRIYVCFGAMKNAFYF-CRPLIGIDGTFLKHAVK 384
Query: 251 GILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQKGL 310
G LL A+A DAN Q +P+A+ V+ E ++W+WF+ L D++ + +V ISD+ KG+
Sbjct: 385 GCLLTAIAHDANNQIYPVAWATVQFENAENWLWFLNQLKHDLELKDGSGYVVISDRCKGI 444
Query: 311 MEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKD 370
+ K + L +EHR CV+H+ N+K + G LL+ + A + + +K + ++
Sbjct: 445 ISAVK-NALPNAEHRPCVKHIVENLKKRHGSLDLLKKFVWNLAWSYSDTQYKANLNEMRA 503
Query: 371 ANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRT 430
S Y+ +M++ ++WCR F + S C+ V NN +E+FNA I+ AR K ++ MME IR
Sbjct: 504 YIMSLYEDVMKEEPNTWCRSWFRIGSYCEDVDNNATESFNATIVKARAKALVPMMETIRR 563
Query: 431 YVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRVVGA-QKYEVRHTITLERH 489
M R + K+ K +I + IL E + G +K+EV
Sbjct: 564 QAMARISKRKAKIGKWKKKI-SEYVSEILKEEWELAVKCEVTKGTHEKFEVWFDGNSNAV 622
Query: 490 VISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLP 549
+ ++ C+C W++ G PC HA +A+ G D + FV + A+ Y+ P+
Sbjct: 623 SLKTEEWDCSCCKWQVTGIPCEHAYAAINDVGKDVEDFVIPMFYTIAWREQYDTGPDPVR 682
Query: 550 GPNQW------IVTP-HDPVCILPPMYKRSAGRPKKLRR-----RDPYENDPNPTKLQRG 597
G W I P DPV PP R G K R P + P K+ +
Sbjct: 683 GQMYWPTGLGLITAPLQDPV---PP--GRKKGEKKNYHRIKGPNESPKKKGPQRLKVGKK 737
Query: 598 GATWKCSRCKQYGHNKRGCKNPVPEEQ 624
G C C + GHN GCK P+E+
Sbjct: 738 GVVMHCKSCGEAGHNAAGCKK-FPKEK 763
>At3g30170 unknown protein
Length = 995
Score = 207 bits (526), Expect = 2e-53
Identities = 163/623 (26%), Positives = 270/623 (43%), Gaps = 38/623 (6%)
Query: 8 EDLESVPTDSEVEDVPRRRFPKFVKEKMGQDFKFSLGMDFSSLLQFKDALHDHFVTIGRE 67
ED P S+ ED R R ++ ++ L +++ FK A+ + + +
Sbjct: 193 EDKIPDPFSSDDEDESRTREVHGHRDAANEEVLLELKKTYNTPDDFKLAVLRYSLKTRYD 252
Query: 68 FEYMKNDSIRCRVKCR-----SKGCPFVILCSKVGNKETFRIKTLVGEHNCARVFDNKSA 122
+ ++++ KC CP+ + CS +I+T V H C R +K
Sbjct: 253 IKLFRSEARIVAAKCSYVDKDGVQCPWKVYCSYEKTMHKMQIRTYVNNHICVRSGHSKML 312
Query: 123 NVRWVKLKLLNKVSTMNKISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQY 182
+ ++ K++ E+ + + + +T + K K ++ + E +
Sbjct: 313 KRSSIAYLFEERLRVNPKLTKYEMAAEILREYNLEVTPDQCAKAKTKVVKARNASHEAHF 372
Query: 183 TLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDG 242
+W + E+ K+ +I + + G K+ RPFIGLDG
Sbjct: 373 ARVWDYQAEVIKQNPWTEFEIETTAG-------------AVIGAKQ------RPFIGLDG 413
Query: 243 CHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVF 302
LK +G LLAAV RD + + P+A+ VVE E D+W WF+ L + +
Sbjct: 414 AFLKWDIKGHLLAAVGRDGDNKIVPIAWAVVEIENDDNWDWFLRHLSASLGLCQMANIAI 473
Query: 303 ISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWK 362
+SD+Q GL++ + H +EHR C +H+ N K + + L+ L A++ +
Sbjct: 474 LSDKQSGLVKAIHTILPH-AEHRQCSKHIMDNWK-RDSHDLELQRLFWKIARSYTVEEFN 531
Query: 363 EKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPIL 422
M+ ++ N AY+ L +W R F + + C+ +NNLSE+FN I AR KPIL
Sbjct: 532 NHMMELQQYNRQAYESLQLTSPVTWSRAFFRIGTCCNDNLNNLSESFNRTIRQARRKPIL 591
Query: 423 TMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRVVGAQKYEVRH 482
M+E IR M R + K + R KR E++ + + + E H
Sbjct: 592 DMLEDIRRQCMVRSAKRFLIAEK----LQTRFTKRAHEEIEKMIVGLRQCERYMARENLH 647
Query: 483 TITLER--HVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNAT 540
I + + + ++ K+C CR ++ G PC HA + + +V D Y + + T
Sbjct: 648 EIYVNNVSYFVDMEQKTCDCRKCQMVGIPCIHATCVIIGKKEKVEDYVSDYYTKVRWRET 707
Query: 541 YENYVTPLPGPNQWIVTPHDPVCILPPMYKR-SAGRPKKLRR---RDPYENDPNPTKLQR 596
Y + P+ G W T PV LPP ++R +AGRP R R+ NP K+ R
Sbjct: 708 YLKGIRPVQGMPLWCRTNRLPV--LPPPWRRGNAGRPSNYARRKGRNEAAAPSNPNKMSR 765
Query: 597 GGATWKCSRCKQYGHNKRGCKNP 619
CS C Q GHNK+ C P
Sbjct: 766 EKRIMTCSNCHQEGHNKQRCNIP 788
>At3g47280 putative protein
Length = 739
Score = 202 bits (514), Expect = 5e-52
Identities = 145/558 (25%), Positives = 268/558 (47%), Gaps = 27/558 (4%)
Query: 67 EFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNC---ARVFDNKSAN 123
+F +K+D R + C + C + + ++ G E++ I+ V H+C R ++ A+
Sbjct: 197 KFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESYVIRKYVSHHSCDSSLRNVSHRQAS 256
Query: 124 VRWVKLKLLNKVSTMN-KISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQY 182
R + + N + ++++ FR +HG+GI +AW+ ++ A + G + +
Sbjct: 257 ARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGINYSKAWRVQKHAAELARGLPDDSF 316
Query: 183 TLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDG 242
+L R+ + ++ G+ +S +F ++ RG+ R I +DG
Sbjct: 317 EVLPRWFHRVQVTNPGSITFFKKDSAN-----KFKYAFLAFGASIRGYKL-MRKVISIDG 370
Query: 243 CHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVF 302
HL +K +G LL A A+D N +P+AF +V+SE SW WF++ L++ I E VF
Sbjct: 371 AHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSENDASWDWFLKCLLNIIPDEND--LVF 428
Query: 303 ISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWK 362
+SD+ + + + H LC HL N++T F G L+ + A++ + +
Sbjct: 429 VSDRAASIASRLSGN-YPLAHHGLCTFHLQKNLETHFRGSSLI-PVYYAASRIYTKTEFD 486
Query: 363 EKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPIL 422
I ++++ Q+L E W R A+S +R +++ +NL+E+ NA + R+ P++
Sbjct: 487 SLFWEITNSDKKLAQYLCEVDVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPVV 545
Query: 423 TMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRV-VGAQKYEVR 481
+ E IR+ + F + E+ +H + K++ D W+ V +++EV+
Sbjct: 546 CLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWLEVCQVNQEEFEVK 605
Query: 482 HTITLERHVISLKDKSCTCRFWELNGFPCRHA-ASALTFNGNDPKQFVDDCYKREAYNAT 540
+ H+++L ++CTC ++++ FPC H ASA N N+ FVD+ + +
Sbjct: 606 G--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASAKHINLNE-NMFVDEFHSTYRWRQA 662
Query: 541 YENYVTPLPGPNQWIV--TPHDPVCILPPMYKRSAGRPKKLRRRDPYENDPNPTKLQRGG 598
Y + P W + T + +C LPP + G+ KK R +E+ + K +
Sbjct: 663 YSESIHPNGDMEYWEIPETISEVIC-LPPSTRVPTGKRKKKRIPSVWEHGRSQPKPK--- 718
Query: 599 ATWKCSRCKQYGHNKRGC 616
KCSRC Q GHNK C
Sbjct: 719 -LHKCSRCGQSGHNKSTC 735
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 199 bits (506), Expect = 4e-51
Identities = 145/560 (25%), Positives = 268/560 (46%), Gaps = 31/560 (5%)
Query: 67 EFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNC---ARVFDNKSAN 123
EF +K+D + + C + C + + ++ G E++ I+ V H+C R ++ A+
Sbjct: 74 EFHTVKSDLTKYVLHCIDENCSWRLRATRAGGSESYVIRKYVSHHSCDSSLRNVSHRQAS 133
Query: 124 VRWVKLKLLNKVSTMN-KISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQY 182
R + + N + ++++ FR +HG+GI +AW+ ++ A + G + +
Sbjct: 134 ARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGINYSKAWRVQEHAAELARGLPDDSF 193
Query: 183 TLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDG 242
+L R+ + ++ G+ +S +F ++ RG+ + I +DG
Sbjct: 194 EVLPRWFHRVQVTNPGSITFFKKDSAN-----KFKYAFLAFGASIRGYKL-MKKVISIDG 247
Query: 243 CHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVF 302
HL +K +G LL A A+D N +P+AF +V+SE SW WF++ L++ I E VF
Sbjct: 248 AHLISKFKGTLLGASAQDGNFNLYPIAFAIVDSENDASWDWFLKCLLNIIPDEND--LVF 305
Query: 303 ISDQQKGLMEVFKED--MLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAA 360
+SD+ + + + H LC HL N++T F G L+ + A++ +
Sbjct: 306 VSDRAASIASGLSGNYPLAHNG---LCTFHLQKNLETHFRGSSLI-PVYYAASRVYTKTE 361
Query: 361 WKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKP 420
+ I ++++ Q+L E W R A+S +R +++ +NL+E+ NA + R+ P
Sbjct: 362 FDSLFWEITNSDKKLAQYLWEVDVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYP 420
Query: 421 ILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRV-VGAQKYE 479
I+ + E IR+ + F + E+ +H + K++ D W+ V +++E
Sbjct: 421 IVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWLEVCQVNQEEFE 480
Query: 480 VRHTITLERHVISLKDKSCTCRFWELNGFPCRHA-ASALTFNGNDPKQFVDDCYKREAYN 538
V+ + H+++L ++CTC ++++ FPC H ASA N N+ FVD+ + +
Sbjct: 481 VKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASAKHINLNE-NMFVDEFHSTYKWR 537
Query: 539 ATYENYVTPLPGPNQWIV--TPHDPVCILPPMYKRSAGRPKKLRRRDPYENDPNPTKLQR 596
Y + P W + T + +C LPP + +GR KK R +E+ + K +
Sbjct: 538 QAYSKSIHPNGDMEYWEIPETISEVIC-LPPSTRVPSGRRKKKRIPSVWEHGRSQPKPK- 595
Query: 597 GGATWKCSRCKQYGHNKRGC 616
KCSRC Q GHN C
Sbjct: 596 ---LHKCSRCGQSGHNNSTC 612
>At2g13970 Mutator-like transposase
Length = 597
Score = 195 bits (496), Expect = 6e-50
Identities = 143/486 (29%), Positives = 224/486 (45%), Gaps = 60/486 (12%)
Query: 140 KISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGN 199
KI+ +VD+ + + + +T + K K + ++E + + ++ +W + E+ + G
Sbjct: 20 KITKKAMVDEIQREYKLTVTEDQCSKAKTIVMRERKATHQEHFSRIWDYQAEIFRTNPGT 79
Query: 200 TCKIN-FESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVA 258
+I P RF R ++C K + CRP IG+DG LK +G LLAA
Sbjct: 80 KFEIETIPGPTIGSLQRFYRLFICFKSQKDSWKQTCRPIIGIDGAFLKWDIKGHLLAATG 139
Query: 259 RDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQKGLMEVFKEDM 318
RD + + P+A+ VVE + D+W WF+ L + +D + R ISD+Q GL + +
Sbjct: 140 RDGDNRIVPIAWAVVEIQNDDNWDWFVRQLSESLDLQDGRSVAIISDKQSGLGKAI-HTV 198
Query: 319 LHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQW 378
+ +EHR RH+ N WK K++++ Q
Sbjct: 199 IPQAEHRQYARHIMDN--------------------------WK------KNSHDMELQR 226
Query: 379 LMEKPTS-SWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFP 437
L K +W R F + S C+ +NNLSE+FN I AR KP+L M+E IR M R
Sbjct: 227 LFWKTNPLTWSRAFFRIGSCCNDNLNNLSESFNRTIRNARRKPLLDMLEDIRRQCMVRNE 286
Query: 438 QMMEKLHKHHGQIMPRPLKRILHEVDHIK---NWIPRVVGAQKYEVRHTITLERHVISLK 494
+ ++ + R I +D W+ R + EVR + + + +
Sbjct: 287 KRFVIAYRLKSRFTKRAHMEIEKMIDGAAVCDRWMAR---HNRIEVRVGLD-DSFSVDMN 342
Query: 495 DKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQW 554
D++C CR W++ G PC HAAS + N + FV D Y + TY + + P+ G W
Sbjct: 343 DRTCGCRKWQMTGIPCIHAASVIIGNRQKVEDFVSDWYTTYMWKQTYYDGIMPVQGKMLW 402
Query: 555 IVTPHDPVCILPPMYKR-SAGRPKKL-RRRDPYENDPNPTKLQRGGATWKCSRCKQYGHN 612
+ + V +LPP ++R + GRPK RR+ YE+ AT SR GHN
Sbjct: 403 PIV--NRVGVLPPPWRRGNPGRPKNHDRRKGIYES-----------ATASTSR---EGHN 446
Query: 613 KRGCKN 618
K GCKN
Sbjct: 447 KAGCKN 452
>At2g29230 Mutator-like transposase
Length = 915
Score = 190 bits (483), Expect = 2e-48
Identities = 157/632 (24%), Positives = 290/632 (45%), Gaps = 40/632 (6%)
Query: 5 YESEDLESVPTDSEVEDVPRRRFPKFVKEKMGQDFKFSLGMDFSSLLQFKDALHDHFVTI 64
+ S + ++ PT E + R +++ + + + +G F + L H +
Sbjct: 308 FASFNRDAAPTLDEQGEEGRVGMELAIRDVVYEGDELFVGRVFKNKQDCNVKLAVH--AL 365
Query: 65 GREFEYMKNDSIR--CRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNCA---RVFDN 119
R F + ++ S + + C S+ CP+ + K+ + + ++I++ EH C R +
Sbjct: 366 NRRFHFRRDRSCKKLMTLTCISETCPWRVYIVKLEDSDNYQIRSANLEHTCTVEERSNYH 425
Query: 120 KSANVRWVKLKLLNKVSTMNK----ISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVE 175
++A R + + +K + ++ I I+ +++ + I+ ++AWK +++A+ +
Sbjct: 426 RAATTRVIGSIIQSKYAGNSRGPRAIDLQRIL---LTDYSVRISYWKAWKSREIAMDSAQ 482
Query: 176 GAIEMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCR 235
G+ +TLL + + LR+ G+ + E RF ++ +GF R
Sbjct: 483 GSAANSFTLLPAYLHVLREANPGSIVDLKTEVDGKGNH-RFKYMFLAFAASIQGFSCMKR 541
Query: 236 PFIGLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPE 295
+ +DG HLK K+ G LL A +DAN Q FP+AFGVV+SE D+W WF +L I
Sbjct: 542 VIV-IDGAHLKGKYGGCLLTASGQDANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIPD- 599
Query: 296 RSRRWVFISDQQK----GLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMG 351
F+SD+ GL V+ + ++H C+ HL N+ T + LL +
Sbjct: 600 -GDNLTFVSDRHSSIYTGLRRVYPK-----AKHGACIVHLQRNIATSYKKKHLLFHVSR- 652
Query: 352 AAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNA 411
AA+A + + + + ++L W R A+ L R +V+ +N++E+ NA
Sbjct: 653 AARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTR-AYFLGKRYNVMTSNVAESLNA 711
Query: 412 AIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPR 471
+ AR+ PI++++E+IRT ++ F E + P+ + + + +
Sbjct: 712 VLKEARELPIISLLEFIRTTLISWFAMRREAARTEASPLPPKMREVVHRNFEKSVRFAVH 771
Query: 472 VVGAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDC 531
+ YE+R HV L +++C+CR ++L PC HA +A G + +
Sbjct: 772 RLDRYDYEIREEGASVYHV-KLMERTCSCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPE 830
Query: 532 YKREAYNATYENYVTPLPGPNQWIVTPHDPVC---ILPPMYKRSAGRPKKLRRRDPYEND 588
Y E++ +Y + P+P P +P+ + PP +R +GRPKK R E
Sbjct: 831 YSVESWRMSYLGTIKPVPEVGDVFALP-EPIASLRLFPPATRRPSGRPKKKRITSRGEFT 889
Query: 589 PNPTKLQRGGATWKCSRCKQYGHNKRGCKNPV 620
++ R CSRC GHN+ CK P+
Sbjct: 890 CPQRQVTR------CSRCTGAGHNRATCKMPI 915
>At2g15150 putative transposase of FARE2.2 (CDS1)
Length = 607
Score = 188 bits (477), Expect = 9e-48
Identities = 131/534 (24%), Positives = 254/534 (47%), Gaps = 21/534 (3%)
Query: 67 EFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNC---ARVFDNKSAN 123
EF +K+D R + C + C + + ++ G E++ I+ V H+C R ++ A+
Sbjct: 74 EFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESYAIRKYVSHHSCDSSLRNVSHRQAS 133
Query: 124 VRWVKLKLLNKVSTMN-KISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQY 182
R + + N + ++++ FR +HG+GI +AW+ ++ A + G + +
Sbjct: 134 ARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGINYSKAWRVQEHAAELARGLPDDSF 193
Query: 183 TLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDG 242
+L R+ + ++ G+ +S +F ++ RG+ R I +DG
Sbjct: 194 EVLPRWFHRVQVTNPGSITFFKKDSAN-----KFKYAFLAFGASIRGYKL-MRKVISIDG 247
Query: 243 CHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVF 302
HL +K +G LL A A+D N +P+AF +V+SE SW WF++ L++ I E VF
Sbjct: 248 AHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSENDASWDWFLKCLLNIIPDEND--LVF 305
Query: 303 ISDQQKGLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWK 362
+S++ + + + H LC HL N++T F G L+ + A++ + +
Sbjct: 306 VSERAASIASGLSGN-YPLAHHGLCTFHLQKNLETHFRGSSLI-PVYYAASRVYTKTEFD 363
Query: 363 EKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPIL 422
I ++++ Q+L E W R A+S +R +++ +NL+E+ NA + R+ PI+
Sbjct: 364 SLFWEITNSDKKLAQYLWEVHVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNREYPIV 422
Query: 423 TMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRV-VGAQKYEVR 481
+ E IR+ + F + E+ +H + K++ D W+ V +++EV+
Sbjct: 423 CLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWLEVCQVNQEEFEVK 482
Query: 482 HTITLERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATY 541
+ H+++L ++CTC ++++ FPC H ++ + FVD+ + + Y
Sbjct: 483 G--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASANHINLNENMFVDEFHSTYRWRQAY 540
Query: 542 ENYVTPLPGPNQWIV--TPHDPVCILPPMYKRSAGRPKKLRRRDPYENDPNPTK 593
+ P W + T + +C LPP + +GR KK R +E+ + K
Sbjct: 541 SESIHPNGDMEYWEIPETISEVIC-LPPSTRVPSGRRKKKRIPSVWEHGRSQPK 593
>At1g21040 hypothetical protein
Length = 904
Score = 176 bits (446), Expect = 4e-44
Identities = 145/592 (24%), Positives = 274/592 (45%), Gaps = 34/592 (5%)
Query: 5 YESEDLESVPTDSEVEDVPRRRFPKFVKEKMGQDFKFSLGMDFSSLLQFKDALHDHFVTI 64
+ S + ++ PT E + R +++ + + + +G F + L H +
Sbjct: 308 FASFNRDAAPTLDEQGEEGRVGMELAIRDVVYEGDELFVGRVFKNKQDCNVKLAVH--AL 365
Query: 65 GREFEYMKNDSIR--CRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNCA---RVFDN 119
R F + ++ S + + C S+ CP+ + K+ + + ++I++ EH C R +
Sbjct: 366 NRRFHFRRDRSCKKLMTLTCISETCPWRVYIVKLEDSDNYQIRSANLEHTCTVEERSNYH 425
Query: 120 KSANVRWVKLKLLNKVSTMNK----ISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVE 175
++A R + + +K + ++ I I+ +++ + I+ ++AWK +++A+ +
Sbjct: 426 RAATTRVIGSIIQSKYAGNSRGPRAIDLQRIL---LTDYSVRISYWKAWKSREIAMDSAQ 482
Query: 176 GAIEMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCR 235
G+ +TLL + + LR+ G+ + E RF ++ +GF R
Sbjct: 483 GSAANSFTLLPAYLHVLREANPGSIVDLKTEVDGKGNH-RFKYMFLAFAASIQGFSCMKR 541
Query: 236 PFIGLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPE 295
+ +DG HLK K+ G LL A +DAN Q FP+AFGVV+SE D+W WF +L I
Sbjct: 542 VIV-IDGAHLKGKYGGCLLTASGQDANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIPD- 599
Query: 296 RSRRWVFISDQQK----GLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMG 351
F+SD+ GL V+ + ++H C+ HL N+ T + LL +
Sbjct: 600 -GDNLTFVSDRHSSIYTGLRRVYPK-----AKHGACIVHLQRNIATSYKKKHLLFHVSR- 652
Query: 352 AAKATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNA 411
AA+A + + + + ++L W R A+ L R +V+ +N++E+ NA
Sbjct: 653 AARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTR-AYFLGERYNVMTSNVAESLNA 711
Query: 412 AIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPR 471
+ AR+ PI++++E+IRT ++ F E + P+ + + + +
Sbjct: 712 VLKEARELPIISLLEFIRTTLISWFAMRREAARTEASPLPPKMREVVHRNFEKSVRFAVH 771
Query: 472 VVGAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDC 531
+ YE+R HV L +++C+CR ++L PC HA +A G + +
Sbjct: 772 RLDRYDYEIREEGASVYHV-KLMERTCSCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPE 830
Query: 532 YKREAYNATYENYVTPLPGPNQWIVTPHDPVC---ILPPMYKRSAGRPKKLR 580
Y E++ +Y + P+P P +P+ + PP +R +GRPKK R
Sbjct: 831 YSVESWRMSYLGTIKPVPEVGDVFALP-EPIASLRLFPPATRRPSGRPKKKR 881
>At1g12720 hypothetical protein
Length = 585
Score = 174 bits (442), Expect = 1e-43
Identities = 134/491 (27%), Positives = 214/491 (43%), Gaps = 49/491 (9%)
Query: 140 KISTNEIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGN 199
K++ E+V + + + + +T + K K ++ + + ++ +W + E+ R +
Sbjct: 20 KMTKYEMVAEIKREYKLEVTPDQCAKAKTKVLKARNASHDTHFSRIWDYQAEVLNRNPNS 79
Query: 200 TCKINFESPRSTLF---PRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAA 256
I E+ T RF R Y+C + K + CRP IG+DG LK +G LLAA
Sbjct: 80 DFDI--ETTARTFIGSKQRFFRLYICFNSQKVSWKQHCRPVIGIDGAFLKWDIKGHLLAA 137
Query: 257 VARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQKGLMEVFKE 316
V RD + + PLA+ VVE E D+W WF++ L + + ISD+Q GL++
Sbjct: 138 VGRDGDNRIVPLAWAVVEIENDDNWDWFLKKLSESLGLCEMVNLALISDKQSGLVKAI-H 196
Query: 317 DMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAY 376
++L +EHR C +H+ N K + + L+ L +++ + M +K N AY
Sbjct: 197 NVLPQAEHRQCSKHIMDNWK-RDSHDMELQRLFWKISRSYTIEEFNTHMANLKSYNPQAY 255
Query: 377 QWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRF 436
L +W I AR KP+L M+E IR M R
Sbjct: 256 ASLQLTSPMTW------------------------TIRQARRKPLLDMLEDIRRQCMVRT 291
Query: 437 PQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRVVGAQKYEVR---HTITLE--RHVI 491
+ + + PR I+ I G +++ R H I + + +
Sbjct: 292 AKRFIIAERLKSRFTPRAHA-------EIEKMIAGSAGCERHLARNNLHEIYVNDVGYFV 344
Query: 492 SLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLPGP 551
+ K+C CR WE+ G PC H + + +V D Y + + TY + + P+ G
Sbjct: 345 DMDKKTCGCRKWEMVGIPCVHTPCVIIGRKEKVEDYVSDYYTKVRWRETYRDGIRPVQGM 404
Query: 552 NQWIVTPHDPVCILPPMYKR-SAGRPKK-LRRRDPYE--NDPNPTKLQRGGATWKCSRCK 607
W PV LPP ++R + GR R++ YE + N K+ R CS CK
Sbjct: 405 PLWPRMSRLPV--LPPPWRRGNPGRQSNYARKKGRYETASSSNKNKMSRANRIMTCSNCK 462
Query: 608 QYGHNKRGCKN 618
Q GHNK CKN
Sbjct: 463 QEGHNKSSCKN 473
>At1g25784 mutator-like transposase, putative
Length = 1127
Score = 173 bits (438), Expect = 3e-43
Identities = 144/535 (26%), Positives = 244/535 (44%), Gaps = 33/535 (6%)
Query: 68 FEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNC---ARVFDNKSANV 124
F+ K+ ++ C + CP+ + V + E+F+I + H C +R + NK AN
Sbjct: 404 FKQPKSCPKTLKMVCVDETCPWQLTARVVKDSESFKITSYATTHTCNIDSRKYYNKHANY 463
Query: 125 RWVKLKLLNKVSTMNKISTN------EIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAI 178
KLL +V ST ++ + + I+ AW+ K++A++ V G
Sbjct: 464 -----KLLGEVVRSRYSSTQGGPRAVDLPQLLLKDLNVRISYSTAWRAKEVAVENVRGDE 518
Query: 179 EMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFI 238
Y L + L+ G +++ +P RF ++ L +G + R +
Sbjct: 519 IANYRFLPTYLYLLQLANPGTITHLHY-TPEDDGKQRFKYVFVSLGASIKGLIY-MRKVV 576
Query: 239 GLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSR 298
+DG L ++G LL A A+D N Q FP+AFGVV+ ET SW WF E L +I P+ S
Sbjct: 577 VVDGTQLVGPYKGCLLIACAQDGNFQIFPIAFGVVDGETDASWAWFFEKLA-EIVPD-SD 634
Query: 299 RWVFISDQQ----KGLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGG-GVLLRDLMMGAA 353
+ +SD+ KGL V+ + H C HL N+ T +G GV L AA
Sbjct: 635 DLMIVSDRHSSIYKGLSVVYPR-----AHHGACAVHLERNLSTYYGKFGV--SALFFSAA 687
Query: 354 KATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAI 413
KA +++ L+++ + ++L + W R A R +++ +N SE+ N +
Sbjct: 688 KAYRVRDFEKYFELLREKSAKCAKYLEDIGFEHWTR-AHCRGERYNIMSSNNSESMNHVL 746
Query: 414 MLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRVV 473
A+ PI+ M+E+IR +M F +K+ + + P +R L E+ + ++
Sbjct: 747 TKAKTYPIVYMIEFIRDVLMRWFASRRKKVARCKSSVTPEVDERFLQELPASGKYAVKMS 806
Query: 474 GAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYK 533
G Y+V HV+ L +CTC + PC HA +A +G DPK V Y
Sbjct: 807 GPWSYQVTSKSGEHFHVV-LDQCTCTCLRYTKLRIPCEHALAAAIEHGIDPKSCVGWWYG 865
Query: 534 REAYNATYENYVTPLPGPNQWIVTPH-DPVCILPPMYKRSAGRPKKLRRRDPYEN 587
+ ++ +++ + P+ P ++ H + ++PP +R GRP R EN
Sbjct: 866 LQTFSDSFQEPILPIADPKDVVIPQHIVDLILIPPYSRRPPGRPLSKRIPSRGEN 920
>At1g34700 hypothetical protein
Length = 990
Score = 169 bits (428), Expect = 4e-42
Identities = 144/531 (27%), Positives = 240/531 (45%), Gaps = 33/531 (6%)
Query: 68 FEYMKNDSIRCRVKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNC---ARVFDNKSANV 124
F+ K+ ++ C + CP+ + V + E+F+I + H C +R NK AN
Sbjct: 404 FKQPKSCPKTLKMVCVDETCPWQLTARVVKDSESFKITSYATTHTCNIDSRKNYNKHANY 463
Query: 125 RWVKLKLLNKVSTMNKISTN------EIVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAI 178
KLL +V ST ++ ++ + I+ AW+ K++A++ V G
Sbjct: 464 -----KLLGEVVRSRYSSTQGGPRAVDLPQLLLNDLNVRISYSTAWRAKEVAVENVRGDE 518
Query: 179 EMQYTLLWRFSNELRKRAAGNTCKINFESPRSTLFPRFSRFYMCLDGCKRGFLAGCRPFI 238
Y L + L+ G +++ +P RF ++ L G L R +
Sbjct: 519 IANYRFLPTYLYLLQLANPGTITHLHY-TPEDDGKQRFKYVFVSL-GASIKCLIYMRKVV 576
Query: 239 GLDGCHLKTKHRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSR 298
+DG L ++G LL A A+D N Q FP+AFGVV+ ET SW WF E L + I S
Sbjct: 577 VVDGTQLVGPYKGCLLIACAQDGNFQIFPIAFGVVDGETDASWAWFFEKLAEIIPD--SD 634
Query: 299 RWVFISDQQ----KGLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGG-GVLLRDLMMGAA 353
+ +SD+ KGL V+ + H C HL N+ T +G GV L AA
Sbjct: 635 DLMIVSDRHSSIYKGLSVVYPR-----AHHGACAVHLERNLSTYYGKFGV--SALFFSAA 687
Query: 354 KATYEAAWKEKMLLIKDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAI 413
KA +++ L+++ + ++L + W R A R +++ +N E+ N +
Sbjct: 688 KAYRVKDFEKYFELLREKSAKCGKYLEDIGFEHWTR-AHCRGERYNIMSSNNFESMNHVL 746
Query: 414 MLARDKPILTMMEWIRTYVMGRFPQMMEKLHKHHGQIMPRPLKRILHEVDHIKNWIPRVV 473
A+ PI+ M+E+IR +M F +K+ + + P +R LHE+ ++ ++
Sbjct: 747 TKAKTYPIVYMIEFIRDVLMRWFASRRKKVARCKSSVTPEVDERFLHELPASGKYVVKMS 806
Query: 474 GAQKYEVRHTITLERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYK 533
G Y+V HV+ L +CTC + PC HA +A +G DPK V Y
Sbjct: 807 GPWSYQVTSKSGEHFHVV-LDQCTCTCLRYTKLRIPCEHALAAAIEHGIDPKSCVGWWYG 865
Query: 534 REAYNATYENYVTPLPGPNQWIVTPHDPVCILPPMYKRSAGRPKKLRRRDP 584
+ ++ +++ + P+ P ++ H IL P Y R R + L +R P
Sbjct: 866 LQTFSDSFQESILPIADPKDVVIPQHIVDLILIPPYSRRPPR-RPLSKRIP 915
>At1g44840 hypothetical protein
Length = 926
Score = 169 bits (427), Expect = 6e-42
Identities = 135/521 (25%), Positives = 233/521 (43%), Gaps = 41/521 (7%)
Query: 80 VKCRSKGCPFVILCSKVGNKETFRIKTLVGEHNCARVFDNKSANVRWVKLKLLNKVSTMN 139
+ C + C + IL + + + IK +H C+ D + ++ K++ V
Sbjct: 413 LSCSDEKCDWRILATLMKGTGYYEIKKASLDHTCS--LDTRGQFMQKATSKVIASVFKAK 470
Query: 140 KISTNE----------IVDDFRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFS 189
+++D R + ++ + W+ ++ A+ V G+ E Y L +
Sbjct: 471 YSDPTSGPVPMDLQQLVLEDLR----VSVSYSKCWRARESALTSVAGSDEESYCYLAEYL 526
Query: 190 NELRKRAAGNTCKINFESP-RSTLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTK 248
+ L+ G I E RF ++ GF R + +DG HLK K
Sbjct: 527 HLLKLTNPGTITHIETERDVEDESKERFLYMFLAFGASIAGF-RHLRRILVVDGTHLKGK 585
Query: 249 HRGILLAAVARDANEQYFPLAFGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQK 308
++G+LL + +DAN Q +PL F VV+SE +SW WF L I S+ +SD+
Sbjct: 586 YKGVLLTSSGQDANFQVYPLGFAVVDSENDESWTWFFTKLERII--ADSKTLTILSDRHS 643
Query: 309 GLMEVFKEDMLHGSEHRLCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLI 368
++ K + + H C+ HL N++TK+ L + L+ A A +KE I
Sbjct: 644 SILVAVKR-VFPQANHGACIIHLCRNIQTKYKNKALTQ-LVKNAGYAFTGTKFKEFYGQI 701
Query: 369 KDANESAYQWLMEKPTSSWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWI 428
+ N++ ++L + ++W RH F R +++ +N++E N A+ R I+ ++ +I
Sbjct: 702 ETTNQNCGKYLHDIGMANWTRHYFR-GQRFNLMTSNIAETLNKALNKGRSSHIVELIRFI 760
Query: 429 RTYVMGRFPQMMEKLHKHHGQIMPRPLKRIL--------HEVDHIKNWIPRVVGAQKYEV 480
R+ + F +K KH G + P K+I +V I NW YE+
Sbjct: 761 RSMLTRWFNARRKKSLKHKGPVPPEVDKQITKTMLTTNGSKVGRITNW--------SYEI 812
Query: 481 RHTITLERHVISLKDKSCTCRFWELNGFPCRHAASALTFNGNDPKQFVDDCYKREAYNAT 540
+ R+V+ L+ K CTC+ ++ PC HA A K VDDC+K A+ ++
Sbjct: 813 NGMLG-GRNVVDLEKKQCTCKRYDKLKIPCSHALVAANSFKISYKALVDDCFKPHAWVSS 871
Query: 541 YENYVTP-LPGPNQWIVTPHDPVCILPPMYKRSAGRPKKLR 580
Y V P G ++ I+ + +LPP +R +GRPK R
Sbjct: 872 YPGAVFPEAIGRDEKILEELEHRSMLPPYTRRPSGRPKVAR 912
>At1g34620 Mutator-like protein
Length = 1034
Score = 165 bits (417), Expect = 8e-41
Identities = 133/558 (23%), Positives = 253/558 (44%), Gaps = 35/558 (6%)
Query: 40 KFSLGMDFSSLLQFKDALHDHFVTIGREFEYMKNDSIRCRVKCRSKGCPFVILCSKVGNK 99
K +G F S K + H + F ++ +KC SK CP+ + SKV
Sbjct: 364 KLFVGRVFKSKSDCKIKIAIHAINRKFHFRTARSTPKFMVLKCISKTCPWRVYASKVDTS 423
Query: 100 ETFRIKTLVGEHNCARVFDNKSANVRWVKLKLLNKVSTMNKISTNE----------IVDD 149
++F+++ H C D + R +++ ++ + ++DD
Sbjct: 424 DSFQVRQANQRHTCT--IDQRRRYHRLATTQVIGELMQSRFLGIKRGPNAAVIRKFLLDD 481
Query: 150 FRSNHGIGITRYRAWKGKQLAIQEVEGAIEMQYTLLWRFSNELRKRAAGNTCKINFESPR 209
+ + I+ ++AW+ +++A+++ G++ Y L+ ++ L++ G+ C ++
Sbjct: 482 YH----VSISYWKAWRAREVAMEKSLGSMAGSYALIPAYAGLLQQANPGSLCFTEYDDD- 536
Query: 210 STLFPRFSRFYMCLDGCKRGFLAGCRPFIGLDGCHLKTKHRGILLAAVARDANEQYFPLA 269
T RF ++ +G+ A R I +DG +K ++ G L++A +D N Q FPLA
Sbjct: 537 PTGPRRFKYQFIAFAASIKGY-AFMRKVIVVDGTSMKGRYGGCLISACCQDGNFQIFPLA 595
Query: 270 FGVVESETKDSWMWFMELLMDDIDPERSRRWVFISDQQK----GLMEVFKEDMLHGSEHR 325
FG+V SE ++ WF + L I + +FISD+ + GL +V+ + + H
Sbjct: 596 FGIVNSENDSAYEWFFQRL--SIIAPDNPDLMFISDRHESIYTGLSKVYTQ-----ANHA 648
Query: 326 LCVRHLYANMKTKFGGGVLLRDLMMGAAKATYEAAWKEKMLLIKDANESAYQWLMEKPTS 385
C HL+ N++ + L R LM AA+A + + L I+ N +L++ S
Sbjct: 649 ACTVHLWRNIRHLYKPKSLCR-LMSEAAQAFHVTDFNRIFLKIQKLNPGCAAYLVDLGFS 707
Query: 386 SWCRHAFSLYSRCDVVMNNLSEAFNAAIMLARDKPILTMMEWIRTYVMGRFPQMMEKLHK 445
W R S R +++ +N+ E++N I AR+ P++ M+E+IRT +M F +
Sbjct: 708 EWTR-VHSKGRRFNIMDSNICESWNNVIREAREYPLICMLEYIRTTLMDWFATRRAQAED 766
Query: 446 HHGQIMPRPLKRILHEVDHIKNWIPRVVGAQKYEVRHTITLERHVISLKDKSCTCRFWEL 505
+ PR +R+ + + + +++V+ T E ++ L + +C+C ++
Sbjct: 767 CPTTLAPRVQERVEENYQSAMSMSVKPICNFEFQVQER-TGECFIVKLDESTCSCLEFQG 825
Query: 506 NGFPCRHAASALTFNGNDPKQFVDDCYKREAYNATYENYVTPLPGPNQWI---VTPHDPV 562
G PC HA +A G + Y E +YE + P+ + +
Sbjct: 826 LGIPCAHAIAAAARIGVPTDSLAANGYFNELVKLSYEEKIYPIHSVGGEVAPEIAAGTTG 885
Query: 563 CILPPMYKRSAGRPKKLR 580
+ PP +R GRP+K+R
Sbjct: 886 ELQPPFVRRPPGRPRKIR 903
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.137 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,587,816
Number of Sequences: 26719
Number of extensions: 695556
Number of successful extensions: 1879
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 97
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1491
Number of HSP's gapped (non-prelim): 152
length of query: 632
length of database: 11,318,596
effective HSP length: 105
effective length of query: 527
effective length of database: 8,513,101
effective search space: 4486404227
effective search space used: 4486404227
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0335a.7


