

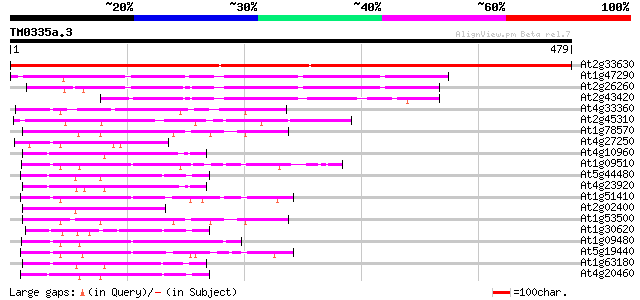
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0335a.3
(479 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g33630 putative steroid dehydrogenase 684 0.0
At1g47290 putative 3-beta-hydroxysteroid dehydrogenase 190 1e-48
At2g26260 3-beta-hydroxysteroid dehydrogenase 186 2e-47
At2g43420 putative sterol dehydrogenase 90 3e-18
At4g33360 putative dihydrokaempferol 4-reductase 60 3e-09
At2g45310 putative nucleotide sugar epimerase 50 2e-06
At1g78570 Similar to dTDP-D-glucose 4,6-dehydratase 50 3e-06
At4g27250 unknown protein 48 1e-05
At4g10960 UDP-galactose 4-epimerase - like protein 48 1e-05
At1g09510 putative cinnamyl alcohol dehydrogenase 48 1e-05
At5g44480 putative protein 47 2e-05
At4g23920 UDPglucose 4-epimerase - like protein 47 2e-05
At1g51410 cinnamyl alcohol dehydrogenase, putative 47 2e-05
At2g02400 putative cinnamoyl-CoA reductase 47 3e-05
At1g53500 dTDP-D-glucose 4,6-dehydratase, putative 47 3e-05
At1g30620 UDP-galactose 4-epimerase-like protein 47 3e-05
At1g09480 putative cinnamyl alcohol dehydrogenase 45 7e-05
At5g19440 cinnamyl-alcohol dehydrogenase - like protein 45 9e-05
At1g63180 uridine diphosphate glucose epimerase like protein 45 1e-04
At4g20460 UDP-glucose 4-epimerase - like protein 44 2e-04
>At2g33630 putative steroid dehydrogenase
Length = 480
Score = 684 bits (1765), Expect = 0.0
Identities = 329/479 (68%), Positives = 398/479 (82%), Gaps = 2/479 (0%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVR 60
MHLSENEG+EG T VVTGGLGFVG+ALCLELVRRGA +VR+FDLR SSPW LK++GVR
Sbjct: 1 MHLSENEGVEGNTFVVTGGLGFVGAALCLELVRRGARQVRSFDLRHSSPWSDDLKNSGVR 60
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG 120
CIQGD+ +K+DV++AL GADCV HLA++GMSGKEML+ R D+VNI GTC+VL+A
Sbjct: 61 CIQGDVTKKQDVDNALDGADCVLHLASYGMSGKEMLRFGRCDEVNINGTCNVLEAAFKHE 120
Query: 121 IKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKND 180
I R+VY STYNVVF G+ I+NGNE LPYFP+D HVD Y R+KSIAEQLVLK+N RPFKN
Sbjct: 121 ITRIVYVSTYNVVFGGKEILNGNEGLPYFPLDDHVDAYSRTKSIAEQLVLKSNGRPFKN- 179
Query: 181 AGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALI 240
G +YTCA+RPAAIYGPGEDRHLPRI+T KLGL LF IG+ +VKSDW++VENLVLA+I
Sbjct: 180 GGKRMYTCAIRPAAIYGPGEDRHLPRIVTLTKLGLALFKIGEPSVKSDWIYVENLVLAII 239
Query: 241 LASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHA 300
LASMGLLDD G+ Q P+AAGQ YF+SDG PVNTFEFL+PLL+SL+Y+LPK +++V A
Sbjct: 240 LASMGLLDDIPGREGQ-PVAAGQPYFVSDGYPVNTFEFLRPLLKSLDYDLPKCTISVPFA 298
Query: 301 LVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSR 360
L LG+I QG YT+LYPWL + WLPQP +LP+EV+KVGVTHYFSYLKAKEE+GYVP SS+
Sbjct: 299 LSLGKIFQGFYTVLYPWLSKSWLPQPLVLPAEVYKVGVTHYFSYLKAKEELGYVPFKSSK 358
Query: 361 EGMASTISYWQQRKMITLDGPTIYTWLFCVIGMISLFCAAFLPDVGIVFLLRATSLFVFR 420
EGMA+TISYWQ+RK +LDGPT++TW+ IGM +LF A +LPD+G V LRA LF FR
Sbjct: 359 EGMAATISYWQERKRRSLDGPTMFTWIAVTIGMSALFAAGWLPDIGPVPFLRAIHLFFFR 418
Query: 421 SMWMIRLVFILATAAHVFEAIYAWYLAKRVDHANARGWFWQTFALGYFSLRFLLKRARK 479
++ +++ VFI+A HV E IYAW+LAKRVD NA GWF QT ALG+FS+RFLLKRA++
Sbjct: 419 TITIVKAVFIVAVVLHVAEGIYAWFLAKRVDPGNAMGWFLQTSALGFFSMRFLLKRAKE 477
>At1g47290 putative 3-beta-hydroxysteroid dehydrogenase
Length = 382
Score = 190 bits (483), Expect = 1e-48
Identities = 131/385 (34%), Positives = 196/385 (50%), Gaps = 36/385 (9%)
Query: 1 MHLSENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDL---------RDSSPWF 51
M ++E E + VVTGG GF L LVR VR DL ++
Sbjct: 3 MEVTETE----RWCVVTGGRGFAARHLVEMLVRYQMFHVRIADLAPAIVLNPHEETGILG 58
Query: 52 AILKDNGVRCIQGDIVRKEDVESALRGADCVFHLAAFGMS-GKEMLQLTRVDQVNITGTC 110
++ V+ + D+ K V +GA+ VFH+AA S LQ + VN+ GT
Sbjct: 59 EAIRSGRVQYVSADLRNKTQVVKGFQGAEVVFHMAAPDSSINNHQLQYS----VNVQGTT 114
Query: 111 HVLDACLHLGIKRLVYCSTYNVVFAGQR-IVNGNESLPYFPIDHHVDPYGRSKSIAEQLV 169
+V+DAC+ +G+KRL+Y S+ +VVF G +N +ESLPY P H D Y +K+ E L+
Sbjct: 115 NVIDACIEVGVKRLIYTSSPSVVFDGVHGTLNADESLPYPP--KHNDSYSATKAEGEALI 172
Query: 170 LKNNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDW 229
LK N R + L TC +RP++I+GPG+ +P ++T A+ G F IGD + D+
Sbjct: 173 LKANGR-------SGLLTCCIRPSSIFGPGDKLMVPSLVTAARAGKSKFIIGDGSNFYDF 225
Query: 230 VFVENLVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYE 289
+VEN+V A + A L A G+ AAGQAYFI++ P+ +EF+ LL L YE
Sbjct: 226 TYVENVVHAHVCAERAL----ASGGEVCAKAAGQAYFITNMEPIKFWEFMSQLLEGLGYE 281
Query: 290 LPKTSLAVDHALVLGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKE 349
P + + + + + Y +L P + + P + PS V + F KAK+
Sbjct: 282 RPSIKIPASLMMPIAYLVELAYKLLGP----YGMKVPVLTPSRVRLLSCNRTFDSSKAKD 337
Query: 350 EIGYVPMVSSREGMASTISYWQQRK 374
+GY P+V +EG+ TI + K
Sbjct: 338 RLGYSPVVPLQEGIKRTIDSFSHLK 362
>At2g26260 3-beta-hydroxysteroid dehydrogenase
Length = 390
Score = 186 bits (472), Expect = 2e-47
Identities = 132/365 (36%), Positives = 190/365 (51%), Gaps = 34/365 (9%)
Query: 15 VVTGGLGFVGSALCLELVRRGAHEVRAFDLR-----DSSPWFAILKDNGVRC-----IQG 64
VVTGG GF L LVR VR DL D +L D G+R I
Sbjct: 13 VVTGGRGFAARHLVEMLVRYEMFCVRIADLAPAIMLDPQEGNGVL-DEGLRSGRVQYISA 71
Query: 65 DIVRKEDVESALRGADCVFHLAAFGMS-GKEMLQLTRVDQVNITGTCHVLDACLHLGIKR 123
D+ K V A +GA+ VFH+AA S LQ + VN+ GT +V+DAC+ +G+KR
Sbjct: 72 DLRDKSQVVKAFQGAEVVFHMAAPDSSINNHQLQYS----VNVQGTQNVIDACVDVGVKR 127
Query: 124 LVYCSTYNVVFAGQR-IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAG 182
L+Y S+ +VVF G I+NG ES+ Y PI H+ D Y +K+ E+L++K N R
Sbjct: 128 LIYTSSPSVVFDGVHGILNGTESMAY-PIKHN-DSYSATKAEGEELIMKANGR------- 178
Query: 183 NCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILA 242
N L TC +RP++I+GPG+ +P ++ A+ G F IGD D+ +VEN+ A + A
Sbjct: 179 NGLLTCCIRPSSIFGPGDRLLVPSLVAAARAGKSKFIIGDGNNLYDFTYVENVAHAHVCA 238
Query: 243 SMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALV 302
L A G AAGQAYFI++ P+ +EF+ LL L YE P + +
Sbjct: 239 ERAL----ASGGDVSTKAAGQAYFITNMEPIKFWEFMSQLLDGLGYERPSIKIPAFIMMP 294
Query: 303 LGRICQGVYTILYPWLDRWWLPQPFILPSEVHKVGVTHYFSYLKAKEEIGYVPMVSSREG 362
+ + + Y +L P + + P + PS V + + F KAK+ +GY P+V +EG
Sbjct: 295 IAHLVELTYKVLGP----YGMTVPQLTPSRVRLLSCSRTFDSTKAKDRLGYAPVVPLQEG 350
Query: 363 MASTI 367
+ TI
Sbjct: 351 IRRTI 355
>At2g43420 putative sterol dehydrogenase
Length = 437
Score = 89.7 bits (221), Expect = 3e-18
Identities = 81/295 (27%), Positives = 138/295 (46%), Gaps = 34/295 (11%)
Query: 78 GADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCSTYNVVFAG- 136
G+ VF++ A + + +V + GT +V+ AC G+++L+Y ST +VVF G
Sbjct: 136 GSYVVFYMGATDLRSHDYFDCYKVI---VQGTRNVISACRESGVRKLIYNSTADVVFDGS 192
Query: 137 QRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAGNCLYTCAVRPAAIY 196
Q I +G+ESL P+ K+ AE L+ N R + L TCA+R + ++
Sbjct: 193 QPIRDGDESLRR-PLKFQ-SMLTDFKAQAEALIKLANNR-------DGLLTCALRSSIVF 243
Query: 197 GPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLVLALILASMGLLDDSAGKGKQ 256
GPG+ +P ++ AK G F +G SD+ + EN+ A I A L Q
Sbjct: 244 GPGDTEFVPFLVNLAKSGYAKFILGSGENISDFTYSENVSHAHICAVKAL-------DSQ 296
Query: 257 RPIAAGQAYFISDGSPVNTFEFLQPLLRSLEYELPKTSLAVDHALVLGRICQGVYTILYP 316
AG+ +FI++ PV ++F+ ++ L Y P L V R+ V+++L
Sbjct: 297 MEFVAGKEFFITNLKPVRFWDFVSHIVEGLGYPRPSIKLPV-------RLVLYVFSLLKW 349
Query: 317 WLDRWWLPQPFILPSEVHKVGV----THYFSYLKAKEEIGYVPMVSSREGMASTI 367
++ L + H+ + T F+ AK+ +GY P+V+ + +I
Sbjct: 350 THEKEGLGSNY---DTAHQYALLASSTRTFNCNAAKKHLGYTPVVTLEVSLLDSI 401
Score = 31.6 bits (70), Expect = 1.0
Identities = 16/32 (50%), Positives = 19/32 (59%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFD 43
KT VV GG GF+G +L L+R G VR D
Sbjct: 13 KTCVVLGGRGFIGRSLVSRLLRLGNWTVRVAD 44
>At4g33360 putative dihydrokaempferol 4-reductase
Length = 344
Score = 59.7 bits (143), Expect = 3e-09
Identities = 65/243 (26%), Positives = 110/243 (44%), Gaps = 36/243 (14%)
Query: 6 NEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF-----DLRDSSPWFAILKDNGVR 60
N E ++VTG G++G+ LC L+RRG H VRA DL D P V
Sbjct: 7 NTETENMKILVTGSTGYLGARLCHVLLRRG-HSVRALVRRTSDLSDLPP--------EVE 57
Query: 61 CIQGDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL- 119
GD+ + A G D VFH AA + + +R VN+ G +VL+A
Sbjct: 58 LAYGDVTDYRSLTDACSGCDIVFHAAA--LVEPWLPDPSRFISVNVGGLKNVLEAVKETK 115
Query: 120 GIKRLVYCSTYNVVFAGQ-RIVNGNE--SLPYFPIDHHVDPYGRSKSIAEQLVLKNNARP 176
+++++Y S++ + + + N N+ + +F + Y RSK++A+++ L
Sbjct: 116 TVQKIIYTSSFFALGSTDGSVANENQVHNERFFCTE-----YERSKAVADKMAL------ 164
Query: 177 FKNDAGNCLYTCAVRPAAIYGPGE---DRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVE 233
N A + + P I+GPG+ + R++ G L IG T + + V+
Sbjct: 165 --NAASEGVPIILLYPGVIFGPGKLTSANMVARMLIERFNGRLPGYIGSGTDRYSFSHVD 222
Query: 234 NLV 236
++V
Sbjct: 223 DVV 225
>At2g45310 putative nucleotide sugar epimerase
Length = 437
Score = 50.4 bits (119), Expect = 2e-06
Identities = 74/307 (24%), Positives = 129/307 (41%), Gaps = 36/307 (11%)
Query: 4 SENEGIEGKTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRD------SSPWFAILKDN 57
S N GI T++VTG GFVG+ + L RRG + + D A+L+ +
Sbjct: 92 STNNGI---TVLVTGAAGFVGTHVSAALKRRGDGVIGLDNFNDYYDPSLKRARRALLERS 148
Query: 58 GVRCIQGDIVRKEDVESALR--GADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDA 115
G+ ++GDI E + + V HLAA M + NI G ++L+
Sbjct: 149 GIFIVEGDINDVELLRKLFKIVSFTHVMHLAAQAGVRYAMENPSSYVHSNIAGFVNLLEI 208
Query: 116 CLHLGIKRLVYCSTYNVVFAGQRIVNG-NESLPYFPIDHHVDP---YGRSKSIAEQLVLK 171
C + + +V+A V G N +P+ D P Y +K E++
Sbjct: 209 CKSVNPQPA-------IVWASSSSVYGLNTKVPFSEKDKTDQPASLYAATKKAGEEI--- 258
Query: 172 NNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKL----GLLLFTIGDQ-TVK 226
A + + G L +R +YGP + T + + +F + TV
Sbjct: 259 --AHTYNHIYG--LSLTGLRFFTVYGPWGRPDMAYFFFTKDILKGKSISIFESANHGTVA 314
Query: 227 SDWVFVENLVLALILASMGLLDDSAGK-GKQRPIAAGQAYFISDGSPVNTFEFLQPLLRS 285
D+ +++++V LA++ + S G GK+R A + + + + SPV + ++ L R
Sbjct: 315 RDFTYIDDIVKG-CLAALDTAEKSTGSGGKKRGPAQLRVFNLGNTSPVPVSDLVRILERQ 373
Query: 286 LEYELPK 292
L+ + K
Sbjct: 374 LKVKAKK 380
>At1g78570 Similar to dTDP-D-glucose 4,6-dehydratase
Length = 669
Score = 50.1 bits (118), Expect = 3e-06
Identities = 59/242 (24%), Positives = 102/242 (41%), Gaps = 29/242 (11%)
Query: 12 KTLVVTGGLGFVGSALCLELVRR-GAHEVRAFDLRDSSPWFAILKDN----GVRCIQGDI 66
K +++TG GF+ S + L+R +++ D D L + + ++GDI
Sbjct: 7 KNILITGAAGFIASHVANRLIRSYPDYKIVVLDKLDYCSNLKNLNPSKHSPNFKFVKGDI 66
Query: 67 VRKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG-IKR 123
+ V L G D + H AA + NI GT +L+AC G I+R
Sbjct: 67 ASADLVNHLLITEGIDTIMHFAAQTHVDNSFGNSFEFTKNNIYGTHVLLEACKVTGQIRR 126
Query: 124 LVYCSTYNVVFAGQR--IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL---KNNARPFK 178
++ ST V +V +E+ P +PY +K+ AE LV+ ++ P
Sbjct: 127 FIHVSTDEVYGETDEDALVGNHEASQLLP----TNPYSATKAGAEMLVMAYGRSYGLPVI 182
Query: 179 NDAGNCLYTCAVRPAAIYGPGE--DRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVENLV 236
GN +YGP + ++ +P+ I A G +L GD + +++ E++
Sbjct: 183 TTRGN----------NVYGPNQFPEKLIPKFILLAMRGQVLPIHGDGSNVRSYLYCEDVA 232
Query: 237 LA 238
A
Sbjct: 233 EA 234
>At4g27250 unknown protein
Length = 354
Score = 48.1 bits (113), Expect = 1e-05
Identities = 46/144 (31%), Positives = 75/144 (51%), Gaps = 14/144 (9%)
Query: 5 ENEGIEGKTLV--VTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDN-GV 59
E +G E KT VTG G++GS L L++RG + V A DL S + + K+N +
Sbjct: 2 ELQGEESKTATYCVTGASGYIGSWLVKSLLQRG-YTVHATLRDLAKSEYFQSKWKENERL 60
Query: 60 RCIQGDIVRKEDVESALRGADCVFHLAA---FGMSGK----EMLQLTRVDQVNITGTCHV 112
R + D+ + A++G D VFH+AA F +S E ++V + + G +V
Sbjct: 61 RLFRADLRDDGSFDDAVKGCDGVFHVAASMEFDISSDHVNLESYVQSKVIEPALKGVRNV 120
Query: 113 LDACL-HLGIKRLVYCSTYNVVFA 135
L +CL +KR+V+ S+ + + A
Sbjct: 121 LSSCLKSKSVKRVVFTSSISTLTA 144
>At4g10960 UDP-galactose 4-epimerase - like protein
Length = 351
Score = 48.1 bits (113), Expect = 1e-05
Identities = 42/167 (25%), Positives = 75/167 (44%), Gaps = 16/167 (9%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNGVRCIQGDIVRKED 71
+ ++V+GG G++GS L+L+ G + V D D+S ++ + + G+ +
Sbjct: 4 RNVLVSGGAGYIGSHTVLQLL-LGGYSVVVVDNLDNSSAVSLQRVKKLAAEHGERLSFHQ 62
Query: 72 VESALRGA----------DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGI 121
V+ R A D V H A G+ + + N+ GT +L+ G
Sbjct: 63 VDLRDRSALEKIFSETKFDAVIHFAGLKAVGESVEKPLLYYNNNLVGTITLLEVMAQHGC 122
Query: 122 KRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
K LV+ S+ V + + + E FPI ++PYGR+K E++
Sbjct: 123 KNLVFSSSATVYGSPKEVPCTEE----FPIS-ALNPYGRTKLFIEEI 164
>At1g09510 putative cinnamyl alcohol dehydrogenase
Length = 322
Score = 47.8 bits (112), Expect = 1e-05
Identities = 75/290 (25%), Positives = 125/290 (42%), Gaps = 44/290 (15%)
Query: 11 GKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNG----VRCIQG 64
GK + VTG G+V S + L+ RG + VRA D D +L +G ++ +
Sbjct: 5 GKMVCVTGASGYVASWIVKLLLLRG-YTVRATVRDPSDEKKTEHLLALDGAKEKLKLFKA 63
Query: 65 DIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-GIKR 123
D++ + E A+ G D VFH A+ Q+ +D + GT +VL C + +KR
Sbjct: 64 DLLEEGSFEQAIEGCDAVFHTASPVSLTVTDPQIELIDPA-VKGTLNVLKTCAKVSSVKR 122
Query: 124 LVYCSTYNVVFAGQRIVNGNE-------SLPYFPIDHHVDPYGRSKSIAEQLVLKNNARP 176
++ S+ V + + N+ S P F + + Y SK++AE + A
Sbjct: 123 VIVTSSMAAVLFREPTLGPNDLVDESCFSDPNFCTEKKL-WYALSKTLAE-----DEAWR 176
Query: 177 FKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKSDW--VFVEN 234
F + G L + P + GP P + + + + L T D + D+ V V +
Sbjct: 177 FAKEKG--LDLVVINPGLVLGP---LLKPSLTFSVNVIVELITGKDNFINKDFRLVDVRD 231
Query: 235 LVLALILASMGLLDDSAGKGKQRPIAAGQAYFISDGSPVNTFEFLQPLLR 284
+ LA I K + P A G+ +I +G PV T ++ +LR
Sbjct: 232 VALAHI------------KAFETPSANGR--YIIEG-PVVTINDIEKILR 266
>At5g44480 putative protein
Length = 436
Score = 47.0 bits (110), Expect = 2e-05
Identities = 47/171 (27%), Positives = 74/171 (42%), Gaps = 16/171 (9%)
Query: 10 EGKT-LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAIL-------KDNGVRC 61
EG T ++VTGG G++GS L L+ R ++ V D A+ + ++
Sbjct: 92 EGVTHVLVTGGAGYIGSHAALRLL-RDSYRVTIVDNLSRGNLGAVKTLQQLFPQTGRLQF 150
Query: 62 IQGDIVRKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL 119
I D+ VE D V H AA G+ L + + T VL+A
Sbjct: 151 IYADLGDPLAVEKIFSENAFDAVMHFAAVAYVGESTLYPLKYYHNITSNTLGVLEAMARH 210
Query: 120 GIKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL 170
+K+L+Y ST + + E P PI +PYG++K +AE ++L
Sbjct: 211 KVKKLIYSSTC-ATYGEPEKMPITEDTPQVPI----NPYGKAKKMAEDMIL 256
>At4g23920 UDPglucose 4-epimerase - like protein
Length = 350
Score = 47.0 bits (110), Expect = 2e-05
Identities = 46/167 (27%), Positives = 75/167 (44%), Gaps = 16/167 (9%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILK------DNGVRCI--Q 63
K+++VTGG G++GS L+L+ G + D D+S ++ + +NG R Q
Sbjct: 3 KSVLVTGGAGYIGSHTVLQLL-EGGYSAVVVDNYDNSSAASLQRVKKLAGENGNRLSFHQ 61
Query: 64 GDIVRKEDVESALRGA--DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGI 121
D+ + +E D V H A G+ + + NI GT +L+ G
Sbjct: 62 VDLRDRPALEKIFSETKFDAVIHFAGLKAVGESVEKPLLYYNNNIVGTVTLLEVMAQYGC 121
Query: 122 KRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
K LV+ S+ V + + ES PI +PYGR+K E++
Sbjct: 122 KNLVFSSSATVYGWPKEVPCTEES----PIS-ATNPYGRTKLFIEEI 163
>At1g51410 cinnamyl alcohol dehydrogenase, putative
Length = 809
Score = 47.0 bits (110), Expect = 2e-05
Identities = 65/249 (26%), Positives = 111/249 (44%), Gaps = 29/249 (11%)
Query: 10 EGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF-----DLRDSSPWFAIL-KDNGVRCIQ 63
E KT+ VTG G++ S + L+ RG + V+A D R + A+ + ++ +
Sbjct: 489 EEKTVCVTGASGYIASWIVKLLLLRG-YTVKASVRDPNDPRKTEHLLALEGAEERLKLFK 547
Query: 64 GDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-GIK 122
+++ + +SA+ G + VFH A+ + Q +D + GT +VL +CL +K
Sbjct: 548 ANLLEEGSFDSAIDGCEGVFHTASPFYHDVKDPQAELLDPA-VKGTINVLSSCLKTSSVK 606
Query: 123 RLVYCSTYNVVFAGQRIVNGNESLPYFPIDH--HVDP-YGRSKS---IAEQLVLKNNARP 176
R+V S+ V NG P +D DP Y R+ + + + +N A
Sbjct: 607 RVVLTSSIAAV-----AFNGMPRTPETIVDETWFADPDYCRASKLWYVLSKTLAENAAWK 661
Query: 177 FKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQTVKS---DWVFVE 233
F + N L ++ PA + GP L + T+ +L G QT + WV V+
Sbjct: 662 FAKE--NNLQLVSINPAMVIGP----LLQPTLNTSAAAVLSLIKGAQTFPNATFGWVNVK 715
Query: 234 NLVLALILA 242
++ A I A
Sbjct: 716 DVANAHIQA 724
>At2g02400 putative cinnamoyl-CoA reductase
Length = 318
Score = 46.6 bits (109), Expect = 3e-05
Identities = 29/125 (23%), Positives = 61/125 (48%), Gaps = 3/125 (2%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAIL---KDNGVRCIQGDIVR 68
+T+ VTG GF+GS + L+ +G ++ A S P + D+ ++ + D++
Sbjct: 4 ETVCVTGANGFIGSWIIRTLIEKGYTKIHASIYPGSDPTHLLQLPGSDSKIKIFEADLLD 63
Query: 69 KEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLGIKRLVYCS 128
+ + A+ G VFH+A+ + + + + GT +VL+A ++R+V S
Sbjct: 64 SDAISRAIDGCAGVFHVASPCTLDPPVDPEKELVEPAVKGTINVLEAAKRFNVRRVVITS 123
Query: 129 TYNVV 133
+ + +
Sbjct: 124 SISAL 128
>At1g53500 dTDP-D-glucose 4,6-dehydratase, putative
Length = 667
Score = 46.6 bits (109), Expect = 3e-05
Identities = 59/245 (24%), Positives = 102/245 (41%), Gaps = 35/245 (14%)
Query: 12 KTLVVTGGLGFVGSALCLELVRR-GAHEVRAF-------DLRDSSPWFAILKDNGVRCIQ 63
K +++TG GF+ S + L+R +++ DL++ P F+ + ++
Sbjct: 9 KNILITGAAGFIASHVANRLIRNYPDYKIVVLDKLDYCSDLKNLDPSFS---SPNFKFVK 65
Query: 64 GDIVRKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG- 120
GDI + V L D + H AA + NI GT +L+AC G
Sbjct: 66 GDIASDDLVNYLLITENIDTIMHFAAQTHVDNSFGNSFEFTKNNIYGTHVLLEACKVTGQ 125
Query: 121 IKRLVYCSTYNVVFAGQR--IVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL---KNNAR 175
I+R ++ ST V V +E+ P +PY +K+ AE LV+ ++
Sbjct: 126 IRRFIHVSTDEVYGETDEDAAVGNHEASQLLP----TNPYSATKAGAEMLVMAYGRSYGL 181
Query: 176 PFKNDAGNCLYTCAVRPAAIYGPGE--DRHLPRIITTAKLGLLLFTIGDQTVKSDWVFVE 233
P GN +YGP + ++ +P+ I A G L GD + +++ E
Sbjct: 182 PVITTRGN----------NVYGPNQFPEKMIPKFILLAMSGKPLPIHGDGSNVRSYLYCE 231
Query: 234 NLVLA 238
++ A
Sbjct: 232 DVAEA 236
>At1g30620 UDP-galactose 4-epimerase-like protein
Length = 419
Score = 46.6 bits (109), Expect = 3e-05
Identities = 52/170 (30%), Positives = 78/170 (45%), Gaps = 23/170 (13%)
Query: 14 LVVTGGLGFVGSALCLELVRRGAHEVRAFD--LRDSSPWFAILKD-----NGVRCIQGDI 66
++VTGG G++GS L L++ ++ V D R + IL++ ++ I D+
Sbjct: 73 VLVTGGAGYIGSHAALRLLKE-SYRVTIVDNLSRGNLAAVRILQELFPEPGRLQFIYADL 131
Query: 67 -----VRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNIT-GTCHVLDACLHLG 120
V K E+A D V H AA G E Q NIT T VL+ G
Sbjct: 132 GDAKAVNKIFTENAF---DAVMHFAAVAYVG-ESTQFPLKYYHNITSNTLVVLETMAAHG 187
Query: 121 IKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL 170
+K L+Y ST + I+ E P PI +PYG++K +AE ++L
Sbjct: 188 VKTLIYSSTC-ATYGEPDIMPITEETPQVPI----NPYGKAKKMAEDIIL 232
>At1g09480 putative cinnamyl alcohol dehydrogenase
Length = 369
Score = 45.4 bits (106), Expect = 7e-05
Identities = 49/196 (25%), Positives = 89/196 (45%), Gaps = 12/196 (6%)
Query: 11 GKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNG----VRCIQG 64
GK + VTG G++ S + L+ RG + V+A DL D +L +G ++ +
Sbjct: 52 GKLVCVTGASGYIASWIVKLLLLRG-YTVKATVRDLTDRKKTEHLLALDGAKERLKLFKA 110
Query: 65 DIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-GIKR 123
D++ + E A+ G D VFH A+ + Q +D + GT +VL+ C ++R
Sbjct: 111 DLLEESSFEQAIEGCDAVFHTASPVFFTVKDPQTELIDPA-LKGTMNVLNTCKETPSVRR 169
Query: 124 LVYCSTYNVVFAGQRIVNGNESL-PYFPIDHHVDPYGRSKSIAEQLVLKNNARPFKNDAG 182
++ S+ V Q V ++ + F D + ++ +++ +N A F D G
Sbjct: 170 VILTSSTAAVLFRQPPVEASDVVDETFFSDPSLCRETKNWYPLSKILAENAAWEFAKDNG 229
Query: 183 NCLYTCAVRPAAIYGP 198
+ + P I+GP
Sbjct: 230 --IDMVVLNPGFIFGP 243
>At5g19440 cinnamyl-alcohol dehydrogenase - like protein
Length = 326
Score = 45.1 bits (105), Expect = 9e-05
Identities = 66/254 (25%), Positives = 107/254 (41%), Gaps = 39/254 (15%)
Query: 10 EGKTLVVTGGLGFVGSALCLELVRRGAHEVRAF--DLRDSSPWFAILKDNGVR----CIQ 63
EGK + VTG G++ S L L+ RG + V+A D D ++ G + +
Sbjct: 6 EGKVVCVTGASGYIASWLVKFLLSRG-YTVKASVRDPSDPKKTQHLVSLEGAKERLHLFK 64
Query: 64 GDIVRKEDVESALRGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL-GIK 122
D++ + +SA+ G VFH A+ + + Q +D + GT +VL++C +K
Sbjct: 65 ADLLEQGSFDSAIDGCHGVFHTASPFFNDAKDPQAELIDPA-VKGTLNVLNSCAKASSVK 123
Query: 123 RLVYCSTYNVVFAGQRIVNGNESLPYFPIDH--HVDP---------YGRSKSIAEQLVLK 171
R+V S+ V NG P +D DP Y SK++AE K
Sbjct: 124 RVVVTSSMAAVG-----YNGKPRTPDVTVDETWFSDPELCEASKMWYVLSKTLAEDAAWK 178
Query: 172 NNARPFKNDAGNCLYTCAVRPAAIYGPGEDRHLPRIITTAKLGLLLFTIGDQT---VKSD 228
+ G + T + PA + GP L + T+ +L G +T +
Sbjct: 179 -----LAKEKGLDIVT--INPAMVIGP----LLQPTLNTSAAAILNLINGAKTFPNLSFG 227
Query: 229 WVFVENLVLALILA 242
WV V+++ A I A
Sbjct: 228 WVNVKDVANAHIQA 241
>At1g63180 uridine diphosphate glucose epimerase like protein
Length = 351
Score = 44.7 bits (104), Expect = 1e-04
Identities = 41/168 (24%), Positives = 72/168 (42%), Gaps = 17/168 (10%)
Query: 12 KTLVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILK---------DNGVRCI 62
+ ++VTGG GF+G+ ++L+ +G +V D D+S A+ + +
Sbjct: 7 QNILVTGGAGFIGTHTVVQLLNQG-FKVTIIDNLDNSVVEAVHRVRELVGPDLSTKLEFN 65
Query: 63 QGDIVRKEDVESALRGA--DCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHLG 120
GD+ K D+E D V H A G+ + R N+ GT ++ +
Sbjct: 66 LGDLRNKGDIEKLFSNQRFDAVIHFAGLKAVGESVGNPRRYFDNNLVGTINLYETMAKYN 125
Query: 121 IKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQL 168
K +V+ S+ V+ IV E ++PYGR+K E++
Sbjct: 126 CKMMVFSSS-ATVYGQPEIVPCVEDFEL----QAMNPYGRTKLFLEEI 168
>At4g20460 UDP-glucose 4-epimerase - like protein
Length = 379
Score = 44.3 bits (103), Expect = 2e-04
Identities = 46/171 (26%), Positives = 75/171 (42%), Gaps = 16/171 (9%)
Query: 10 EGKT-LVVTGGLGFVGSALCLELVRRGAHEVRAFDLRDSSPWFAILKDNG-------VRC 61
EG T ++VTGG G++GS L L+ + ++ V D A+ G ++
Sbjct: 35 EGVTHVLVTGGAGYIGSHAALRLL-KDSYRVTIVDNLSRGNLGAVKVLQGLFPEPGRLQF 93
Query: 62 IQGDIVRKEDVESAL--RGADCVFHLAAFGMSGKEMLQLTRVDQVNITGTCHVLDACLHL 119
I D+ + V+ D V H AA G+ L + + T VL+A
Sbjct: 94 IYADLGDAKAVDKIFSENAFDAVMHFAAVAYVGESTLDPLKYYHNITSNTLVVLEAVARH 153
Query: 120 GIKRLVYCSTYNVVFAGQRIVNGNESLPYFPIDHHVDPYGRSKSIAEQLVL 170
+K+L+Y ST + + E P PI +PYG++K +AE ++L
Sbjct: 154 KVKKLIYSSTC-ATYGEPDKMPIVEVTPQVPI----NPYGKAKKMAEDMIL 199
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,679,016
Number of Sequences: 26719
Number of extensions: 457264
Number of successful extensions: 1256
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 40
Number of HSP's that attempted gapping in prelim test: 1175
Number of HSP's gapped (non-prelim): 77
length of query: 479
length of database: 11,318,596
effective HSP length: 103
effective length of query: 376
effective length of database: 8,566,539
effective search space: 3221018664
effective search space used: 3221018664
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0335a.3


