

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0334a.8
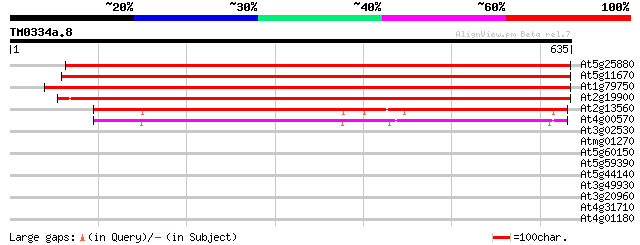
(635 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g25880 malate dehydrogenase - like protein 954 0.0
At5g11670 NADP dependent malic enzyme - like protein 946 0.0
At1g79750 malate oxidoreductase like protein 932 0.0
At2g19900 malate oxidoreductase (malic enzyme) 932 0.0
At2g13560 malate oxidoreductase (malic enzyme) 431 e-121
At4g00570 putative malate oxidoreductase 430 e-120
At3g02530 putative chaperonin 31 2.4
Atmg01270 -mitochondrial genome- ribosomal protein S7 (RPS7) 30 3.1
At5g60150 putative protein 30 3.1
At5g59390 transcriptional regulator - like protein 30 5.4
At5g44140 prohibitin 29 7.0
At3g49930 zinc-finger-like protein 29 7.0
At3g20960 cytochrome P450 like protein 29 7.0
At4g31710 putative protein (AtGLR2.4) 29 9.2
At4g01180 hypothetical protein 29 9.2
>At5g25880 malate dehydrogenase - like protein
Length = 588
Score = 954 bits (2466), Expect = 0.0
Identities = 464/571 (81%), Positives = 523/571 (91%)
Query: 64 NSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKERDAHYLRG 123
+S GG+ D+YGEDSAT DQL+TPW SVASG TL+RDPRYNKGLAFT+KERDAHY+ G
Sbjct: 17 SSGVGGGISDVYGEDSATLDQLVTPWVTSVASGYTLMRDPRYNKGLAFTDKERDAHYITG 76
Query: 124 LLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVY 183
LLPP V +Q++QE++ MHN+RQY VPL +YMA+MDLQERNERLFYK+LIDNVEELLP+VY
Sbjct: 77 LLPPVVLSQDVQERKVMHNLRQYTVPLQRYMALMDLQERNERLFYKLLIDNVEELLPVVY 136
Query: 184 TPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDL 243
TP VGEACQKYGSIY+RPQGLYISLKEKGKILEVLKNWP++ IQVIVVTDGERILGLGDL
Sbjct: 137 TPTVGEACQKYGSIYRRPQGLYISLKEKGKILEVLKNWPQRGIQVIVVTDGERILGLGDL 196
Query: 244 GCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYE 303
GCQGMGIPVGKLSLYTALGG+RPS+CLP+TIDVGTNNE LLN+EFY GL+QKRA G+EY
Sbjct: 197 GCQGMGIPVGKLSLYTALGGIRPSACLPITIDVGTNNEKLLNNEFYIGLKQKRANGEEYA 256
Query: 304 ELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLL 363
E L EFM AVKQNYGEKVL+QFEDFANH+AF LL +Y SSHLVFNDDIQGTASVVLAGL+
Sbjct: 257 EFLQEFMCAVKQNYGEKVLVQFEDFANHHAFELLSKYCSSHLVFNDDIQGTASVVLAGLI 316
Query: 364 ASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIV 423
A+ K++G +LADHTFLFLGAGEAGTGIAELIAL+ISK+T P++ETRKK+WLVDSKGLIV
Sbjct: 317 AAQKVLGKSLADHTFLFLGAGEAGTGIAELIALKISKETGKPIDETRKKIWLVDSKGLIV 376
Query: 424 RSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEK 483
R ESLQHFK+PWAH+H+PVK LL AV AIKPTVLIG+SGVGKTFTKEVVEAMA+LNEK
Sbjct: 377 SERKESLQHFKQPWAHDHKPVKELLAAVNAIKPTVLIGTSGVGKTFTKEVVEAMATLNEK 436
Query: 484 PLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANNAYIFPGF 543
PLILALSNPTSQ+ECTAEEAYTW+KG+AIFASGSPFDPV+Y GK + PGQANN YIFPG
Sbjct: 437 PLILALSNPTSQAECTAEEAYTWTKGRAIFASGSPFDPVQYDGKKFTPGQANNCYIFPGL 496
Query: 544 GLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKV 603
GLGLI+SGAIRVRD+MLLAASEALA+QV++EN+ GLIYPPF NIRKISA+IAASV AK
Sbjct: 497 GLGLIMSGAIRVRDDMLLAASEALASQVTEENFANGLIYPPFANIRKISANIAASVGAKT 556
Query: 604 YELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
YELGLAS+LPRPKDLVK AESCMYSP YR++
Sbjct: 557 YELGLASNLPRPKDLVKMAESCMYSPVYRNF 587
>At5g11670 NADP dependent malic enzyme - like protein
Length = 588
Score = 946 bits (2445), Expect = 0.0
Identities = 462/578 (79%), Positives = 524/578 (89%), Gaps = 2/578 (0%)
Query: 59 GEDFKNSEES--GGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEKER 116
GED ++ GG+ D+YGEDSAT DQL+TPW SVASG TL+RDPRYNKGLAFT+KER
Sbjct: 10 GEDVADNRSGVGGGISDVYGEDSATLDQLVTPWVTSVASGYTLMRDPRYNKGLAFTDKER 69
Query: 117 DAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDNVE 176
DAHYL GLLPP + +Q++QE++ MHN+RQY VPL +YMA+MDLQERNERLFYK+LIDNVE
Sbjct: 70 DAHYLTGLLPPVILSQDVQERKVMHNLRQYTVPLQRYMALMDLQERNERLFYKLLIDNVE 129
Query: 177 ELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDGER 236
ELLP+VYTP VGEACQKYGSI+++PQGLYISL EKGKILEVLKNWP++ IQVIVVTDGER
Sbjct: 130 ELLPVVYTPTVGEACQKYGSIFRKPQGLYISLNEKGKILEVLKNWPQRGIQVIVVTDGER 189
Query: 237 ILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKR 296
ILGLGDLGCQGMGIPVGKLSLYTALGG+RPS+CLP+TIDVGTNNE LLNDEFY GL+Q+R
Sbjct: 190 ILGLGDLGCQGMGIPVGKLSLYTALGGIRPSACLPITIDVGTNNEKLLNDEFYIGLKQRR 249
Query: 297 ATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGTAS 356
ATG+EY E L EFM AVKQNYGEKVL+QFEDFANHNAF+LL +YS SHLVFNDDIQGTAS
Sbjct: 250 ATGQEYAEFLHEFMCAVKQNYGEKVLVQFEDFANHNAFDLLSKYSDSHLVFNDDIQGTAS 309
Query: 357 VVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVWLV 416
VVLAGL+A+ K++G LADHTFLFLGAGEAGTGIAELIAL+ISK+T AP+ ETRKK+WLV
Sbjct: 310 VVLAGLIAAQKVLGKKLADHTFLFLGAGEAGTGIAELIALKISKETGAPITETRKKIWLV 369
Query: 417 DSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEA 476
DSKGLIV SR ESLQHFK+PWAHEH+PVK L+ AV AIKPTVLIG+SGVG+TFTKEVVEA
Sbjct: 370 DSKGLIVSSRKESLQHFKQPWAHEHKPVKDLIGAVNAIKPTVLIGTSGVGQTFTKEVVEA 429
Query: 477 MASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQANN 536
MA+ NEKPLILALSNPTSQ+ECTAE+AYTW+KG+AIF SGSPFDPV Y GK Y PGQANN
Sbjct: 430 MATNNEKPLILALSNPTSQAECTAEQAYTWTKGRAIFGSGSPFDPVVYDGKTYLPGQANN 489
Query: 537 AYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIA 596
YIFPG GLGLI+SGAIRVRD+MLLAASEALAAQV++E+Y GLIYPPF+NIR+ISA+IA
Sbjct: 490 CYIFPGLGLGLIMSGAIRVRDDMLLAASEALAAQVTEEHYANGLIYPPFSNIREISANIA 549
Query: 597 ASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
A VAAK Y+LGLAS+LPR KDLVK+AES MYSP YR+Y
Sbjct: 550 ACVAAKTYDLGLASNLPRAKDLVKFAESSMYSPVYRNY 587
>At1g79750 malate oxidoreductase like protein
Length = 646
Score = 932 bits (2410), Expect = 0.0
Identities = 456/595 (76%), Positives = 523/595 (87%)
Query: 40 GEPKLNIAAMSRNGGEGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTL 99
G + A S + E + S SGGV+D+YGED+ATED ITPW+ SVASG TL
Sbjct: 51 GSVLIETTATSSSSLETSAADIVPKSTVSGGVQDVYGEDAATEDMPITPWSLSVASGYTL 110
Query: 100 LRDPRYNKGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDL 159
LRDP +NKGLAF+ +ERDAHYLRGLLPP V +Q+LQ K+ MH +RQY+VPL KYMAMMDL
Sbjct: 111 LRDPHHNKGLAFSHRERDAHYLRGLLPPTVISQDLQVKKIMHTLRQYQVPLQKYMAMMDL 170
Query: 160 QERNERLFYKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLK 219
QE NERLFYK+LID+VEELLP++YTP VGEACQKYGSI+ RPQGL+ISLKEKGKI EVL+
Sbjct: 171 QETNERLFYKLLIDHVEELLPVIYTPTVGEACQKYGSIFLRPQGLFISLKEKGKIHEVLR 230
Query: 220 NWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTN 279
NWPEK IQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPS+CLPVTIDVGTN
Sbjct: 231 NWPEKNIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSACLPVTIDVGTN 290
Query: 280 NETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDR 339
NE LLNDEFY GLRQ+RATG+EY EL+ EFM AVKQNYGEKV+IQFEDFANHNAF+LL +
Sbjct: 291 NEKLLNDEFYIGLRQRRATGEEYSELMHEFMTAVKQNYGEKVVIQFEDFANHNAFDLLAK 350
Query: 340 YSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEIS 399
Y ++HLVFNDDIQGTASVVLAGL+A+L+ VGG+L+DH FLFLGAGEAGTGIAELIALEIS
Sbjct: 351 YGTTHLVFNDDIQGTASVVLAGLIAALRFVGGSLSDHRFLFLGAGEAGTGIAELIALEIS 410
Query: 400 KQTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVL 459
K++ P+EE RK +WLVDSKGLIV SR ES+QHFKKPWAH+HEP++ L+DAVKAIKPTVL
Sbjct: 411 KKSHIPLEEARKNIWLVDSKGLIVSSRKESIQHFKKPWAHDHEPIRELVDAVKAIKPTVL 470
Query: 460 IGSSGVGKTFTKEVVEAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPF 519
IG+SGVG+TFT++VVE MA LNEKP+IL+LSNPTSQSECTAEEAYTWS+G+AIFASGSPF
Sbjct: 471 IGTSGVGQTFTQDVVETMAKLNEKPIILSLSNPTSQSECTAEEAYTWSQGRAIFASGSPF 530
Query: 520 DPVKYGGKVYAPGQANNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKG 579
PV+Y GK + PGQANNAYIFPGFGLGLI+SG IRV D+MLLAASEALA ++ +E+Y+KG
Sbjct: 531 APVEYEGKTFVPGQANNAYIFPGFGLGLIMSGTIRVHDDMLLAASEALAEELMEEHYEKG 590
Query: 580 LIYPPFTNIRKISAHIAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
+IYPPF NIRKISA IAA VAAK YELGLA+ LP+PK+L + AES MYSP YRSY
Sbjct: 591 MIYPPFRNIRKISARIAAKVAAKAYELGLATRLPQPKELEQCAESSMYSPSYRSY 645
>At2g19900 malate oxidoreductase (malic enzyme)
Length = 581
Score = 932 bits (2409), Expect = 0.0
Identities = 462/580 (79%), Positives = 520/580 (89%), Gaps = 1/580 (0%)
Query: 55 EGVVGEDFKNSEESGGVRDLYGEDSATEDQLITPWTFSVASGNTLLRDPRYNKGLAFTEK 114
E V D K+S + GGV D+YGEDSAT + ITPW+ SV+SG +LLRDPRYNKGLAFTEK
Sbjct: 2 EKVTNSDLKSSVD-GGVVDVYGEDSATIEHNITPWSLSVSSGYSLLRDPRYNKGLAFTEK 60
Query: 115 ERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVPLHKYMAMMDLQERNERLFYKILIDN 174
ERD HYLRGLLPP V Q+LQEKR ++NIRQY+ PL KYMA+ +LQERNERLFYK+LIDN
Sbjct: 61 ERDTHYLRGLLPPVVLDQKLQEKRLLNNIRQYQFPLQKYMALTELQERNERLFYKLLIDN 120
Query: 175 VEELLPIVYTPVVGEACQKYGSIYKRPQGLYISLKEKGKILEVLKNWPEKTIQVIVVTDG 234
VEELLPIVYTP VGEACQK+GSI++RPQGL+ISLK+KGKIL+VLKNWPE+ IQVIVVTDG
Sbjct: 121 VEELLPIVYTPTVGEACQKFGSIFRRPQGLFISLKDKGKILDVLKNWPERNIQVIVVTDG 180
Query: 235 ERILGLGDLGCQGMGIPVGKLSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQ 294
ERILGLGDLGCQGMGIPVGKL+LY+ALGGVRPS+CLPVTIDVGTNNE LLNDEFY GLRQ
Sbjct: 181 ERILGLGDLGCQGMGIPVGKLALYSALGGVRPSACLPVTIDVGTNNEKLLNDEFYIGLRQ 240
Query: 295 KRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLDRYSSSHLVFNDDIQGT 354
KRATG+EY ELL+EFM AVKQNYGEKVLIQFEDFANHNAF LL +YS +HLVFNDDIQGT
Sbjct: 241 KRATGQEYSELLNEFMSAVKQNYGEKVLIQFEDFANHNAFELLAKYSDTHLVFNDDIQGT 300
Query: 355 ASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETRKKVW 414
ASVVLAGL+++ KL LA+HTFLFLGAGEAGTGIAELIAL +SKQ A VEE+RKK+W
Sbjct: 301 ASVVLAGLVSAQKLTNSPLAEHTFLFLGAGEAGTGIAELIALYMSKQMNASVEESRKKIW 360
Query: 415 LVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVV 474
LVDSKGLIV SR +SLQ FKKPWAHEHEPVK LL A+KAIKPTVLIGSSGVG++FTKEV+
Sbjct: 361 LVDSKGLIVNSRKDSLQDFKKPWAHEHEPVKDLLGAIKAIKPTVLIGSSGVGRSFTKEVI 420
Query: 475 EAMASLNEKPLILALSNPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAPGQA 534
EAM+S+NE+PLI+ALSNPT+QSECTAEEAYTWSKG+AIFASGSPFDPV+Y GKV+ QA
Sbjct: 421 EAMSSINERPLIMALSNPTTQSECTAEEAYTWSKGRAIFASGSPFDPVEYEGKVFVSTQA 480
Query: 535 NNAYIFPGFGLGLIISGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAH 594
NNAYIFPGFGLGL+ISGAIRV D+MLLAA+EALA QVS+ENY+KG+IYP F++IRKISA
Sbjct: 481 NNAYIFPGFGLGLVISGAIRVHDDMLLAAAEALAGQVSKENYEKGMIYPSFSSIRKISAQ 540
Query: 595 IAASVAAKVYELGLASHLPRPKDLVKYAESCMYSPGYRSY 634
IAA+VA K YELGLA LPRPKD+VK AES MYSP YR Y
Sbjct: 541 IAANVATKAYELGLAGRLPRPKDIVKCAESSMYSPTYRLY 580
>At2g13560 malate oxidoreductase (malic enzyme)
Length = 623
Score = 431 bits (1109), Expect = e-121
Identities = 242/572 (42%), Positives = 348/572 (60%), Gaps = 38/572 (6%)
Query: 96 GNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYEVP------ 149
G +L DP +NKG AFT ER+ LRGLLPP V E Q R M ++++ E
Sbjct: 46 GLDILHDPWFNKGTAFTMTERNRLDLRGLLPPNVMDSEQQIFRFMTDLKRLEEQARDGPS 105
Query: 150 ----LHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLY 205
L K+ + L +RNE ++YK+LI+N+EE PIVYTP VG CQ Y +++RP+G+Y
Sbjct: 106 DPNALAKWRILNRLHDRNETMYYKVLINNIEEYAPIVYTPTVGLVCQNYSGLFRRPRGMY 165
Query: 206 ISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVR 265
S +++G+++ ++ NWP + + +IVVTDG RILGLGDLG G+GI VGKL LY A G+
Sbjct: 166 FSAEDRGEMMSMVYNWPAEQVDMIVVTDGSRILGLGDLGVHGIGIAVGKLDLYVAAAGIN 225
Query: 266 PSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQF 325
P LPV IDVGTNNE L ND Y GL+Q+R +Y +++DEFM AV + V++QF
Sbjct: 226 PQRVLPVMIDVGTNNEKLRNDPMYLGLQQRRLEDDDYIDVIDEFMEAVYTRW-PHVIVQF 284
Query: 326 EDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLADH---TFLFLG 382
EDF + AF LL RY ++ +FNDD+QGTA V +AGLL +++ G + D + G
Sbjct: 285 EDFQSKWAFKLLQRYRCTYRMFNDDVQGTAGVAIAGLLGAVRAQGRPMIDFPKMKIVVAG 344
Query: 383 AGEAGTGIAELIALEISK---QTKAPVEETRKKVWLVDSKGLIVRSRLESLQHFKKPWAH 439
AG AG G+ +++ T+ + + + W+VD++GLI R E++ +P+A
Sbjct: 345 AGSAGIGVLNAARKTMARMLGNTETAFDSAQSQFWVVDAQGLITEGR-ENIDPEAQPFAR 403
Query: 440 EHEPVK--------ALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAM-ASLNEKPLILALS 490
+ + ++ L++ V+ +KP VL+G S VG F+KEV+EAM S + +P I A+S
Sbjct: 404 KTKEMERQGLKEGATLVEVVREVKPDVLLGLSAVGGLFSKEVLEAMKGSTSTRPAIFAMS 463
Query: 491 NPTSQSECTAEEAYTWSKGKAIFASGSPFDPVKYG-GKVYAPGQANNAYIFPGFGLGLII 549
NPT +ECT ++A++ IFASGSPF V++G G V Q NN Y+FPG GLG ++
Sbjct: 464 NPTKNAECTPQDAFSILGENMIFASGSPFKNVEFGNGHVGHCNQGNNMYLFPGIGLGTLL 523
Query: 550 SGAIRVRDEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLA 609
SGA V D ML AASE LAA +S+E +G+IYPP + IR I+ IAA+V + E L
Sbjct: 524 SGAPIVSDGMLQAASECLAAYMSEEEVLEGIIYPPISRIRDITKRIAAAVIKEAIEEDLV 583
Query: 610 SHLPR----------PKDLVKYAESCMYSPGY 631
+ L++Y E+ M++P Y
Sbjct: 584 EGYREMDAREIQKLDEEGLMEYVENNMWNPEY 615
>At4g00570 putative malate oxidoreductase
Length = 607
Score = 430 bits (1106), Expect = e-120
Identities = 236/566 (41%), Positives = 336/566 (58%), Gaps = 34/566 (6%)
Query: 96 GNTLLRDPRYNKGLAFTEKERDAHYLRGLLPPAVFTQELQEKRTMHNIRQYE-------- 147
G +L DP +NK F ERD +RGLLPP V T Q R + + R E
Sbjct: 39 GADILHDPWFNKDTGFPLTERDRLGIRGLLPPRVMTCVQQCDRFIESFRSLENNTKGEPE 98
Query: 148 --VPLHKYMAMMDLQERNERLFYKILIDNVEELLPIVYTPVVGEACQKYGSIYKRPQGLY 205
V L K+ + L +RNE L+Y++LIDN+++ PI+YTP VG CQ Y +Y+RP+G+Y
Sbjct: 99 NVVALAKWRMLNRLHDRNETLYYRVLIDNIKDFAPIIYTPTVGLVCQNYSGLYRRPRGMY 158
Query: 206 ISLKEKGKILEVLKNWPEKTIQVIVVTDGERILGLGDLGCQGMGIPVGKLSLYTALGGVR 265
S K+KG+++ ++ NWP + +IV+TDG RILGLGDLG QG+GIP+GKL +Y A G+
Sbjct: 159 FSAKDKGEMMSMIYNWPAPQVDMIVITDGSRILGLGDLGVQGIGIPIGKLDMYVAAAGIN 218
Query: 266 PSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQF 325
P LP+ +DVGTNNE LL ++ Y G+RQ R G+EY E++DEFM A + K ++QF
Sbjct: 219 PQRVLPIMLDVGTNNEKLLQNDLYLGVRQPRLEGEEYLEIIDEFMEAAFTRW-PKAVVQF 277
Query: 326 EDFANHNAFNLLDRYSSSHLVFNDDIQGTASVVLAGLLASLKLVGGTLAD---HTFLFLG 382
EDF AF L+RY +FNDD+QGTA V LAGLL +++ G ++D + +G
Sbjct: 278 EDFQAKWAFGTLERYRKKFCMFNDDVQGTAGVALAGLLGTVRAQGRPISDFVNQKIVVVG 337
Query: 383 AGEAGTGIAELIALEISKQTKAPVEETRKKVWLVDSKGLIVRSRLE----SLQHFKKPWA 438
AG AG G+ ++ +++ E K +L+D GL+ R + ++ K P
Sbjct: 338 AGSAGLGVTKMAVQAVARMAGISESEATKNFYLIDKDGLVTTERTKLDPGAVLFAKNP-- 395
Query: 439 HEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNE-KPLILALSNPTSQSE 497
E ++++ VK ++P VL+G SGVG F +EV++AM + KP I A+SNPT +E
Sbjct: 396 AEIREGASIVEVVKKVRPHVLLGLSGVGGIFNEEVLKAMRESDSCKPAIFAMSNPTLNAE 455
Query: 498 CTAEEAYTWSKGKAIFASGSPFDPVKY-GGKVYAPGQANNAYIFPGFGLGLIISGAIRVR 556
CTA +A+ + G +FASGSPF+ V+ GKV QANN Y+FPG GLG ++SGA V
Sbjct: 456 CTAADAFKHAGGNIVFASGSPFENVELENGKVGHVNQANNMYLFPGIGLGTLLSGARIVT 515
Query: 557 DEMLLAASEALAAQVSQENYDKGLIYPPFTNIRKISAHIAASVAAKVYELGLA------- 609
D ML AASE LA+ ++ E KG++YP NIR I+A + A+V +A
Sbjct: 516 DGMLQAASECLASYMTDEEVQKGILYPSINNIRHITAEVGAAVLRAAVTDDIAEGHGDVG 575
Query: 610 ----SHLPRPKDLVKYAESCMYSPGY 631
SH+ + +D V Y M+ P Y
Sbjct: 576 PKDLSHMSK-EDTVNYITRNMWFPVY 600
>At3g02530 putative chaperonin
Length = 535
Score = 30.8 bits (68), Expect = 2.4
Identities = 25/101 (24%), Positives = 42/101 (40%), Gaps = 14/101 (13%)
Query: 351 IQGTASVVLAGLLASLKLVGGTLADHTFLFLGAGEAGTGIAELIALEISKQTKAPVEETR 410
IQ ++++A + + G T +F+G E+ KQ++ ++E
Sbjct: 68 IQNPTAIMIARTAVAQDDISGDGTTSTVIFIG--------------ELMKQSERCIDEGM 113
Query: 411 KKVWLVDSKGLIVRSRLESLQHFKKPWAHEHEPVKALLDAV 451
LVD + R+ L+ L FK P EP K +L V
Sbjct: 114 HPRVLVDGFEIAKRATLQFLDTFKTPVVMGDEPDKEILKMV 154
>Atmg01270 -mitochondrial genome- ribosomal protein S7 (RPS7)
Length = 148
Score = 30.4 bits (67), Expect = 3.1
Identities = 17/50 (34%), Positives = 25/50 (50%)
Query: 419 KGLIVRSRLESLQHFKKPWAHEHEPVKALLDAVKAIKPTVLIGSSGVGKT 468
+G R R Q F +P E + +K ++DAV+ IKP + GV T
Sbjct: 22 EGKRTRVRAIVYQTFHRPARTERDVIKLMVDAVENIKPICEVAKVGVAGT 71
>At5g60150 putative protein
Length = 1195
Score = 30.4 bits (67), Expect = 3.1
Identities = 29/95 (30%), Positives = 37/95 (38%), Gaps = 1/95 (1%)
Query: 438 AHEHEPVKALLDAVKAIKPTVLIGSSGVGKTFTKEVVEAMASLNEKPLILALSNPT-SQS 496
A E LLDA KP +I F+ E + A+ + E+ L L S +
Sbjct: 1098 ASEGNDKMILLDAKLEKKPDPIIVKPPNAVPFSDEWLAAIEAAGEEILTLKSGRVQHSPT 1157
Query: 497 ECTAEEAYTWSKGKAIFASGSPFDPVKYGGKVYAP 531
E +A E WS K PFD KY K P
Sbjct: 1158 EKSAPEPGPWSPVKKSNQVVGPFDCTKYNNKGLPP 1192
>At5g59390 transcriptional regulator - like protein
Length = 561
Score = 29.6 bits (65), Expect = 5.4
Identities = 13/36 (36%), Positives = 23/36 (63%)
Query: 281 ETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQN 316
ET L+++ T R++RAT EY+++L E ++ N
Sbjct: 373 ETALHEKMMTLARKERATNDEYQDVLKEMIQVWNAN 408
>At5g44140 prohibitin
Length = 278
Score = 29.3 bits (64), Expect = 7.0
Identities = 21/68 (30%), Positives = 33/68 (47%), Gaps = 4/68 (5%)
Query: 279 NNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQNYGEKVLIQFEDFANHNAFNLLD 338
N++T ND T R + + L E R + QNYGE+VL N ++
Sbjct: 86 NSQTGSND-LQTVTIGLRVLTRPMGDRLPEIYRTLGQNYGERVL---PSIINETLKAVVA 141
Query: 339 RYSSSHLV 346
+Y++SHL+
Sbjct: 142 QYNASHLI 149
>At3g49930 zinc-finger-like protein
Length = 215
Score = 29.3 bits (64), Expect = 7.0
Identities = 13/29 (44%), Positives = 19/29 (64%)
Query: 258 YTALGGVRPSSCLPVTIDVGTNNETLLND 286
Y ALGG + S PV++DV +N T+ N+
Sbjct: 106 YQALGGHKTSHRKPVSVDVNNSNGTVTNN 134
>At3g20960 cytochrome P450 like protein
Length = 418
Score = 29.3 bits (64), Expect = 7.0
Identities = 20/77 (25%), Positives = 42/77 (53%), Gaps = 3/77 (3%)
Query: 104 RYNKGLAFTEKERDAHYLRGLLPPA-VFTQELQEKRTMHNI-RQYEVPLHKYMAMMDLQE 161
R + G +F+E+ +A +RGL+ + +++ + + ++ +PL K +MD+
Sbjct: 92 RMSMGRSFSEENGEAERVRGLVAESYALAKKIFFASVLRRLLKKLRIPLFK-KDIMDVSN 150
Query: 162 RNERLFYKILIDNVEEL 178
R L KIL+++ E+L
Sbjct: 151 RFNELLEKILVEHNEKL 167
>At4g31710 putative protein (AtGLR2.4)
Length = 898
Score = 28.9 bits (63), Expect = 9.2
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 1/70 (1%)
Query: 255 LSLYTALGGVRPSSCLPVTIDVGTNNETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVK 314
L L+ A V + L I N +LNDEFY G R K K E + ++ V+
Sbjct: 819 LLLFVAAATVCTLALLKFVICFLIQNRIILNDEFYRGKRMKEMWLKFMESDGESYISRVR 878
Query: 315 QNYGEKVLIQ 324
+VLIQ
Sbjct: 879 STC-PQVLIQ 887
>At4g01180 hypothetical protein
Length = 554
Score = 28.9 bits (63), Expect = 9.2
Identities = 13/36 (36%), Positives = 22/36 (61%)
Query: 281 ETLLNDEFYTGLRQKRATGKEYEELLDEFMRAVKQN 316
ET L+++ T R++RAT EY++ E ++ K N
Sbjct: 363 ETALHEKMMTLARKERATNDEYQDARKEMIKVWKAN 398
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,185,139
Number of Sequences: 26719
Number of extensions: 635215
Number of successful extensions: 1631
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1607
Number of HSP's gapped (non-prelim): 16
length of query: 635
length of database: 11,318,596
effective HSP length: 105
effective length of query: 530
effective length of database: 8,513,101
effective search space: 4511943530
effective search space used: 4511943530
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0334a.8


